DNA Methylation Mediates lncRNA2919 Regulation of Hair Follicle Regeneration
Abstract
:1. Introduction
2. Results
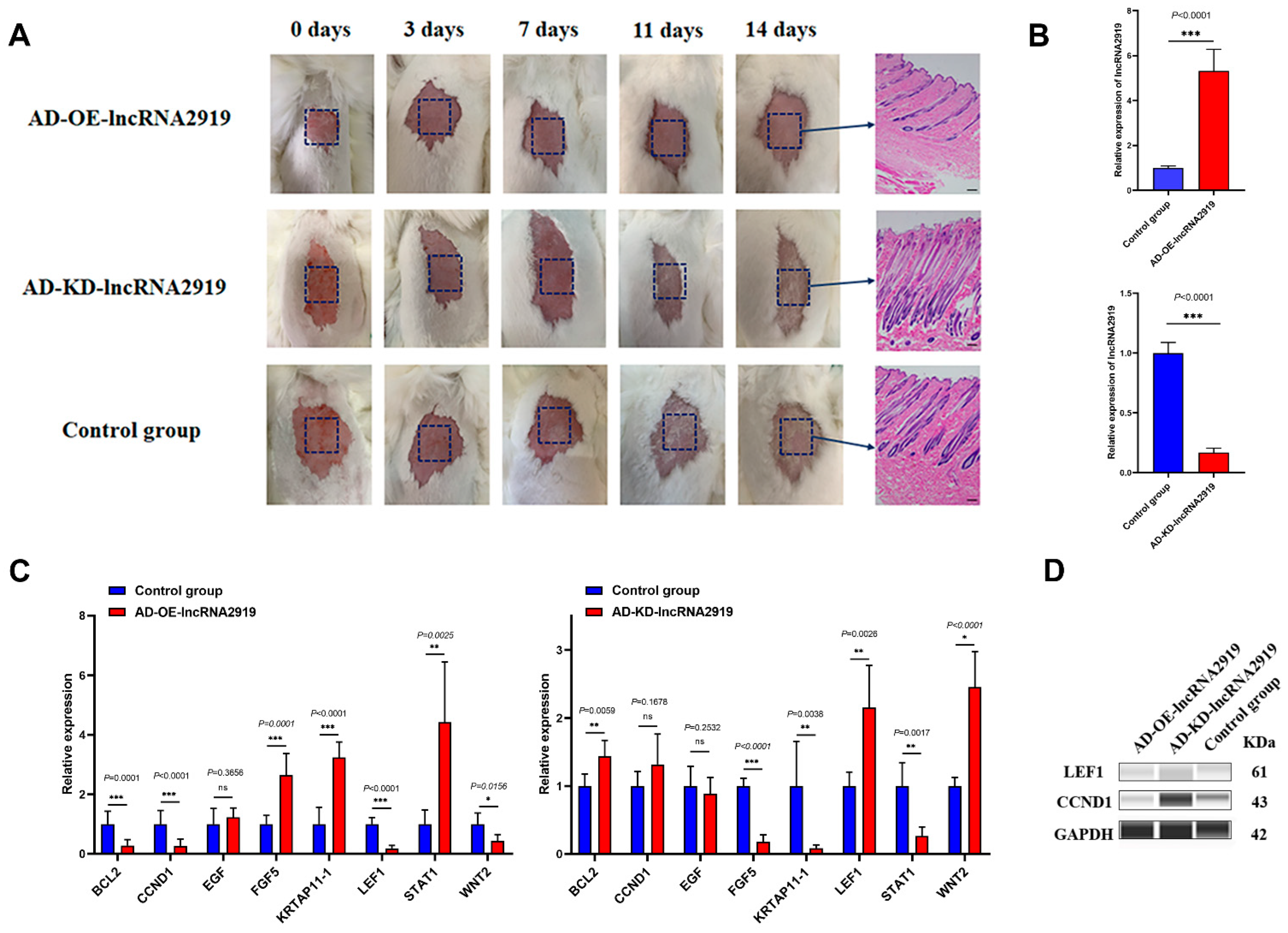
2.1. LncRNA2919 Regulates HF Regeneration in the In Vivo Rabbit Model
2.2. DNA Methylation Level of LncRNA2919 during the HF Cycle
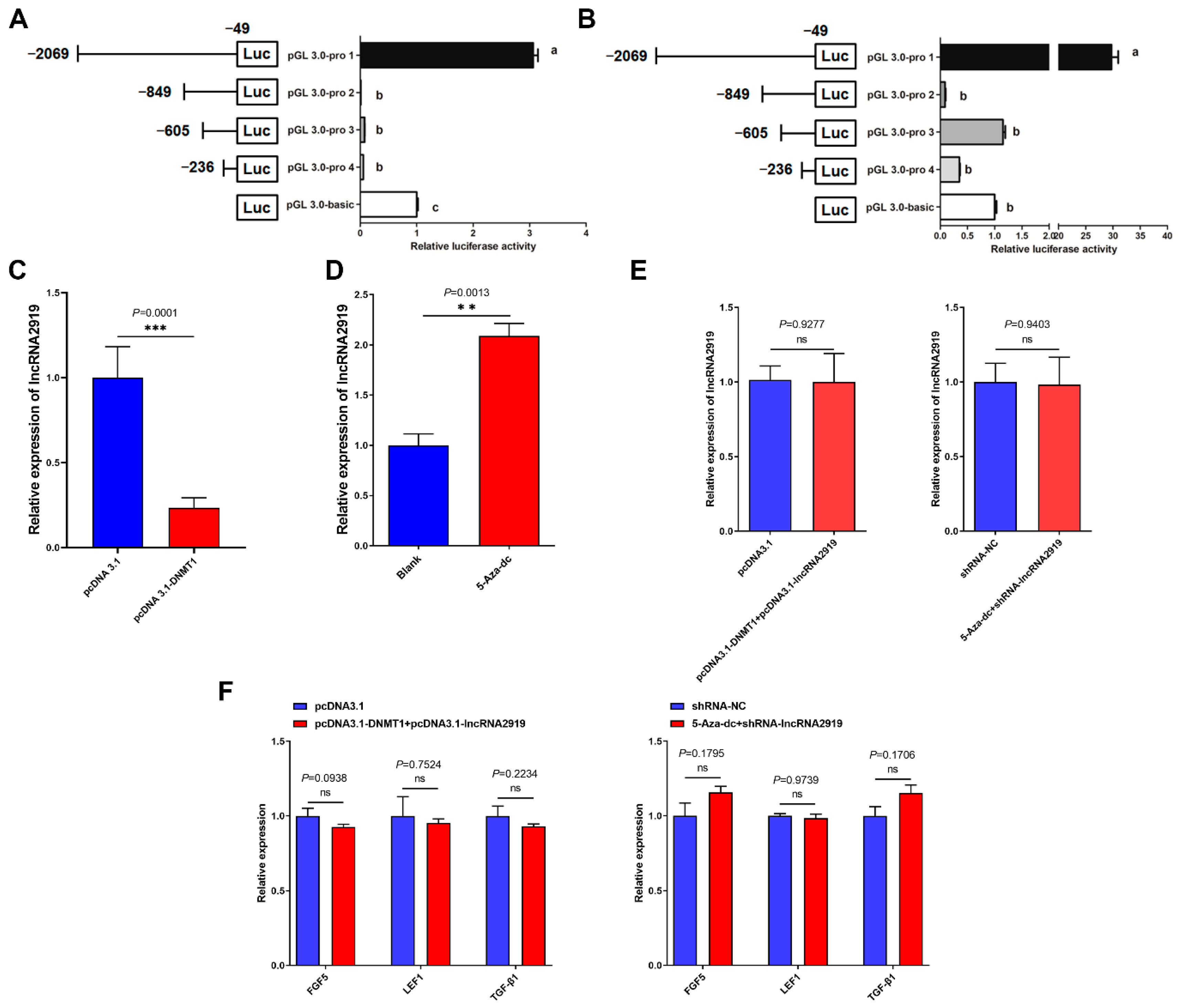
2.3. DNA Methylation Regulates LncRNA2919 Transcriptional Expression
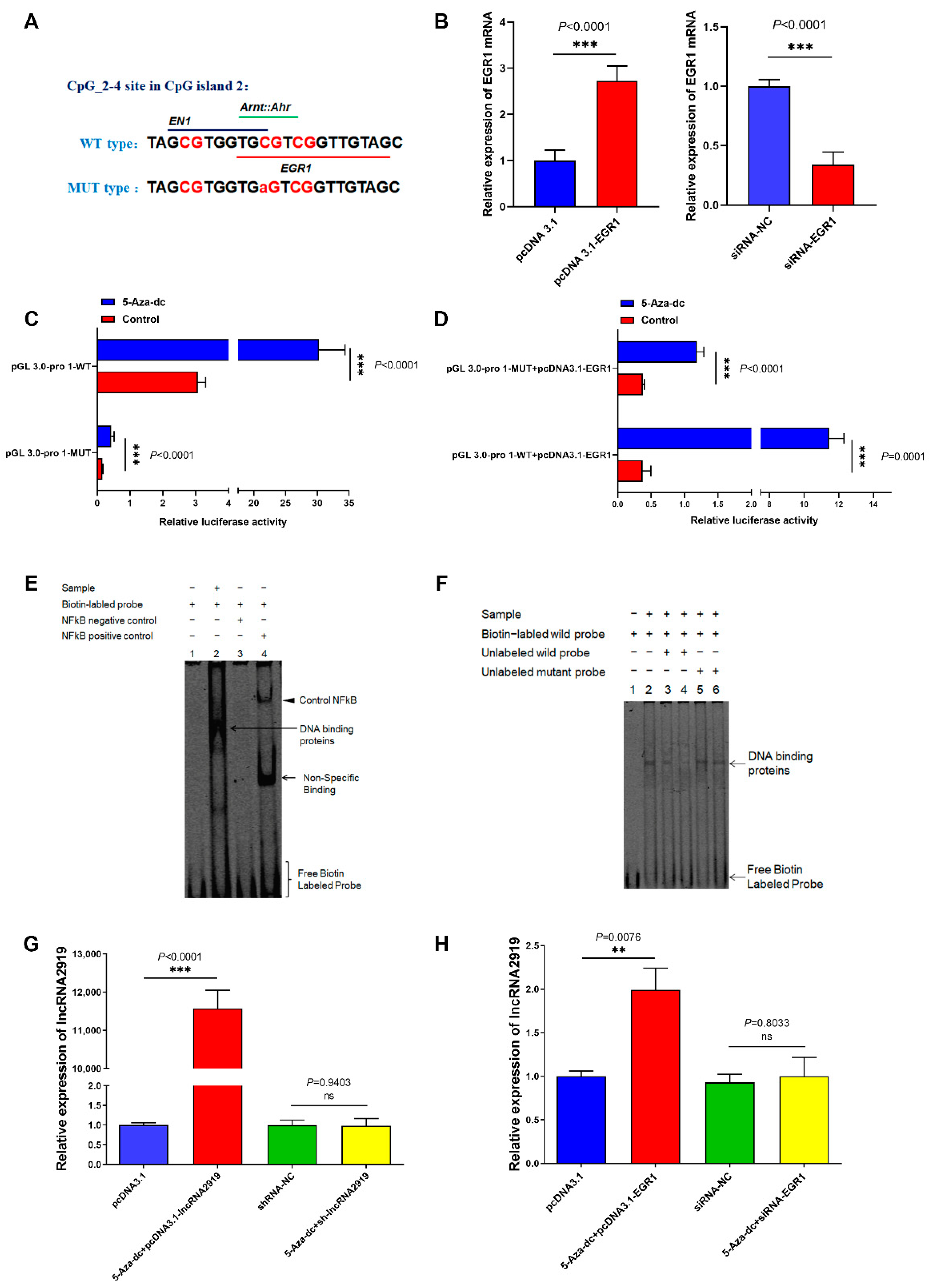
2.4. TF EGR1 Regulates LncRNA2919 Transcriptional Expression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Culture
4.3. Plasmid Preparation, Small Interfering RNA, Short Hairpin RNA, Adenovirus Plasmids, Transfection
4.4. Subcutaneous Injection of Adenovirus in Angora Rabbit
4.5. RNA Isolation and Quantitative RT-PCR Analysis
4.6. Protein Simple Wes Western Blotting System
4.7. DNA Extraction and Bisulfite Sequencing PCR
4.8. Dual-Luciferase Assay Verification of the Promoter Region
4.9. 5-Aza-Deoxycytidine-Induced Demethylation
4.10. Electrophoretic Mobility Shift Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. 2009, 19, R132–R142. [Google Scholar] [CrossRef] [PubMed]
- Stenn, K.; Paus, R. Controls of hair follicle cycling. Physiol. Rev. 2001, 81, 449–494. [Google Scholar] [CrossRef] [PubMed]
- Philpott, M.P.; Sanders, D.; Westgate, G.E.; Kealey, T. Human hair growth in vitro: A model for the study of hair follicle biology. J. Dermatol. Sci. 1994, 7, S55–S72. [Google Scholar] [CrossRef]
- Al-Nuaimi, Y.; Baier, G.; Watson, R.E.; Chuong, C.M.; Paus, R. The cycling hair follicle as an ideal systems biology research model. Exp. Dermatol. 2010, 19, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding rnas. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncrna promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Ferre, F.; Colantoni, A.; Helmer-Citterich, M. Revealing protein–lncrna interaction. Brief. Bioinform. 2016, 17, 106–116. [Google Scholar] [CrossRef]
- Si, Y.; Bai, J.; Wu, J.; Li, Q.; Mo, Y.; Fang, R.; Lai, W. Lncrna plncrna-1 regulates proliferation and differentiation of hair follicle stem cells through tgf-β1-mediated wnt/β-catenin signal pathway. Mol. Med. Rep. 2018, 17, 1191–1197. [Google Scholar] [CrossRef]
- Lin, B.-J.; Lin, G.-Y.; Zhu, J.-Y.; Yin, G.-Q.; Huang, D.; Yan, Y.-Y. Lncrna-pcat1 maintains characteristics of dermal papilla cells and promotes hair follicle regeneration by regulating mir-329/wnt10b axis. Exp. Cell Res. 2020, 394, 112031. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Hu, S.; Yang, N.; Wang, M.; Liu, M.; Li, J.; Xiao, Y.; Wu, X. Systematic analysis of non-coding rnas involved in the angora rabbit (oryctolagus cuniculus) hair follicle cycle by rna sequencing. Front. Genet. 2019, 10, 407. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, A.; Liu, S.; Lu, F.; Guo, Y.; Zhang, G.; Xu, F.; Shi, Y.; Shen, S.; Liang, J. Aberrant methylation-mediated silencing of lncrna meg3 functions as a cerna in esophageal cancer. Mol. Cancer Res. 2017, 15, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.; Feng, M.; Cheng, Z.; Zhang, Q.; Li, H.; Xu, J.; Zhang, H.; Li, Y.; Qian, L. Altered DNA methylation of long noncoding rna uc.167 inhibits cell differentiation in heart development. BioMed Res. Int. 2018, 2018, 4658024. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wei, Y.; Wang, X.; Zhang, Z.; Yin, J.; Li, W.; Chen, L.; Lyu, X.; Shi, Z.; Yan, W. DNA-methylation-mediated activating of lncrna snhg12 promotes temozolomide resistance in glioblastoma. Mol. Cancer 2020, 19, 1–19. [Google Scholar] [CrossRef]
- Wang, S.; Li, F.; Liu, J.; Zhang, Y.; Zheng, Y.; Ge, W.; Qu, L.; Wang, X. Integrative analysis of methylome and transcriptome reveals the regulatory mechanisms of hair follicle morphogenesis in cashmere goat. Cells 2020, 9, 969. [Google Scholar] [CrossRef]
- Zhu, Y.B.; Wang, Z.Y.; Yin, R.H.; Jiao, Q.; Zhao, S.J.; Cong, Y.Y.; Xue, H.L.; Guo, D.; Wang, S.Q.; Zhu, Y.X. A lncrna-h19 transcript from secondary hair follicle of liaoning cashmere goat: Identification, regulatory network and expression regulated potentially by its promoter methylation. Gene 2018, 641, 78–85. [Google Scholar] [CrossRef]
- Jiao, Q.; Yin, R.H.; Zhao, S.J.; Wang, Z.Y.; Zhu, Y.B.; Wang, W.; Zheng, Y.Y.; Yin, X.B.; Guo, D.; Wang, S.Q. Identification and molecular analysis of a lncrna-hotair transcript from secondary hair follicle of cashmere goat reveal integrated regulatory network with the expression regulated potentially by its promoter methylation. Gene 2019, 688, 182–192. [Google Scholar] [CrossRef]
- Millar, S.E. Molecular mechanisms regulating hair follicle development. J. Investig. Dermatol. 2002, 118, 216–225. [Google Scholar] [CrossRef]
- Amoh, Y.; Li, L.; Yang, M.; Moossa, A.; Katsuoka, K.; Penman, S.; Hoffman, R.M. Nascent blood vessels in the skin arise from nestin-expressing hair-follicle cells. Proc. Natl. Acad. Sci. USA 2004, 101, 13291–13295. [Google Scholar] [CrossRef]
- Paus, R.; Arck, P.; Tiede, S. (Neuro-) endocrinology of epithelial hair follicle stem cells. Mol. Cell. Endocrinol. 2008, 288, 38–51. [Google Scholar] [CrossRef]
- Ji, S.; Zhu, Z.; Sun, X.; Fu, X. Functional hair follicle regeneration: An updated review. Signal Transduct. Target. Ther. 2021, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Sheng, S.D.; Hui, T.Y.; Yue, C.; Sun, J.M.; Guo, D.; Guo, S.L.; Li, B.J.; Xue, H.L.; Wang, Z.Y. An integrated analysis of cashmere fineness lncrnas in cashmere goats. Genes 2019, 10, 266. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Lin, E.; Zhang, H.; Liu, Y.; Cao, G.; Fu, C.; Chen, L.; Zeng, Y.; Cai, B.; Yuan, Y. Lncrna h19 overexpression activates wnt signaling to maintain the hair follicle regeneration potential of dermal papilla cells. Front. Genet. 2020, 11, 694. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Zheng, Y.; Ma, S.; Xing, Q.; Wang, X.; Yang, B.; Yin, G.; Guan, F. Long non-coding rna regulates hair follicle stem cell proliferation and differentiation through pi3k/akt signal pathway. Mol. Med. Rep. 2018, 17, 5477–5483. [Google Scholar] [CrossRef]
- Fujimoto, S.; Takase, T.; Kadono, N.; Maekubo, K.; Hirai, Y. Krtap11-1, a hair keratin-associated protein, as a possible crucial element for the physical properties of hair shafts. J. Dermatol. Sci. 2014, 74, 39–47. [Google Scholar] [CrossRef]
- Gong, H.; Zhou, H.; Dyer, J.M.; Hickford, J.G. Identification of the ovine kap11-1 gene (krtap11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011, 38, 5429–5433. [Google Scholar] [CrossRef]
- Sato, N.; Leopold, P.L.; Crystal, R.G. Effect of adenovirus-mediated expression of sonic hedgehog gene on hair regrowth in mice with chemotherapy-induced alopecia. J. Natl. Cancer Inst. 2001, 93, 1858–1864. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, K.; Yang, K.; Ye, J.; Xing, Y.; Guo, H.; Deng, F.; Lian, X.; Yang, T. Adenovirus-mediated wnt10b overexpression induces hair follicle regeneration. J. Investig. Dermatol. 2013, 133, 42–48. [Google Scholar] [CrossRef]
- Hébert, J.M.; Rosenquist, T.; Götz, J.; Martin, G.R. Fgf5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell 1994, 78, 1017–1025. [Google Scholar] [CrossRef]
- Ota, Y.; Saitoh, Y.; Suzuki, S.; Ozawa, K.; Kawano, M.; Imamura, T. Fibroblast growth factor 5 inhibits hair growth by blocking dermal papilla cell activation. Biochem. Biophys. Res. Commun. 2002, 290, 169–176. [Google Scholar] [CrossRef]
- Wang, Z.; Qi, Q.-Q.; Chen, Y.-L. Analysis on single nucleotide polymorphisms of keratin-associated proteins gene in plateau tibetan sheep. China Anim. Husb. Vet. Med. 2010, 37, 120–124. [Google Scholar]
- Calò, V.; Migliavacca, M.; Bazan, V.; Macaluso, M.; Buscemi, M.; Gebbia, N.; Russo, A. Stat proteins: From normal control of cellular events to tumorigenesis. J. Cell. Physiol. 2003, 197, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Driskell, R.R.; Luo, M.; Abbott, D.; Filali, M.; Cheng, N.; Sigmund, C.D.; Engelhardt, J.F. Characterization of lef-1 promoter segments that facilitate inductive developmental expression in skin. J. Investig. Dermatol. 2004, 123, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Bayle, J.; Fitch, J.; Jacobsen, K.; Kumar, R.; Lafyatis, R.; Lemaire, R. Increased expression of wnt2 and sfrp4 in tsk mouse skin: Role of wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J. Investig. Dermatol. 2008, 128, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Müller-Röver, S.; Rossiter, H.; Lindner, G.; Peters, E.M.; Kupper, T.S.; Paus, R. Hair follicle apoptosis and bcl-2. J. Investig. Dermatol. Symp. Proc. 1999, 4, 272–277. [Google Scholar] [CrossRef]
- Stenn, K.; Lawrence, L.; Veis, D.; Korsmeyer, S.; Seiberg, M. Expression of the bcl-2 protooncogene in the cycling adult mouse hair follicle. J. Investig. Dermatol. 1994, 103, 107–111. [Google Scholar] [CrossRef]
- Xu, X.; Lyle, S.; Liu, Y.; Solky, B.; Cotsarelis, G. Differential expression of cyclin d1 in the human hair follicle. Am. J. Pathol. 2003, 163, 969–978. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, H.; Wang, H. Long noncoding rnas in DNA methylation: New players stepping into the old game. Cell Biosci. 2016, 6, 45. [Google Scholar] [CrossRef]
- Heilmann, K.; Toth, R.; Bossmann, C.; Klimo, K.; Plass, C.; Gerhauser, C. Genome-wide screen for differentially methylated long noncoding rnas identifies esrp2 and lncrna esrp2-as regulated by enhancer DNA methylation with prognostic relevance for human breast cancer. Oncogene 2017, 36, 6446–6461. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Z.; Zhu, Y.; Wang, W.; Bai, M.; Jiao, Q.; Wang, Y.; Zhao, S.; Yin, X.; Guo, D. Lncrna-000133 from secondary hair follicle of cashmere goat: Identification, regulatory network and its effects on inductive property of dermal papilla cells. Anim. Biotechnol. 2020, 31, 122–134. [Google Scholar] [CrossRef]
- Sarver, A.L.; Li, L.; Subramanian, S. Microrna mir-183 functions as an oncogene by targeting the transcription factor egr1 and promoting tumor cell migration. Cancer Res. 2010, 70, 9570–9580. [Google Scholar] [CrossRef] [PubMed]
- Baron, V.; Adamson, E.D.; Calogero, A.; Ragona, G.; Mercola, D. The transcription factor egr1 is a direct regulator of multiple tumor suppressors including tgf β 1, pten, p53, and fibronectin. Cancer Gene Ther. 2006, 13, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Svaren, J.; Ehrig, T.; Abdulkadir, S.A.; Ehrengruber, M.U.; Watson, M.A.; Milbrandt, J. Egr1 target genes in prostate carcinoma cells identified by microarray analysis. J. Biol. Chem. 2000, 275, 38524–38531. [Google Scholar] [CrossRef] [PubMed]
- Geng, R.; Yuan, C.; Chen, Y. Exploring differentially expressed genes by rna-seq in cashmere goat (capra hircus) skin during hair follicle development and cycling. PLoS ONE 2013, 8, e62704. [Google Scholar] [CrossRef]
- Li, J.; Zhao, B.; Zhang, C.; Zhang, X.; Dai, Y.; Hu, S.; Yang, N.; Chen, Y.; Wu, X. Establishment and Functional Characterization of Immortalized Rabbit Dermal Papilla Cell Lines. 2021. Available online: https://assets.researchsquare.com/files/rs-1105281/v2/a6a5ae45-071a-42be-bec3-9c2c6334bb6f.pdf?c=1649770637 (accessed on 12 April 2022).
- Zhao, B.; Li, J.; Liu, M.; Hu, S.; Yang, N.; Liang, S.; Zhang, X.; Dai, Y.; Bao, Z.; Chen, Y.; et al. Lncrna2919 suppresses rabbit dermal papilla cell proliferation via trans-regulatory actions. Cells 2022, 11, 2443. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time pcr data by the comparative c t method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Harris, V.M. Protein detection by simple western™ analysis. In Western Blotting; Springer: Berlin, Germany, 2015; pp. 465–468. [Google Scholar]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; Van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D. Jaspar 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Li, J.; Liu, M.; Yang, N.; Bao, Z.; Zhang, X.; Dai, Y.; Cai, J.; Chen, Y.; Wu, X. DNA Methylation Mediates lncRNA2919 Regulation of Hair Follicle Regeneration. Int. J. Mol. Sci. 2022, 23, 9481. https://doi.org/10.3390/ijms23169481
Zhao B, Li J, Liu M, Yang N, Bao Z, Zhang X, Dai Y, Cai J, Chen Y, Wu X. DNA Methylation Mediates lncRNA2919 Regulation of Hair Follicle Regeneration. International Journal of Molecular Sciences. 2022; 23(16):9481. https://doi.org/10.3390/ijms23169481
Chicago/Turabian StyleZhao, Bohao, Jiali Li, Ming Liu, Naisu Yang, Zhiyuan Bao, Xiyu Zhang, Yingying Dai, Jiawei Cai, Yang Chen, and Xinsheng Wu. 2022. "DNA Methylation Mediates lncRNA2919 Regulation of Hair Follicle Regeneration" International Journal of Molecular Sciences 23, no. 16: 9481. https://doi.org/10.3390/ijms23169481
APA StyleZhao, B., Li, J., Liu, M., Yang, N., Bao, Z., Zhang, X., Dai, Y., Cai, J., Chen, Y., & Wu, X. (2022). DNA Methylation Mediates lncRNA2919 Regulation of Hair Follicle Regeneration. International Journal of Molecular Sciences, 23(16), 9481. https://doi.org/10.3390/ijms23169481





