Systematic Analysis and Functional Characterization of R2R3-MYB Genes in Scutellaria baicalensis Georgi
Abstract
1. Introduction
2. Results
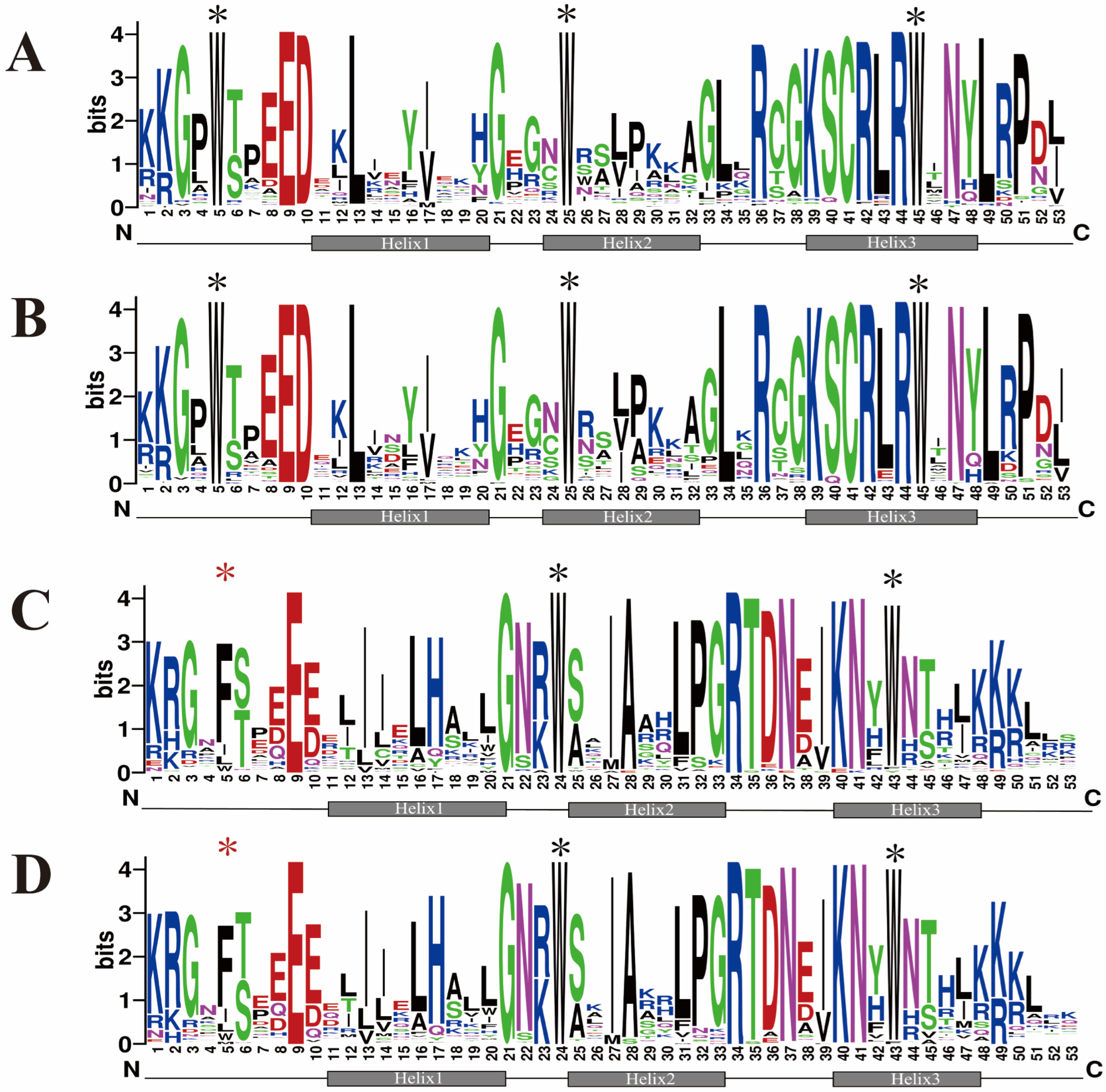
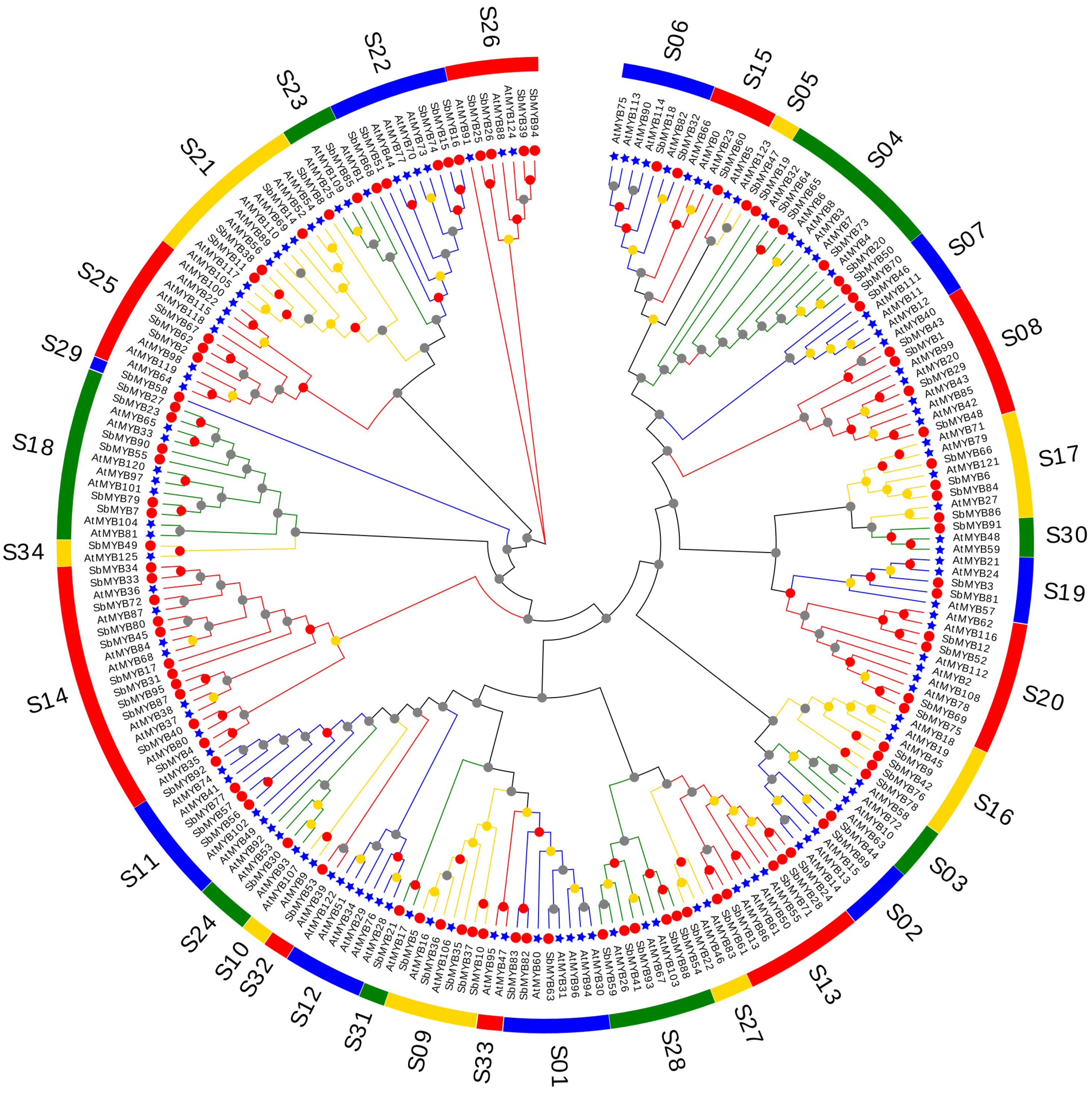
2.1. Identification, Sequence Feature, and Phylogenetic Analysis of the R2R3-SbMYB Members
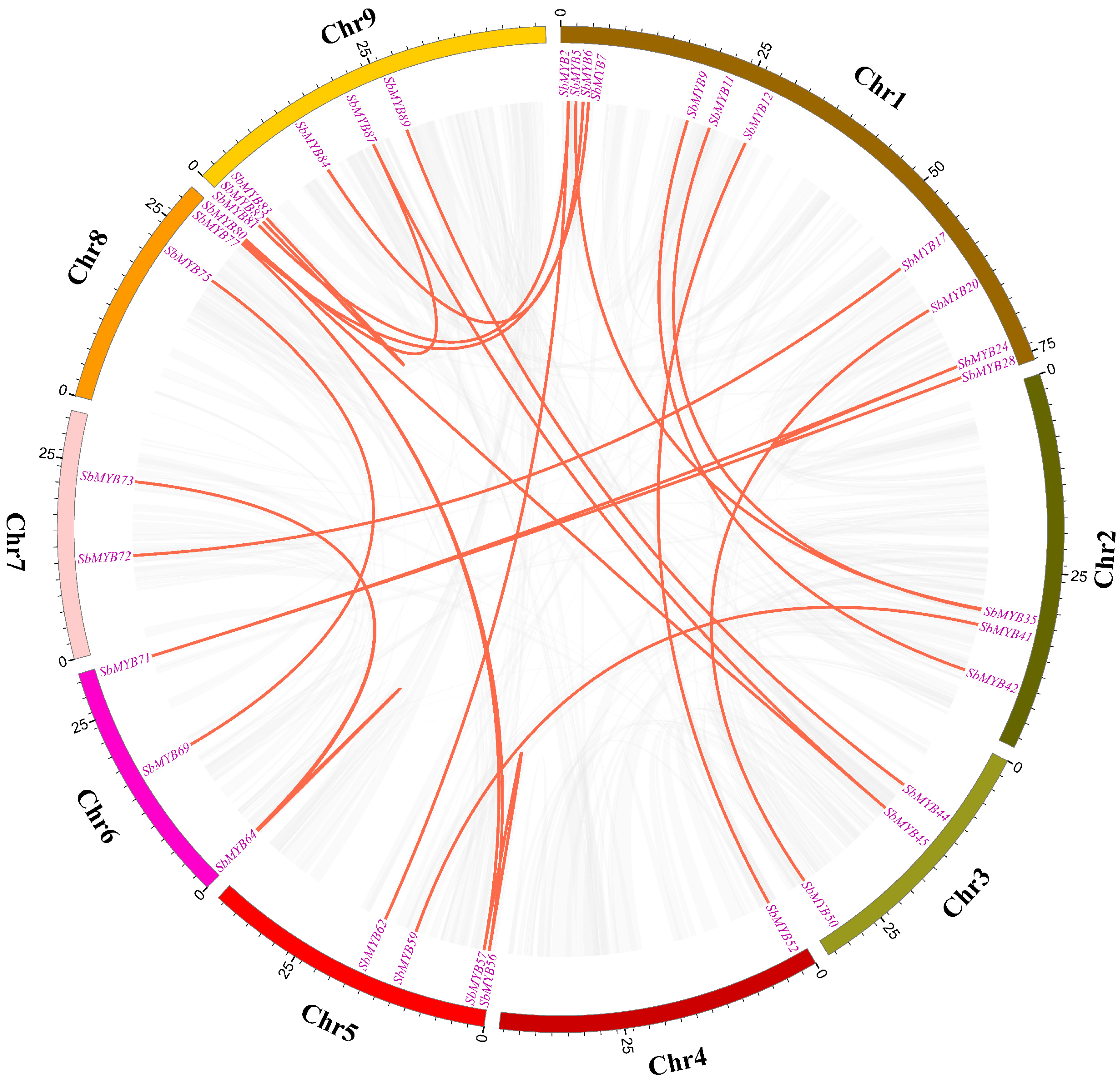
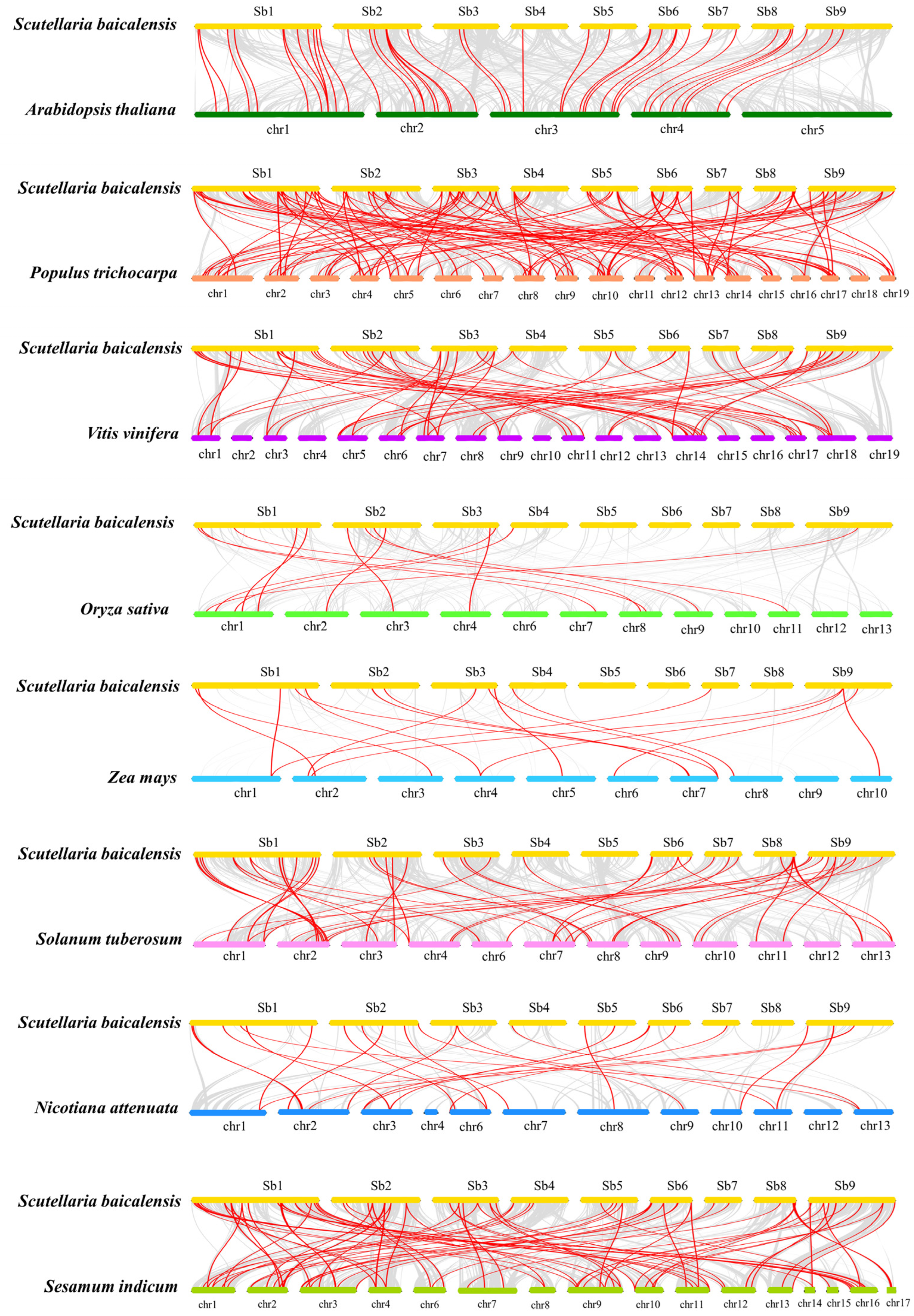
2.2. Analyses of Chromosome Distribution, Gene Duplication, and Synteny for R2R3-SbMYB Genes
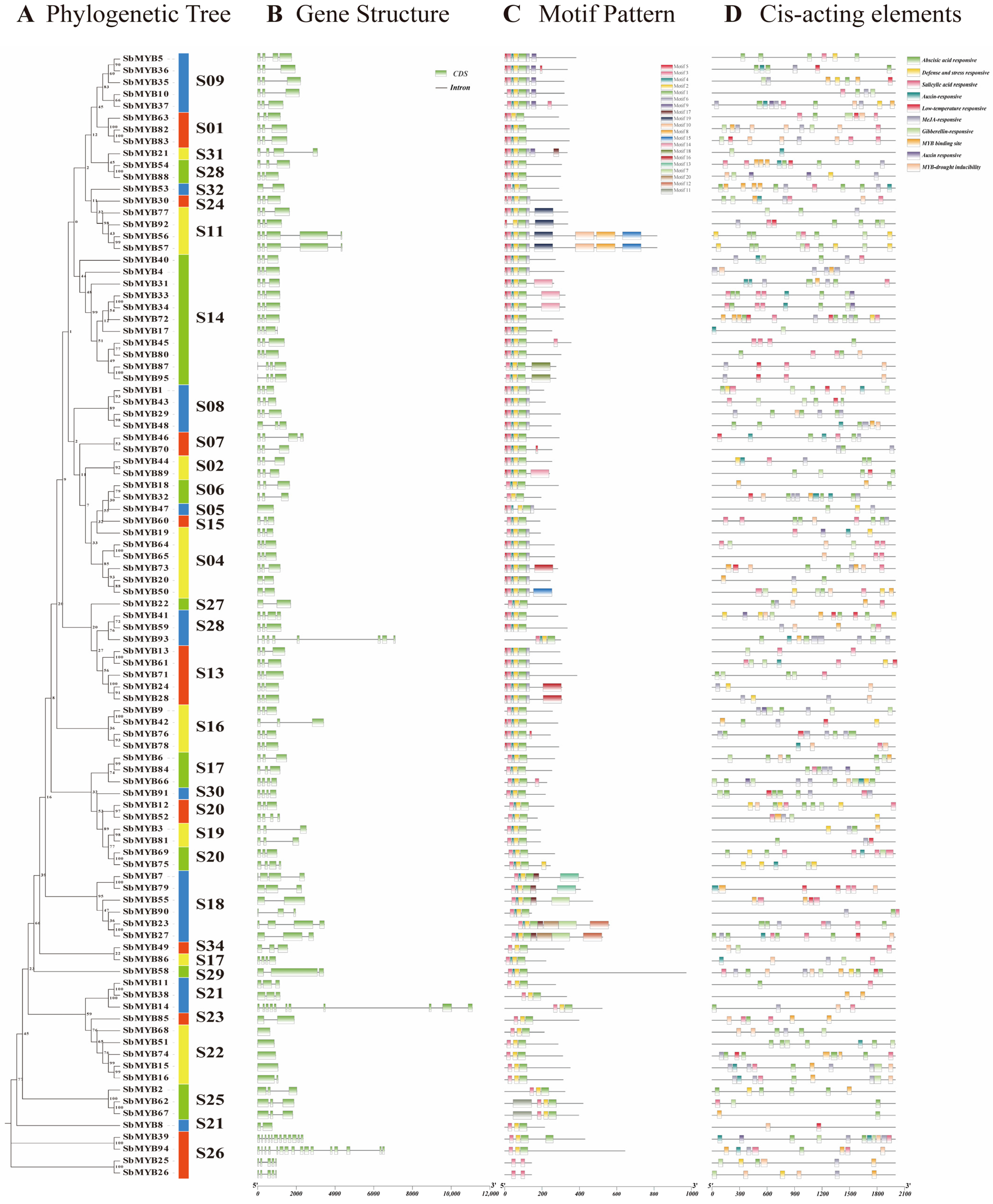
2.3. Gene Structure, Motif Composition, and Cis-Elements Analysis
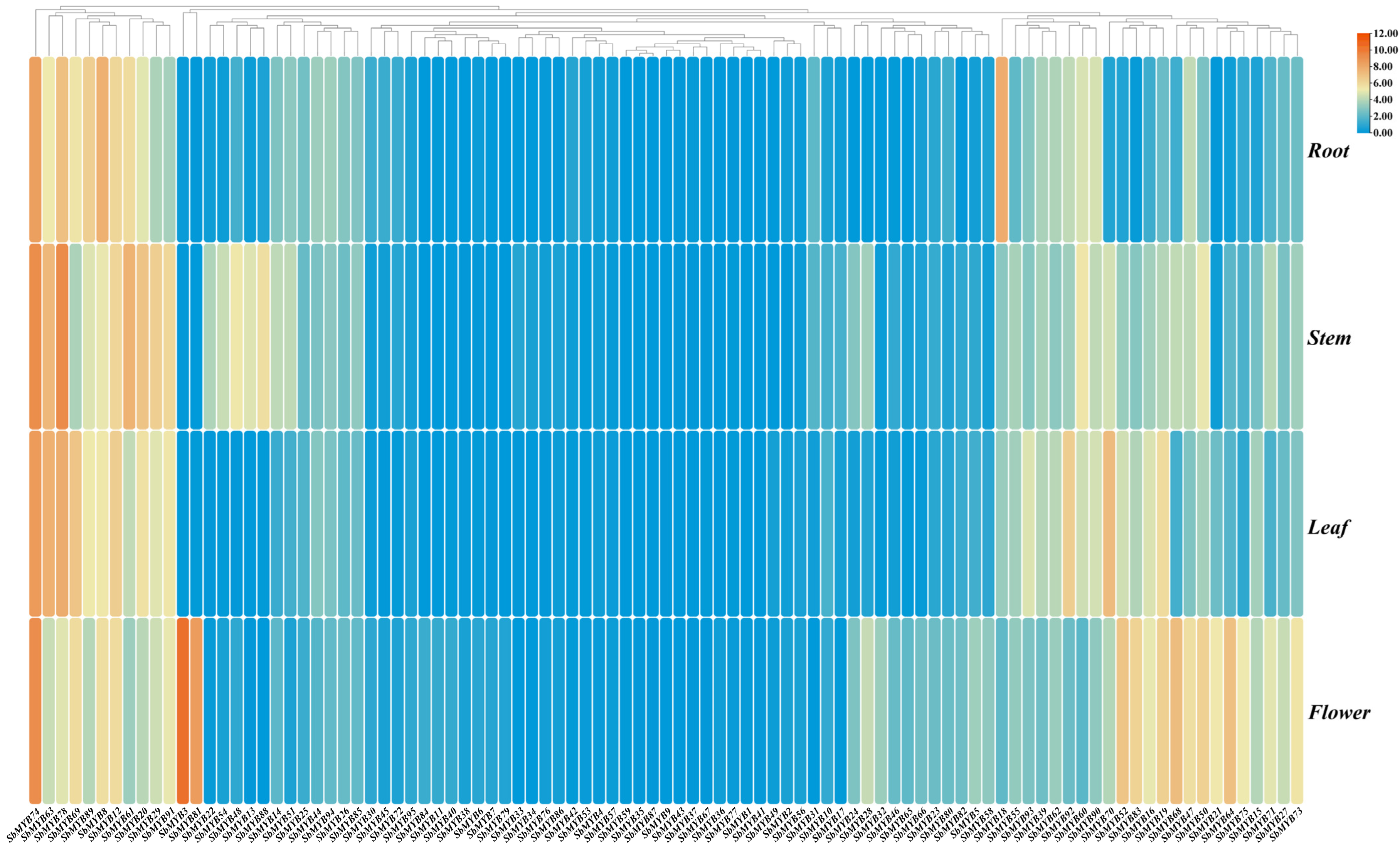
2.4. RNA-Seq Analysis of R2R3-SbMYB Expression in Various Tissues
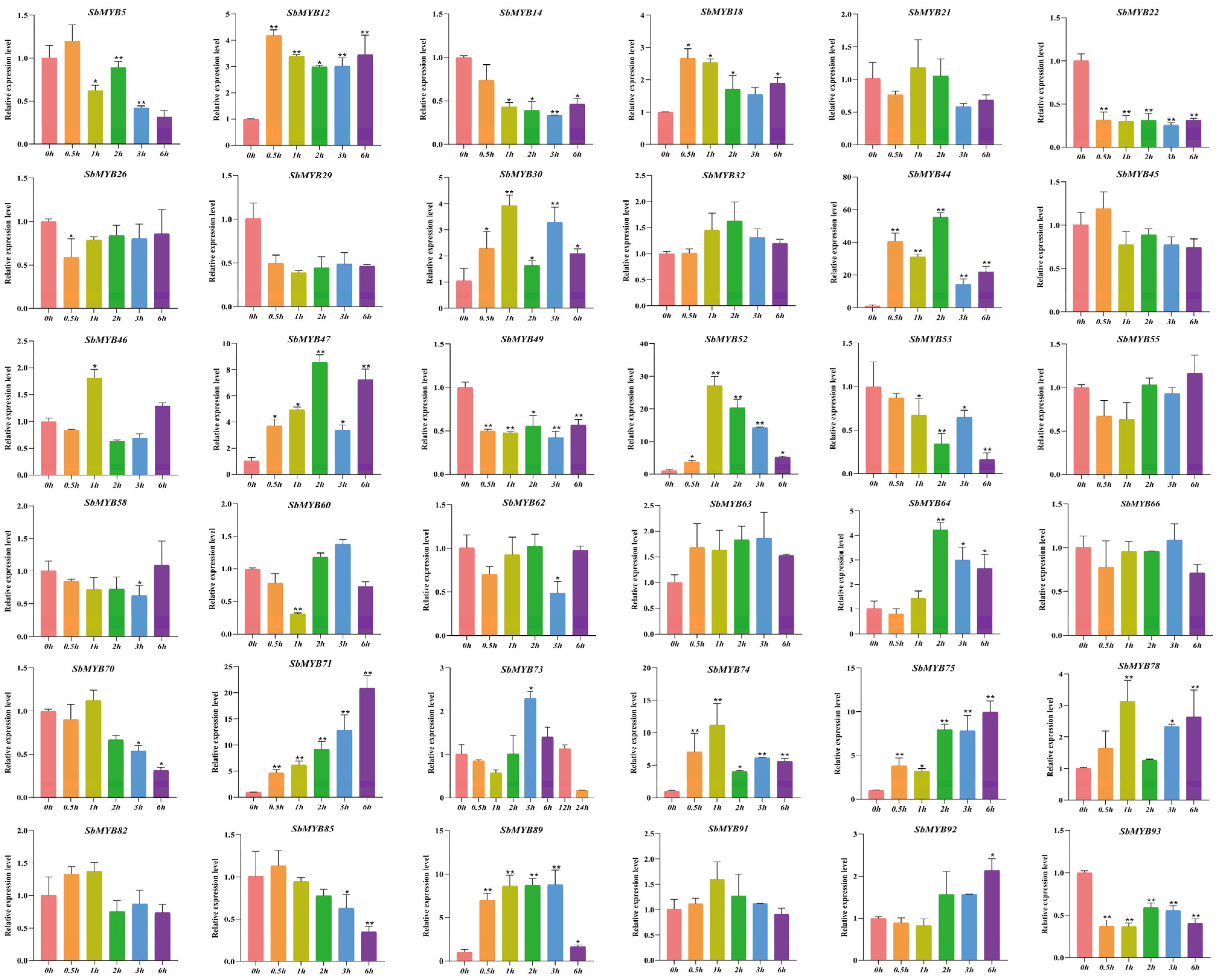
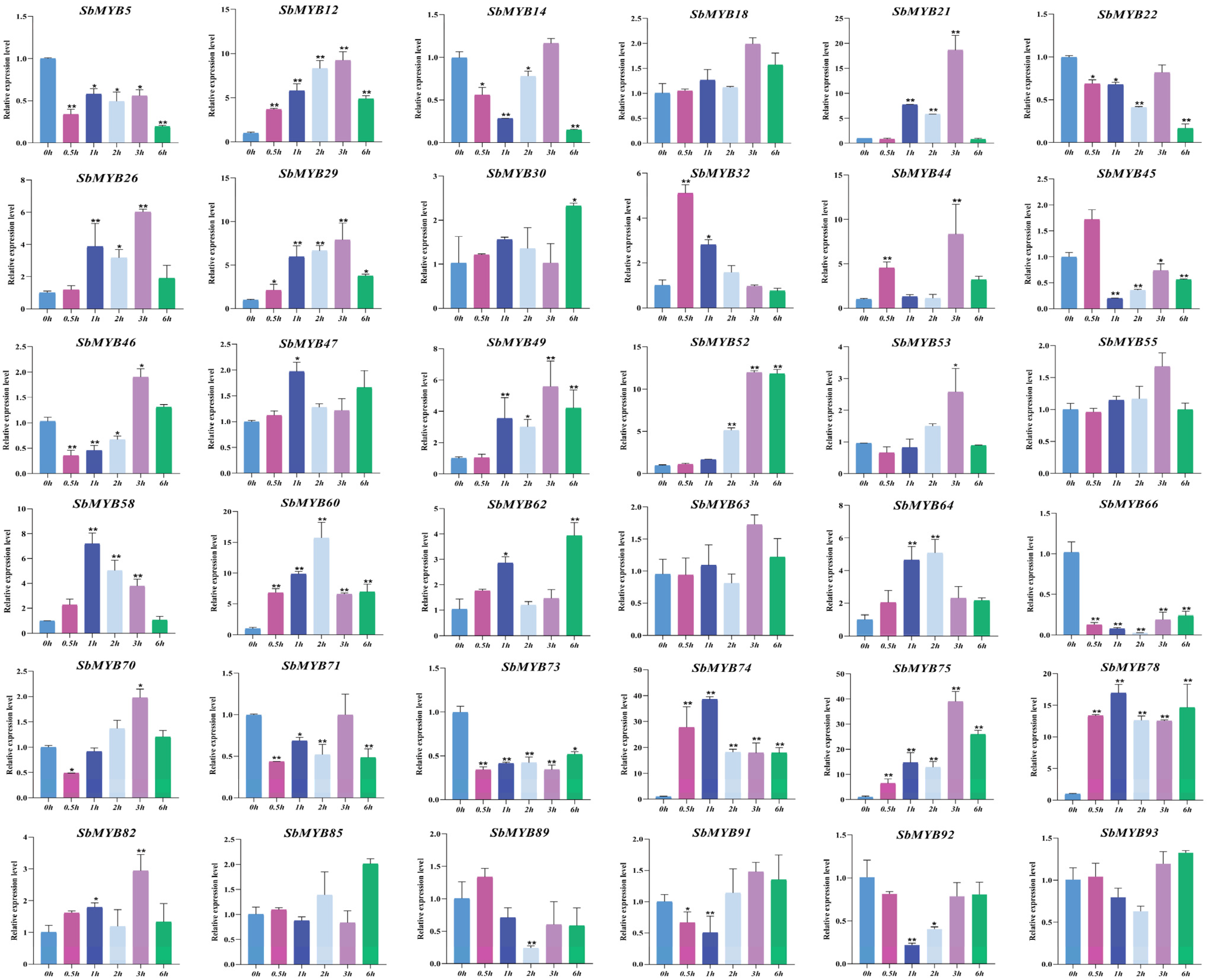
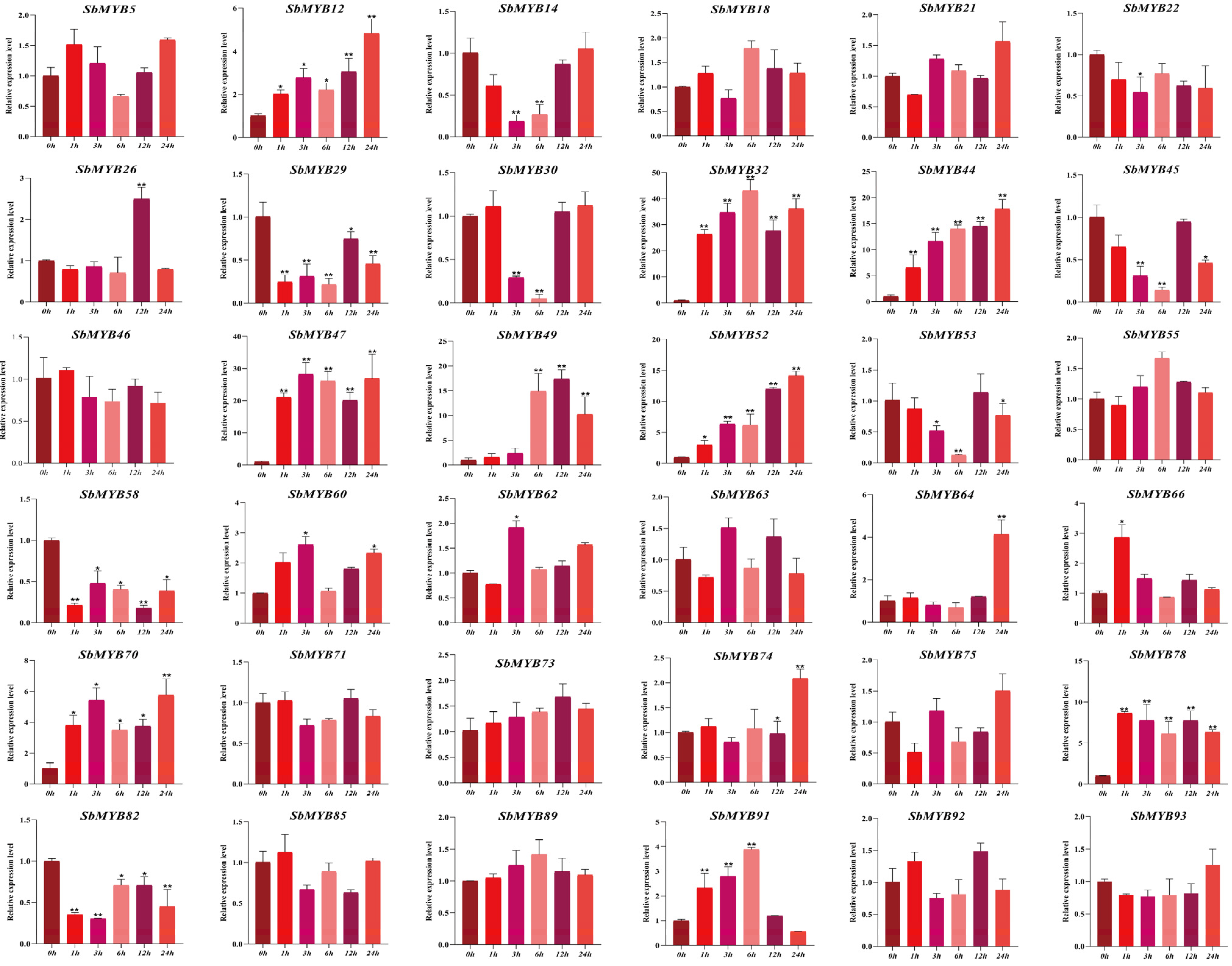
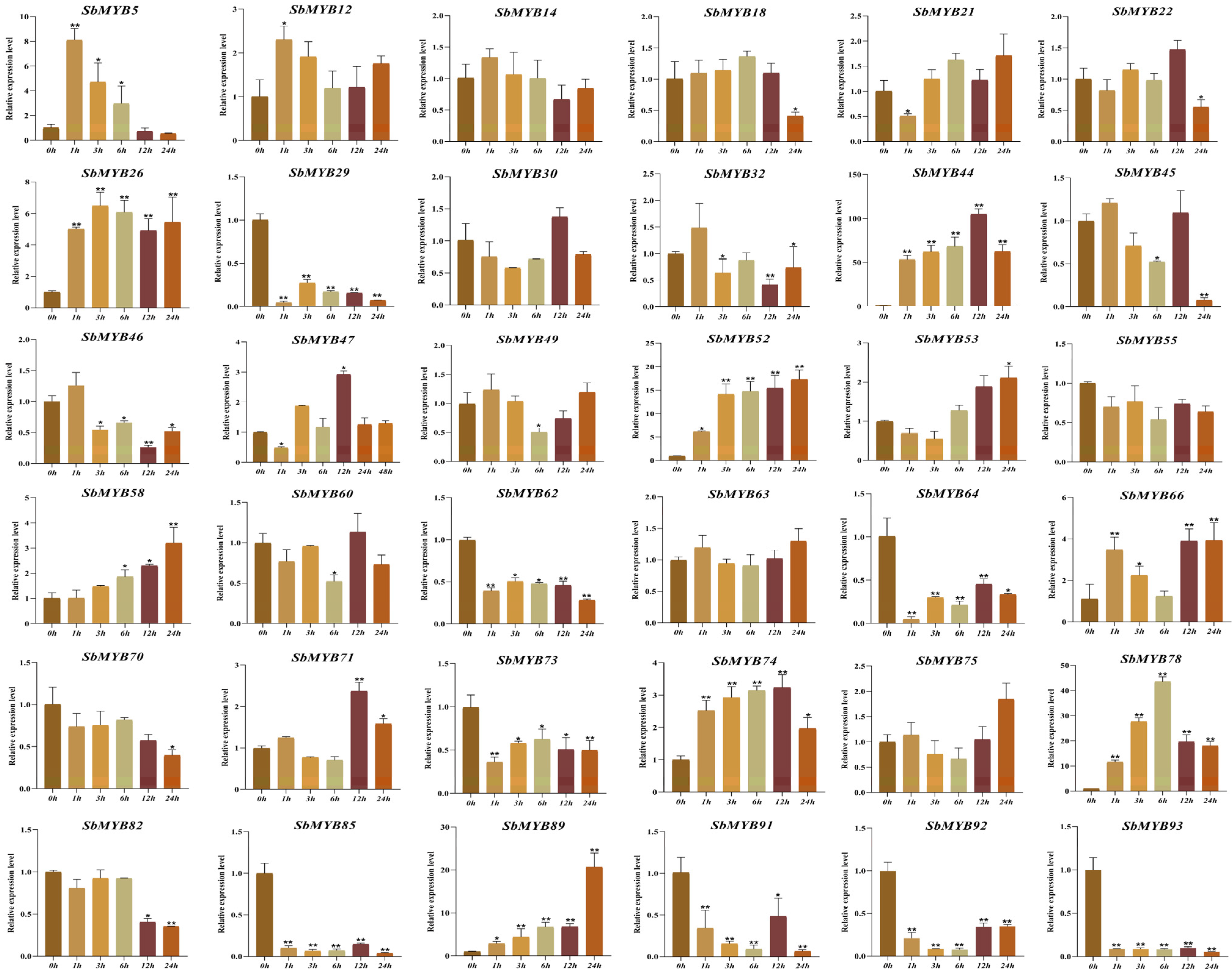
2.5. Expression Patterns of R2R3-SbMYBs under Hormone Stimuli and Abiotic Stresses
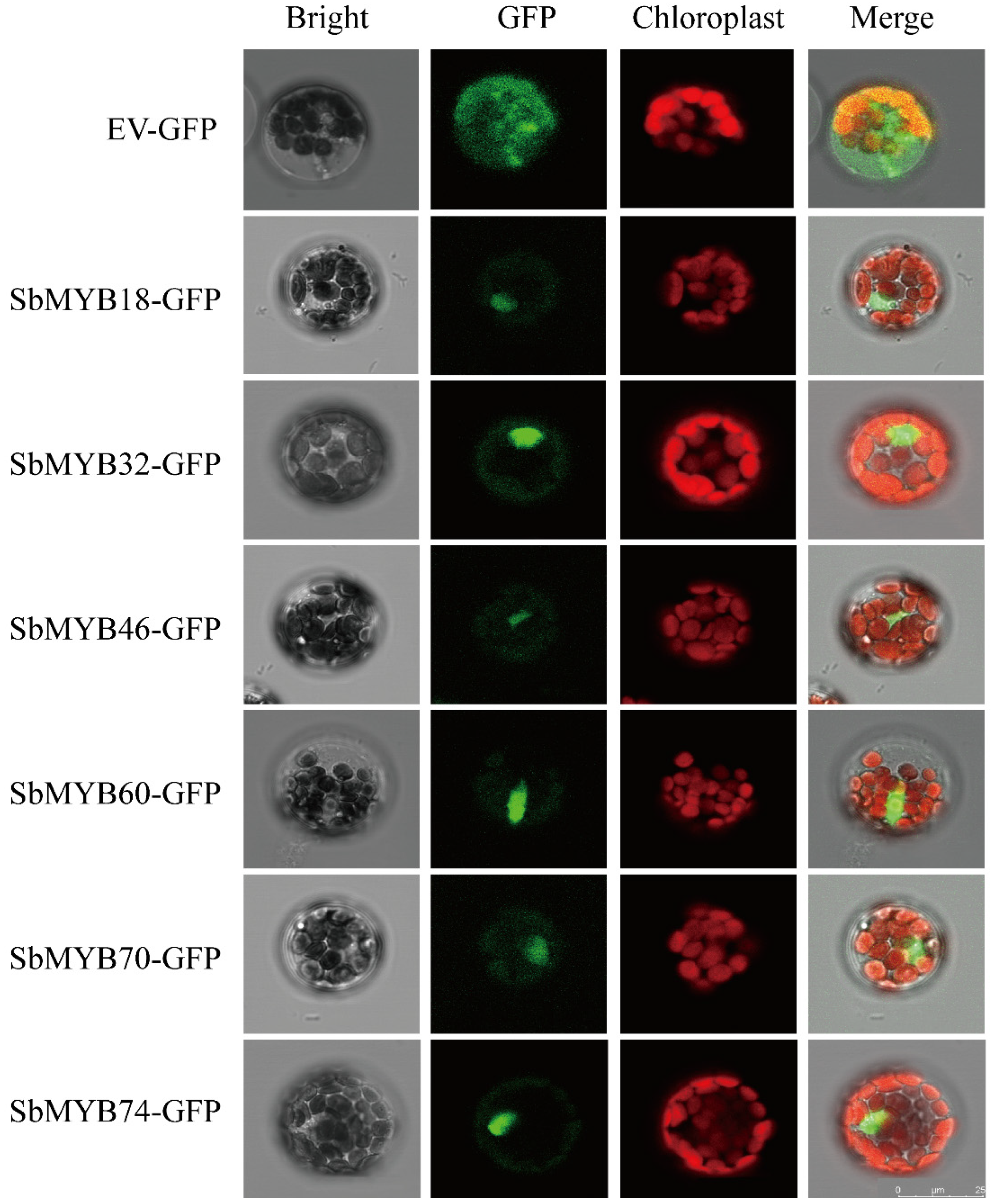
2.6. Subcellular Localization and Trans-Activating Assays of R2R3-SbMYBs
3. Discussion
4. Materials and Methods
4.1. Identification of R2R3-MYB Transcriptional Factors in S. baicalensis
4.2. Chromosome Distribution, Gene Duplication, Synteny, and Phylogenetic Analyses of R2R3-SbMYB Genes
4.3. Gene Structure, Motif Composition, and Cis-Elements Analysis
4.4. Plant Materials and Various Treatments
4.5. RNA-Seq Data and qRT-PCR Analysis
4.6. Subcellular Location Analyses of SbMYBs
4.7. Transcriptional Activation Assays of SbMYBs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef] [PubMed]
- Millard, P.S.; Kragelund, B.; Burow, M. R2R3 MYB Transcription Factors—Functions outside the DNA-Binding Domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Guo, Y.; Sun, Q.; Zeng, W.; Li, J.; Li, X.; Xu, W. Genome-Wide Identification of R2R3-MYB Transcription Factors Regulating Secondary Cell Wall Thickening in Cotton Fiber Development. Plant Cell Physiol. 2019, 60, 687–701. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Hai; Du; Bo-Run; Feng; Si-Si; Yang; Yu-Bi; Huang; Yi-Xiong; Tang, The R2R3-MYB Transcription Factor Gene Family in Maize. PLoS ONE 2012, 7, e37463. [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genom. 2014, 15, 277. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 2020, 145, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Hai, D.; Si-Si, Y.; Zhe, L.; Bo-Run, F.; Lei, L.; Yu-Bi, H.; Yi-Xiong, T. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Guo, T.; Guo, B.; An, X. Comprehensive analysis of the R2R3-MYB transcription factor gene family in Populus trichocarpa. Ind. Crops Prod. 2021, 168, 113614. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, T.; Wang, M.; Li, T.; Wang, G.; Fu, F.F.; Cao, F. Systematic investigation and expression profiles of the GbR2R3-MYB transcription factor family in ginkgo (Ginkgo biloba L.). Int. J. Biol. Macromol. 2021, 172, 250–262. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Shen, X.; Li, H.; Liu, X.; Niu, C.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Denekamp, M.; Smeekens, S.C. Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol. 2003, 132, 1415–1423. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2010, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2010, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2010, 49, 414–427. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N.; et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef]
- Castillejo, C.; Waurich, V.; Wagner, H.; Ramos, R.; Oiza, N.; Muñoz, P.; Triviño, J.C.; Caruana, J.; Liu, Z.; Cobo, N.; et al. Allelic Variation of MYB10 Is the Major Force Controlling Natural Variation in Skin and Flesh Color in Strawberry (Fragaria spp.) Fruit. Plant Cell 2020, 32, 3723–3749. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Kaili, C.; Hongli, L.; Qian, L.; Yali, L. Ectopic Expression of the Grape Hyacinth (Muscari armeniacum) R2R3-MYB Transcription Factor Gene, MaAN2, Induces Anthocyanin Accumulation in Tobacco. Front. Plant Sci. 2017, 8, 965. [Google Scholar] [CrossRef]
- Huang, W.J.; Khaidun, A.B.M.; Chen, J.J.; Zhang, C.J.; Yuan, H.Y. A R2R3-MYB Transcription Factor Regulates the Flavonol Biosynthetic Pathway in a Traditional Chinese Medicinal Plant, Epimedium sagittatum. Front. Plant Sci. 2016, 7, 1089. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lv, H.; Wang, Y. Functional Characterization of a Novel R2R3-MYB Transcription Factor Modulating the Flavonoid Biosynthetic Pathway from Epimedium sagittatum. Front. Plant Sci. 2017, 8, 1274. [Google Scholar] [CrossRef] [PubMed]
- Hemm, M.R.; Herrmann, K.M.; Chapple, C. AtMYB4: A transcription factor general in the battle against UV. Trends Plant Sci. 2001, 6, 135–136. [Google Scholar] [CrossRef]
- Ding, T.; Zhang, R.; Zhang, H.; Zhou, Z.; Liu, C.; Wu, M.; Wang, H.; Dong, H.; Liu, J.; Yao, J.L.; et al. Identification of gene co-expression networks and key genes regulating flavonoid accumulation in apple (Malus × domestica) fruit skin. Plant Sci. 2021, 304, 110747. [Google Scholar] [CrossRef]
- Pérez-Díaz, J.R.; Pérez-Díaz, J.; Madrid-Espinoza, J.; González-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76. [Google Scholar] [CrossRef]
- Yan, J.; Wang, B.; Zhong, Y.; Yao, L.; Cheng, L.; Wu, T. The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 2015, 89, 35–48. [Google Scholar] [CrossRef]
- Liu, R.X.; Song, G.H.; Wu, P.G.; Zhang, X.W.; Hu, H.J.; Liu, J.; Miao, X.S.; Hou, Z.Y.; Wang, W.Q.; Wei, S.L. Distribution patterns of the contents of five biologically activate ingredients in the root of Scutellaria baicalensis. Chin. J. Nat. Med. 2017, 15, 152–160. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, X.Y.; Martin, C. Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci. Bull. 2016, 61, 1391–1398. [Google Scholar] [CrossRef]
- Pei, T.; Yan, M.; Huang, Y.; Wei, Y.; Martin, C.; Zhao, Q. Specific Flavonoids and Their Biosynthetic Pathway in Scutellaria baicalensis. Front. Plant Sci. 2022, 13, 866282. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yao, S.; Zhao, W.; Li, M.; Xu, Y. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv 2020. bioRxiv:2020.04.13.038687. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, Y.; Wang, G.; Hill, L.; Weng, J.K.; Chen, X.Y.; Xue, H.; Martin, C. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2016, 2, e1501780. [Google Scholar] [CrossRef]
- Zhao, Q.; Cui, M.Y.; Levsh, O.; Yang, D.; Liu, J.; Li, J.; Hill, L.; Yang, L.; Hu, Y.; Weng, J.K.; et al. Two CYP82D Enzymes Function as Flavone Hydroxylases in the Biosynthesis of Root-Specific 4’-Deoxyflavones in Scutellaria baicalensis. Mol. Plant 2018, 11, 135–148. [Google Scholar] [CrossRef]
- Bai, C.; Yang, J.; Cao, B.; Xue, Y.; Gao, P.; Liang, H.; Li, G. Growth years and post-harvest processing methods have critical roles on the contents of medicinal active ingredients of Scutellaria baicalensis. Ind. Crops Prod. 2020, 158, 112985. [Google Scholar] [CrossRef]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef]
- Song, X.; Yang, Q.; Liu, Y.; Li, J.; Chang, X.; Xian, L.; Zhang, J. Genome-wide identification of Pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis. Int. J. Biol. Macromol. 2021, 192, 16–27. [Google Scholar] [CrossRef]
- Huang, D.; Ming, R.; Xu, S.; Yao, S.; Li, L.; Huang, R.; Tan, Y. Genome-Wide Identification of R2R3-MYB Transcription Factors: Discovery of a “Dual-Function” Regulator of Gypenoside and Flavonol Biosynthesis in Gynostemma pentaphyllum. Front. Plant Sci. 2021, 12, 796248. [Google Scholar] [CrossRef]
- Lotkowska, M.E.; Tohge, T.; Fernie, A.R.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. The Arabidopsis Transcription Factor MYB112 Promotes Anthocyanin Formation during Salinity and under High Light Stress. Plant Physiol. 2015, 169, 1862–1880. [Google Scholar] [CrossRef]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-Wide Identification, Evolution and Functional Divergence of MYB Transcription Factors in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Cheng, Z.; Xie, L.; Li, X.; Li, J.; Mu, S.; Gao, J. The R2R3MYB Gene Family in Phyllostachys edulis: Genome-Wide Analysis and Identification of Stress or Development-Related R2R3MYBs. Front. Plant Sci. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jin, X.; Ma, Z.; Chen, H.; Liu, M. Basic helix-loop-helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 2020, 155, 1478–1490. [Google Scholar] [CrossRef]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef]
- Yang, M. The FOUR LIPS (FLP) and MYB88 genes conditionally suppress the production of nonstomatal epidermal cells in Arabidopsis cotyledons. Am. J. Bot. 2016, 103, 1559–1566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shin, B.; Choi, G.; Yi, H.; Yang, S.; Cho, I.; Kim, J.; Lee, S.; Paek, N.C.; Kim, J.H.; Song, P.S.; et al. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 2002, 30, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 2020, 228, 1864–1879. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Liao, X.; Luo, X.; Zhu, Y.; Feng, S.; Yu, C.; Lu, J.; Shen, C.; Wang, H. Comparative Metabolomic and Proteomic Analyses Reveal the Regulation Mechanism Underlying MeJA-Induced Bioactive Compound Accumulation in Cutleaf Groundcherry (Physalis angulata L.) Hairy Roots. J. Agric. Food Chem. 2018, 66, 6336–6347. [Google Scholar] [CrossRef]
- Deng, C.; Wang, Y.; Huang, F.; Lu, S.; Zhao, L.; Ma, X.; Kai, G. SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 2020, 62, 1688–1702. [Google Scholar] [CrossRef]
- Cheng, L.; Han, M.; Yang, L.M.; Yang, L.; Sun, Z.; Zhang, T. Changes in the physiological characteristics and baicalin biosynthesis metabolism of Scutellaria baicalensis Georgi under drought stress. Ind. Crops Prod. 2018, 122, 473–482. [Google Scholar] [CrossRef]
- Bhatia, C.; Pandey, A.; Gaddam, S.R.; Hoecker, U.; Trivedi, P.K. Low Temperature-Enhanced Flavonol Synthesis Requires Light-Associated Regulatory Components in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2099–2112. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Cui, Y.; Cui, Z.; Xu, J.; Hao, D.; Shi, J.; Wang, D.; Xiao, H.; Duan, X.; Chen, R.; Li, W. NG-Circos: Next-generation Circos for data visualization and interpretation. NAR Genom. Bioinform. 2020, 2, lqaa069. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Nystrom, S.L.; McKay, D.J. Memes: A motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, W.; Wang, D.; Hu, S.; Zhang, Q.; Wang, Z.; Cui, L. Genome-Wide Identification and Characterization of the WRKY Gene Family in Scutellaria baicalensis Georgi under Diverse Abiotic Stress. Int. J. Mol. Sci. 2022, 23, 4225. [Google Scholar] [CrossRef]
- Wang, W.; Hu, S.; Cao, Y.; Chen, R.; Cao, X. Selection and evaluation of reference genes for qRT-PCR of Scutellaria baicalensis Georgi under different experimental conditions. Mol. Biol. Rep. 2021, 48, 1115–1126. [Google Scholar] [CrossRef]
- Reece-Hoyes, J.S.; Walhout, A.J.M. Gateway Recombinational Cloning. Cold Spring Harb. Protoc. 2018, 2018, pdb.top094912. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Hu, S.; Zhang, C.; Yang, J.; Zhang, T.; Wang, D.; Cao, X.; Wang, Z. Systematic Analysis and Functional Characterization of R2R3-MYB Genes in Scutellaria baicalensis Georgi. Int. J. Mol. Sci. 2022, 23, 9342. https://doi.org/10.3390/ijms23169342
Wang W, Hu S, Zhang C, Yang J, Zhang T, Wang D, Cao X, Wang Z. Systematic Analysis and Functional Characterization of R2R3-MYB Genes in Scutellaria baicalensis Georgi. International Journal of Molecular Sciences. 2022; 23(16):9342. https://doi.org/10.3390/ijms23169342
Chicago/Turabian StyleWang, Wentao, Suying Hu, Caijuan Zhang, Jing Yang, Tong Zhang, Donghao Wang, Xiaoyan Cao, and Zhezhi Wang. 2022. "Systematic Analysis and Functional Characterization of R2R3-MYB Genes in Scutellaria baicalensis Georgi" International Journal of Molecular Sciences 23, no. 16: 9342. https://doi.org/10.3390/ijms23169342
APA StyleWang, W., Hu, S., Zhang, C., Yang, J., Zhang, T., Wang, D., Cao, X., & Wang, Z. (2022). Systematic Analysis and Functional Characterization of R2R3-MYB Genes in Scutellaria baicalensis Georgi. International Journal of Molecular Sciences, 23(16), 9342. https://doi.org/10.3390/ijms23169342





