Single Nucleotide Polymorphisms of Toll-like Receptor 4 in Hepatocellular Carcinoma—A Single-Center Study
Abstract
1. Introduction
2. Results
2.1. Patients’ Characteristics
2.2. TLR4 Genotyping
2.3. TLR4 SNPs and HCC Occurrence
2.4. TLR4 SNPs and Secondary Endpoints
2.4.1. All-Cause and Liver-Related Deaths
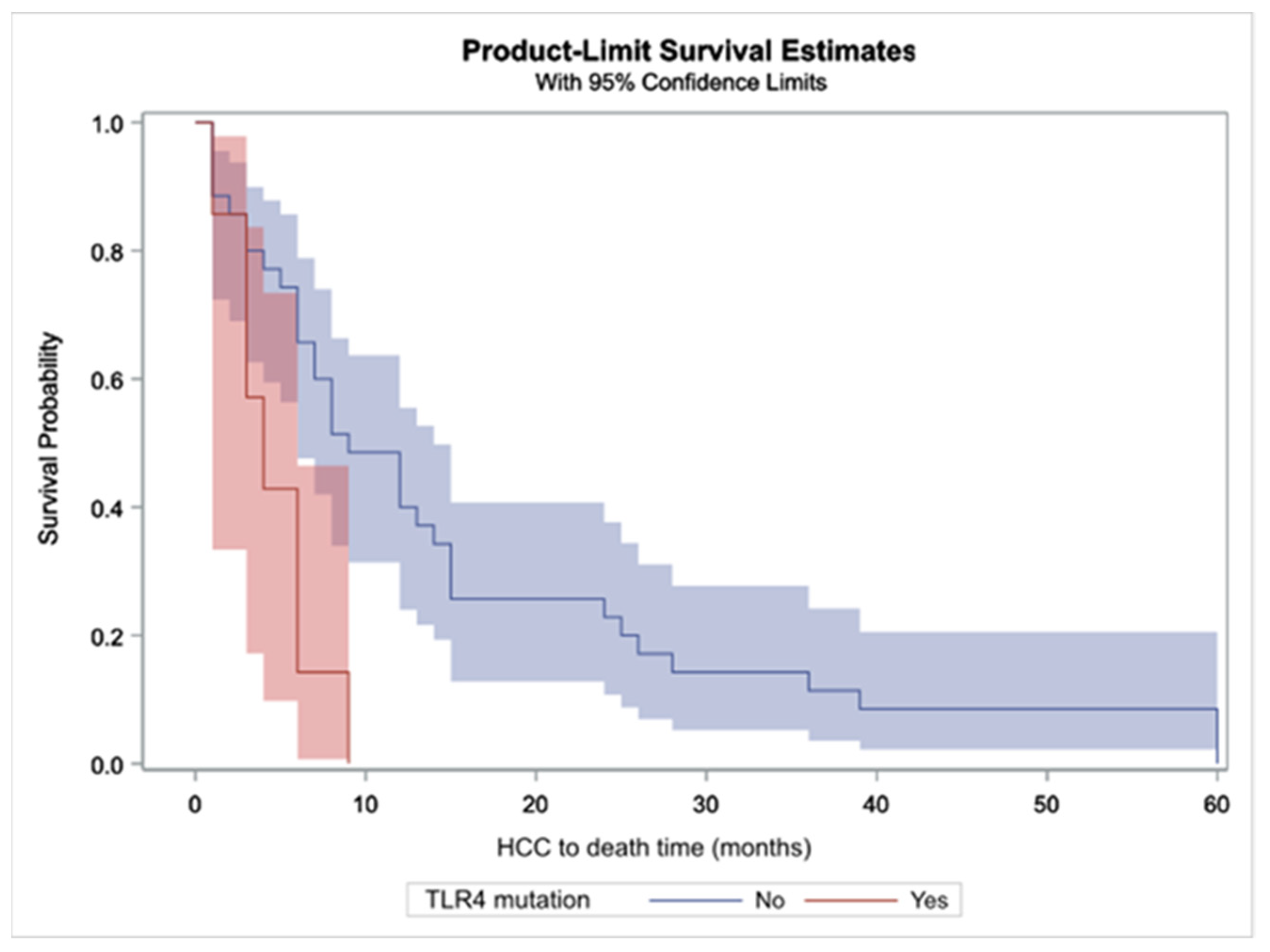
2.4.2. Causes of Death in HCC Patients and Duration between HCC Diagnosis and All-Cause Death
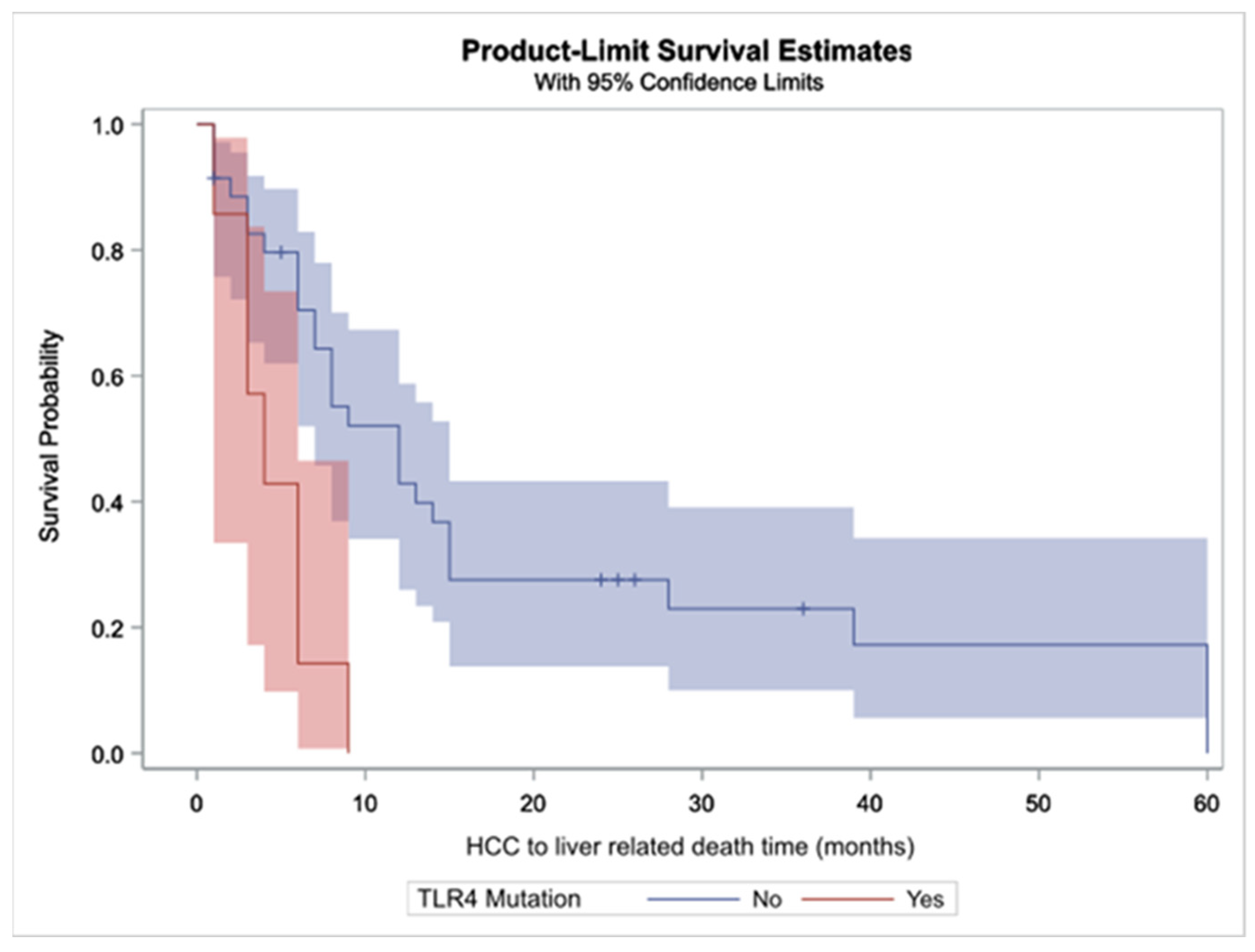
2.4.3. Duration between HCC Diagnosis and Liver Related Death
2.4.4. Duration between Diagnosis of Cirrhosis and HCC Development or Death
3. Discussion
4. Materials and Methods
4.1. Inclusion Criteria and Definitions
4.2. TLR Genotyping
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIH | autoimmune hepatitis |
| ALP | alkaline phosphatase |
| ALT | alanine aminotransferase |
| ASH | alcoholic steatohepatitis |
| AST | aspartate aminotransferase |
| CTP | Child–Turcotte–Pugh |
| gGT | gamma glutamyl-transferase |
| HBV | hepatitis B virus |
| HCV HR | hepatitis C virus hazard ratio |
| INR | international normalized ratio |
| MELD | model for end stage liver disease |
| n | number |
| NASH | non-alcoholic steatohepatitis |
| PBC | primary biliary cholangitis |
| SNP | single nucleotide polymorphism |
| TLR4 | Toll-like receptor-4 |
| HCC | hepatocellular carcinoma |
References
- International Agency for Research on Cancer, World Health Organization. Cancer Today. Available online: https://gco.iarc.fr/today/online-analysis-table (accessed on 29 April 2022).
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Robinson, M.W.; Harmon, C.; O’Farrelly, C. Liver immunology and its role in inflammation and homeostasis. Cell Mol. Immunol. 2016, 13, 267–276. [Google Scholar] [CrossRef]
- Del Campo, J.A.; Gallego, P.; Grande, L. Role of inflammatory response in liver diseases: Therapeutic strategies. World J. Hepatol. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Balkwill, F.; Charles, K.A.; Mantovani, A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005, 7, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.S.; Seki, E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J. Gastroenterol. Hepatol. 2013, 28 (Suppl. 1), 38–42. [Google Scholar] [CrossRef] [PubMed]
- Elsharkawy, A.M.; Mann, D.A. Nuclear factor-kappaB and the hepatic inflammation-fibrosis-cancer axis. Hepatology 2007, 46, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Blasius, A.L.; Beutler, B. Intracellular Toll-like receptors. Immunity 2010, 32, 305–315. [Google Scholar] [CrossRef]
- Kiziltas, S. Toll-like receptors in pathophysiology of liver diseases. World J. Hepatol. 2016, 8, 1354–1369. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Y.; Liu, D.; Liu, J. LPS promotes epithelial-mesenchymal transition and activation of TLR4/JNK signaling. Tumour Biol. 2014, 35, 10429–10435. [Google Scholar] [CrossRef] [PubMed]
- Song, I.J.; Yang, Y.M.; Inokuchi-Shimizu, S.; Roh, Y.S.; Yang, L.; Seki, E. The contribution of Toll-like receptor signaling to the development of liver fibrosis and cancer in hepatocyte-specific TAK1-deleted mice. Int. J. Cancer 2018, 142, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, J.; Lin, H.; Hua, F.; Wang, X.; Liu, H.; Lv, X.; Yu, J.; Mi, S.; Wang, J.; et al. Toll-like receptor 4 activity protects against hepatocellular tumorigenesis and progression by regulating expression of DNA repair protein Ku70 in mice. Hepatology 2013, 57, 1869–1881. [Google Scholar] [CrossRef]
- Nguyen, J.; Jiao, J.; Smoot, K.; Watt, G.P.; Zhao, C.; Song, X.; Stevenson, H.L.; McCormick, J.B.; Fisher-Hoch, S.P.; Zhang, J.; et al. Toll-like receptor 4: A target for chemoprevention of hepatocellular carcinoma in obesity and steatohepatitis. Oncotarget 2018, 9, 29495–29507. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, N.; Tan, H.Y.; Guo, W.; Chen, F.; Zhong, Z.; Man, K.; Tsao, S.W.; Lao, L.; Feng, Y. Direct inhibition of the TLR4/MyD88 pathway by geniposide suppresses HIF-1α-independent VEGF expression and angiogenesis in hepatocellular carcinoma. Br. J. Pharmacol. 2020, 177, 3240–3257. [Google Scholar] [CrossRef]
- Liu, W.T.; Jing, Y.Y.; Gao, L.; Li, R.; Yang, X.; Pan, X.R.; Yang, Y.; Meng, Y.; Hou, X.J.; Zhao, Q.D.; et al. Lipopolysaccharide induces the differentiation of hepatic progenitor cells into myofibroblasts constitutes the hepatocarcinogenesis-associated microenvironment. Cell Death Differ. 2020, 27, 85–101. [Google Scholar] [CrossRef]
- Jing, Y.Y.; Han, Z.P.; Sun, K.; Zhang, S.S.; Hou, J.; Liu, Y.; Li, R.; Gao, L.; Zhao, X.; Zhao, Q.D.; et al. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Med. 2012, 10, 98. [Google Scholar] [CrossRef]
- Liu, W.T.; Jing, Y.Y.; Yu, G.F.; Han, Z.P.; Yu, D.D.; Fan, Q.M.; Ye, F.; Li, R.; Gao, L.; Zhao, Q.D.; et al. Toll like receptor 4 facilitates invasion and migration as a cancer stem cell marker in hepatocellular carcinoma. Cancer Lett. 2015, 358, 136–143. [Google Scholar] [CrossRef]
- Wei, X.; Liu, H.; Li, X.; Liu, X. Over-expression of MiR-122 promotes apoptosis of hepatocellular carcinoma via targeting TLR4. Ann. Hepatol. 2019, 18, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Eiró, N.; Altadill, A.; Juárez, L.M.; Rodríguez, M.; González, L.O.; Atienza, S.; Bermúdez, S.; Fernandez-Garcia, B.; Fresno-Forcelledo, M.F.; Rodrigo, L.; et al. Toll-like receptors 3, 4 and 9 in hepatocellular carcinoma: Relationship with clinicopathological characteristics and prognosis. Hepatol. Res. 2014, 44, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.B.; Pimentel-Nunes, P.; Afonso, L.; Rolanda, C.; Lopes, P.; Roncon-Albuquerque, R., Jr.; Gonçalves, N.; Boal-Carvalho, I.; Pardal, F.; Lopes, S.; et al. Increased hepatic expression of TLR2 and TLR4 in the hepatic inflammation-fibrosis-carcinoma sequence. Innate Immun. 2012, 18, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Zahran, A.M.; Zahran, Z.A.M.; El-Badawy, O.; Abdel-Rahim, M.H.; Ali, W.A.M.; Rayan, A.; Abbas El-Masry, M.; Abozaid, M.A.A.; Hetta, H.F. Prognostic impact of Toll-like receptors 2 and 4 expression on monocytes in Egyptian patients with hepatocellular carcinoma. Immunol. Res. 2019, 67, 157–165. [Google Scholar] [CrossRef]
- Guo, J.; Loke, J.; Zheng, F.; Hong, F.; Yea, S.; Fukata, M.; Tarocchi, M.; Abar, O.T.; Huang, H.; Sninsky, J.J.; et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology 2009, 49, 960–968. [Google Scholar] [CrossRef]
- Plantinga, T.S.; Ioana, M.; Alonso, S.; Izagirre, N.; Hervella, M.; Joosten, L.A.; van der Meer, J.W.; de la Rúa, C.; Netea, M.G. The evolutionary history of TLR4 polymorphisms in Europe. J. Innate Immun. 2012, 4, 168–175. [Google Scholar] [CrossRef]
- Zhang, K.; Zhou, B.; Wang, Y.; Rao, L.; Zhang, L. The TLR4 gene polymorphisms and susceptibility to cancer: A systematic review and meta-analysis. Eur. J. Cancer 2013, 49, 946–954. [Google Scholar] [CrossRef]
- Budulac, S.E.; Boezen, H.M.; Hiemstra, P.S.; Lapperre, T.S.; Vonk, J.M.; Timens, W.; Postma, D.S.; GLUCOLD study group. Toll-like receptor (TLR2 and TLR4) polymorphisms and chronic obstructive pulmonary disease. PLoS ONE 2012, 7, e43124. [Google Scholar] [CrossRef]
- Fan, J.; Liang, R. Quantitative assessment of TLR4 gene polymorphisms and T2DM risk: A meta-analysis. Mol. Genet. Genomic. Med. 2020, 8, e1466. [Google Scholar] [CrossRef]
- Martinez-Chamorro, A.; Moreno, A.; Gómez-García, M.; Cabello, M.J.; Martin, J.; Lopez-Nevot, M.Á. Epistatic interaction between TLR4 and NOD2 in patients with Crohn’s Disease: Relation with risk and phenotype in a Spanish cohort. Immunobiology 2016, 221, 927–933. [Google Scholar] [CrossRef]
- Cai, X.; Fu, Y.; Chen, Q. Association between TLR4 A299G polymorphism and pneumonia risk: A meta-analysis. Med. Sci. Monit. 2015, 21, 625–629. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, H.; Zhou, S.; Zhang, J.; Lei, S.; Zhou, J. Correlations between TLR polymorphisms and inflammatory bowel disease: A meta-analysis of 49 case-control studies. Immunol. Res. 2019, 67, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Singh, J.; Jindal, S.K.; Birbian, N.; Singla, N. Role of TLR4 C>1196T (Thr399Ile) and TLR4 A>896G (Asp299Gly) polymorphisms in a North Indian population with asthma: A case-control study. Int. J. Immunogenet. 2014, 41, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Schröder, N.W.; Schumann, R.R. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet. Infect. Dis. 2005, 5, 156–164. [Google Scholar] [CrossRef]
- Neamatallah, M.; El-Bendary, M.; Elalfy, H.; Besheer, T.; El-Maksoud, M.A.; Elhammady, D.; Abed, S.; Elegezy, M.; Kandeel, L.; Eldeib, D.; et al. Impact of Toll-like Receptors 2(TLR2) and TLR 4 Gene Variations on HCV Susceptibility, Response to Treatment and Development of Hepatocellular Carcinoma in Cirrhotic HCV Patients. Immunol. Invest. 2020, 49, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Salum, G.M.; Dawood, R.M.; Abd El-Meguid, M.; Ibrahim, N.E.; Abdel Aziz, A.O.; El Awady, M.K. Correlation between IL28B/TLR4 genetic variants and HCC development with/without DAAs treatment in chronic HCV patients. Genes Dis. 2019, 7, 392–400. [Google Scholar] [CrossRef]
- Iliadi, A.; Makrythanasis, P.; Tzetis, M.; Tsipi, M.; Traeger-Synodinos, J.; Ioannou, P.C.; Rapti, A.; Kanavakis, E.; Christopoulos, T.K. Association of TLR4 single-nucleotide polymorphisms and sarcoidosis in Greek patients. Genet. Test Mol. Biomark. 2009, 13, 849–853. [Google Scholar] [CrossRef]
- Karananou, P.; Tramma, D.; Katafigiotis, S.; Alataki, A.; Lambropoulos, A.; Papadopoulou-Alataki, E. The Role of TLR4 Asp299Gly and TLR4 Thr399Ile Polymorphisms in the Pathogenesis of Urinary Tract Infections: First Evaluation in Infants and Children of Greek Origin. J. Immunol. Res. 2019, 2019, 6503832. [Google Scholar] [CrossRef] [PubMed]
- Liadaki, K.; Petinaki, E.; Skoulakis, C.; Tsirevelou, P.; Klapsa, D.; Germenis, A.E.; Speletas, M. Toll-like receptor 4 gene (TLR4), but not TLR2, polymorphisms modify the risk of tonsillar disease due to Streptococcus pyogenes and Haemophilus influenzae. Clin. Vaccine Immunol. 2011, 18, 217–222. [Google Scholar] [CrossRef]
- Papadopoulos, A.I.; Ferwerda, B.; Antoniadou, A.; Sakka, V.; Galani, L.; Kavatha, D.; Panagopoulos, P.; Poulakou, G.; Kanellakopoulou, K.; van der Meer, J.W.; et al. Association of Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms with increased infection risk in patients with advanced HIV-1 infection. Clin. Infect. Dis. 2010, 51, 242–247. [Google Scholar] [CrossRef]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology 2004, 127 (5 Suppl. 1), S35–S50. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Sghaier, I.; Zidi, S.; Mouelhi, L.; Ghazoueni, E.; Brochot, E.; Almawi, W.Y.; Loueslati, B.Y. TLR3 and TLR4 SNP variants in the liver disease resulting from hepatitis B virus and hepatitis C virus infection. Br. J. Biomed. Sci. 2019, 76, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Agúndez, J.A.; García-Martín, E.; Devesa, M.J.; Carballo, M.; Martínez, C.; Lee-Brunner, A.; Fernández, C.; Díaz-Rubio, M.; Ladero, J.M. Polymorphism of the TLR4 gene reduces the risk of hepatitis C virus-induced hepatocellular carcinoma. Oncology 2012, 82, 35–40. [Google Scholar] [CrossRef] [PubMed]
- . Zhu, L.; Yuan, H.; Jiang, T.; Wang, R.; Ma, H.; Zhang, S. Association of TLR2 and TLR4 polymorphisms with risk of cancer: A meta-analysis. PLoS ONE 2013, 8, e82858. [Google Scholar] [CrossRef]
- Shi, G.; Wang, C.; Zhang, P.; Ji, L.; Xu, S.; Tan, X.; Li, H. Donor Polymorphisms of Toll-like Receptor 4 rs1927914 Associated with the Risk of Hepatocellular Carcinoma Recurrence Following Liver Transplantation. Arch. Med. Res. 2017, 48, 553–560. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Peng, L.J.; Cao, Y.R.; Zeng, Z.P.; Wu, Y.J.; Shi, H.; Cheng, S.Y.; Wang, J.Y.; Friedman, S.L.; Sninsky, J.J.; et al. Risk Factors for Hepatocellular Carcinoma in Cirrhotic Patients with Chronic Hepatitis B. Genet. Test Mol. Biomark. 2016, 20, 535–543. [Google Scholar] [CrossRef]
- Minmin, S.; Xiaoqian, X.; Hao, C.; Baiyong, S.; Xiaxing, D.; Junjie, X.; Xi, Z.; Jianquan, Z.; Songyao, J. Single nucleotide polymorphisms of Toll-like receptor 4 decrease the risk of development of hepatocellular carcinoma. PLoS ONE 2011, 6, e19466. [Google Scholar] [CrossRef]
- Alvarado-Tapias, E.; Guarner-Argente, C.; Oblitas, E.; Sánchez, E.; Vidal, S.; Román, E.; Concepción, M.; Poca, M.; Gely, C.; Pavel, O.; et al. Toll-like receptor 4 polymorphisms and bacterial infections in patients with cirrhosis and ascites. World J. Hepatol. 2018, 10, 124–133. [Google Scholar] [CrossRef]
- Dinya, T.; Tornai, T.; Vitalis, Z.; Tornai, I.; Balogh, B.; Tornai, D.; Antal-Szalmas, P.; Sumegi, A.; Andrikovics, H.; Bors, A.; et al. Functional polymorphisms of innate immunity receptors are not risk factors for the non-SBP type bacterial infections in cirrhosis. Liver Int. 2018, 38, 1242–1252. [Google Scholar] [CrossRef]
- Agnese, D.M.; Calvano, J.E.; Hahm, S.J.; Coyle, S.M.; Corbett, S.A.; Calvano, S.E.; Lowry, S.F. Human Toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J. Infect. Dis. 2002, 18, 1522–1525. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, E.; Mira, J.P.; Frees, K.L.; Schwartz, D.A. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch. Intern. Med. 2002, 162, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Feterowski, C.; Emmanuilidis, K.; Miethke, T.; Gerauer, K.; Rump, M.; Ulm, K.; Holzmann, B.; Weighardt, H. Effects of functional Toll-like receptor-4 mutations on the immune response to human and experimental sepsis. Immunology 2003, 109, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Ahmad-Nejad, P.; Denz, C.; Zimmer, W.; Wacker, J.; Bugert, P.; Weiss, C.; Quintel, M.; Neumaier, M. The presence of functionally relevant Toll-like receptor polymorphisms does not significantly correlate with development or outcome of sepsis. Genet. Test Mol. Biomark. 2011, 15, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Kumpf, O.; Giamarellos-Bourboulis, E.J.; Koch, A.; Hamann, L.; Mouktaroudi, M.; Oh, D.Y.; Latz, E.; Lorenz, E.; Schwartz, D.A.; Ferwerda, B.; et al. Influence of genetic variations in TLR4 and TIRAP/Mal on the course of sepsis and pneumonia and cytokine release: An observational study in three cohorts. Crit. Care 2010, 14, R103. [Google Scholar] [CrossRef]
- van der Windt, D.J.; Sud, V.; Zhang, H.; Varley, P.R.; Goswami, J.; Yazdani, H.O.; Tohme, S.; Loughran, P.; O’Doherty, R.M.; Minervini, M.I.; et al. Neutrophil extracellular traps promote inflammation and development of hepatocellular carcinoma in nonalcoholic steatohepatitis. Hepatology 2018, 68, 1347–1360. [Google Scholar] [CrossRef]
- . Wang, Z.; Lin, H.; Hua, F.; Hu, Z.W. Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy 2013, 9, 925–927. [Google Scholar] [CrossRef]
- Hsu, C.M.; Yang, M.D.; Chang, W.S.; Jeng, L.B.; Lee, M.H.; Lu, M.C.; Chang, S.C.; Tsai, C.W.; Tsai, Y.; Tsai, F.J.; et al. The contribution of XRCC6/Ku70 to hepatocellular carcinoma in Taiwan. Anticancer Res. 2013, 33, 529–535. [Google Scholar]
- Burgess, D.J. Senescence: Tumorigenesis under surveillance. Nat. Rev. Cancer 2011, 12, 6. [Google Scholar] [CrossRef]
- Kim, W.R.; Biggins, S.W.; Kremers, W.K.; Wiesner, R.H.; Kamath, P.S.; Benson, J.T.; Edwards, E.; Therneau, T.M. Hyponatremia and mortality among patients on the liver-transplant waiting list. N. Engl. J. Med. 2008, 359, 1018–1026. [Google Scholar] [CrossRef]
- Child, C.G.; Turcotte, J.G. Surgery and portal hypertension. Major Probl. Clin. Surg. 1964, 1, 85. [Google Scholar]
- Konstantinidou, M.K.; Goutas, N.; Vlachodimitropoulos, D.; Chaidaroglou, A.; Stefanou, D.; Poumpouridou, N.; Mastorakou, R.; Gazouli, M.; Kyparissopoulos, D.; Spiliopoulou, C. TLR-4 and CD14 Genotypes and Soluble CD14: Could They Predispose to Coronary Atherosclerosis? J. Cardiovasc. Dev. Dis. 2016, 3, 9. [Google Scholar] [CrossRef] [PubMed]
| Patient Characteristic | Value |
|---|---|
| Age in years, median (range) | 65 (25–78) |
| Gender male, n (%) | 175 (67.3) |
| AST (IU/mL), median (range) | 47 (29–75) |
| ALT (IU/mL), median (range) | 32 (20–52) |
| gGT (IU/mL), median (range) | 71 (38–156) |
| ALP (IU/mL), median (range) | 116 (83–179) |
| Total bilirubin (mg/dL), median (range) | 1.35 (0.86–2.94) |
| INR median (range) | 1.34 (1.2–1.6) |
| MELD score, median (range) | 12 (9–18) |
| CTP score, median (range) | 7 (6–9) |
| CTP staging (A/B/C), n (%) | 105/96/59, (40.4/37/22.6) |
| HBV-induced cirrhosis, n (%) | 40 (15.4) |
| HCV-induced cirrhosis, n (%) | 49 (18.9) |
| NASH-induced cirrhosis, n (%) | 34 (13.1) |
| ASH-induced cirrhosis, n (%) | 83 (31.9) |
| PBC-induced cirrhosis, n (%) | 17 (6.5) |
| AIH-induced cirrhosis, n (%) | 8 (3.1) |
| PBC/AIH-induced cirrhosis, n (%) | 7 (2.7) |
| Cryptogenic cirrhosis, n (%) | 10 (3.8) |
| TLR4 299 heterozygous/homozygous SNP, n (%) | 49/13, (18.9/5.0) |
| TLR4 399 SNP heterozygous/homozygous SNP, n (%) | 8/0 (3.1/0) |
| Univariable Analysis | Multivariable Analysis | |||
|---|---|---|---|---|
| Variable | OR (95% CI) | p Value | OR (95% CI) | p Value |
| Age | 1.03 (1–1.05) | 0.0692 | 1.03 (1–1.07) | 0.0348 |
| Gender (Male) | 2.4 (1.12–4.99) | 0.0234 | 2.7 (1.07–6.83) | 0.0358 |
| NASH | 1.64 (0.38–7.11) | 0.3204 | 1.44 (0.31–6.78) | 0.5915 |
| Alcohol | 1.08 (0.28–4.22) | 0.8873 | 0.9 (0.21–3.87) | 0.468 |
| Viral | 1.88 (0.5–7.03) | 0.0653 | 1.88 (0.46–7.66) | 0.1137 |
| Autoimmune | 0.55 (0.1–3.06) | 0.1624 | 0.88 (0.13–5.89) | 0.6357 |
| MELD | 1.01 (0.98–1.05) | 0.4620 | 1 (0.93–1.07) | 0.8892 |
| CTP | 1.07 (0.95–1.21) | 0.2761 | 1.13 (0.89–1.43) | 0.3137 |
| TLR4 Mutations (both) | 0.6 (0.3–1.31) | 0.2071 | 0.08 (0–3.2) | 0.1812 |
| TLR4 299 Mutation (any) | 0.5 (0.23–1.17) | 0.1141 | 5.29 (0.15–183.4) | 0.3575 |
| TLR4 399 Mutation (any) | 2.4 (0.6–10.7) | 0.2231 | 17.7 (0.76–414.14) | 0.0741 |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Parameter | Hazard Ratio | p Value | Hazard Ratio | p Value |
| TLR4 mutation | 0.802 | 0.2690 | 0.717 | 0.1374 |
| MELD | 1.008 | 0.3694 | 0.993 | 0.6746 |
| CTP | 1.024 | 0.5024 | 1.078 | 0.2582 |
| HCC | 1.382 | 0.0936 | 1.314 | 0.1832 |
| Age | 1.007 | 0.3044 | 1.004 | 0.5434 |
| Gender (female) | 0.647 | 0.0276 | 0.555 | 0.0251 |
| Cause of cirrhosis | ||||
| NASH | 0.768 | 0.2275 | 0.645 | 0.4233 |
| Alcohol | 0.905 | 0.6224 | 0.437 | 0.1067 |
| Viral | 1.287 | 0.1851 | 0.441 | 0.1125 |
| Autoimmune | 1.096 | 0.7273 | 0.712 | 0.5662 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Androutsakos, T.; Bakasis, A.-D.; Pouliakis, A.; Gazouli, M.; Vallilas, C.; Hatzis, G. Single Nucleotide Polymorphisms of Toll-like Receptor 4 in Hepatocellular Carcinoma—A Single-Center Study. Int. J. Mol. Sci. 2022, 23, 9430. https://doi.org/10.3390/ijms23169430
Androutsakos T, Bakasis A-D, Pouliakis A, Gazouli M, Vallilas C, Hatzis G. Single Nucleotide Polymorphisms of Toll-like Receptor 4 in Hepatocellular Carcinoma—A Single-Center Study. International Journal of Molecular Sciences. 2022; 23(16):9430. https://doi.org/10.3390/ijms23169430
Chicago/Turabian StyleAndroutsakos, Theodoros, Athanasios-Dimitrios Bakasis, Abraham Pouliakis, Maria Gazouli, Christos Vallilas, and Gregorios Hatzis. 2022. "Single Nucleotide Polymorphisms of Toll-like Receptor 4 in Hepatocellular Carcinoma—A Single-Center Study" International Journal of Molecular Sciences 23, no. 16: 9430. https://doi.org/10.3390/ijms23169430
APA StyleAndroutsakos, T., Bakasis, A.-D., Pouliakis, A., Gazouli, M., Vallilas, C., & Hatzis, G. (2022). Single Nucleotide Polymorphisms of Toll-like Receptor 4 in Hepatocellular Carcinoma—A Single-Center Study. International Journal of Molecular Sciences, 23(16), 9430. https://doi.org/10.3390/ijms23169430








