Abstract
Achromobacter spp. can establish occasional or chronic lung infections in patients with cystic fibrosis (CF). Chronic colonization has been associated with worse prognosis highlighting the need to identify markers of bacterial persistence. To this purpose, we analyzed phenotypic features of 95 Achromobacter spp. isolates from 38 patients presenting chronic or occasional infection. Virulence was tested in Galleria mellonella larvae, cytotoxicity was tested in human bronchial epithelial cells, biofilm production in static conditions was measured by crystal violet staining and susceptibility to selected antibiotics was tested by the disk diffusion method. The presence of genetic loci associated to the analyzed phenotypic features was evaluated by a genome-wide association study. Isolates from occasional infection induced significantly higher mortality of G. mellonella larvae and showed a trend for lower cytotoxicity than chronic infection isolates. No significant difference was observed in biofilm production among the two groups. Additionally, antibiotic susceptibility testing showed that isolates from chronically-infected patients were significantly more resistant to sulfonamides and meropenem than occasional isolates. Candidate genetic biomarkers associated with antibiotic resistance or sensitivity were identified. Achromobacter spp. strains isolated from people with chronic and occasional lung infection exhibit different virulence and antibiotic susceptibility features, which could be linked to persistence in CF lungs. This underlines the possibility of identifying predictive biomarkers of persistence that could be useful for clinical purposes.
1. Introduction
Achromobacter spp. are opportunistic pathogens that can colonize the lungs of patients with cystic fibrosis (CF), causing chronic or occasional infections. In particular, chronic colonization has been associated with a decline in respiratory function, increased frequency of exacerbations and lung inflammation []. Although virulence factors supporting invasiveness and survival have been described (e.g., swimming motility, biofilm formation, lipopolysaccharide, type III secretion system, phospholipase C, proteases) [,,,,,,,,,,,], virulence features related to the ability of Achromobacter spp. to colonize the lungs of CF patients chronically or occasionally are still not fully clear.
Additionally, multidrug resistance strongly contributes to Achromobacter spp. persistence in CF patients. These bacteria show an innate resistance to many classes of antibiotics, especially to those relevant to CF lung infection treatment such as aminoglycosides, aztreonam, tetracyclines, penicillins and cephalosporins [,,]. Moreover, clinical isolates exhibit acquired resistance, especially to β-lactams. The most active agents against Achromobacter spp. are, among others, trimethoprim–sulfamethoxazole, ceftazidime, piperacillin and carbapenems []; however, mutations of genes related to antibiotic resistance may occur, causing resistance to these antibiotics []. Indeed, isolates from long-term chronic infection tend to be resistant to more antibiotics than earlier or occasional isolates [].
The variation in virulence factors and antibiotic resistance among Achromobacter spp. isolates highlights the necessity to better understand the involvement of these features in the pathogenic potential and mechanisms of colonization of these microorganisms. To this purpose, we evaluated virulence, cytotoxicity, biofilm formation and antibiotic susceptibility of clinical isolates causing occasional and chronic CF infections.
2. Results
Ninety-five Achromobacter spp. isolates were analyzed in this study. Seventy-nine isolates (range = 1–11 successively collected isolates, mean = 3.3 isolates/patient) were recovered from 24 chronically-infected CF patients with a mean time delay of 469 days (range = 21–1825 days). One isolate was recovered from each occasionally-infected CF patient (n = 14), except for two patients, P06 and P12, from whom we recovered 2 isolates with a time delay of 112 days and 155 days, respectively. We compared phenotypic features such as virulence, biofilm formation, cytotoxicity and antibiotic susceptibility between chronic and occasional and between early and late chronic isolates.
2.1. Virulence
Virulence is an important feature for bacterial pathogenicity, invasion and interactions with the host. Virulence testing (Figure 1 and Figure S1) in a G. mellonella larvae model showed that isolates from occasionally-infected patients induced significantly higher mortality of larvae than chronic infection isolates (Kaplan–Meier survival estimate p-value = 0.02; Cox hazard ratio = 1.32; 95% CI = 1.04–1.66). When comparing early against late chronic isolates, no significant difference was observed.
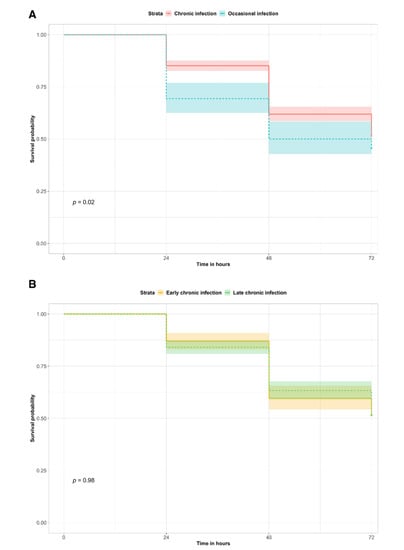
Figure 1.
G. mellonella survival curve for virulence testing. The survival probabilities of G. mellonella larvae infected with chronic or occasional infection isolates (A) and with early or late chronic infection isolates (B) are reported at each time point. The p-values of the survival curve comparisons are indicated with p.
Biofilm formation plays an important role in the persistence of bacteria in CF chronic lung infections, protecting pathogens against environmental stress and increasing tolerance towards antibiotics and host defenses. No significant difference was observed in biofilm production among chronic and occasional isolates and between early and late chronic isolates (Figure 2A,B).
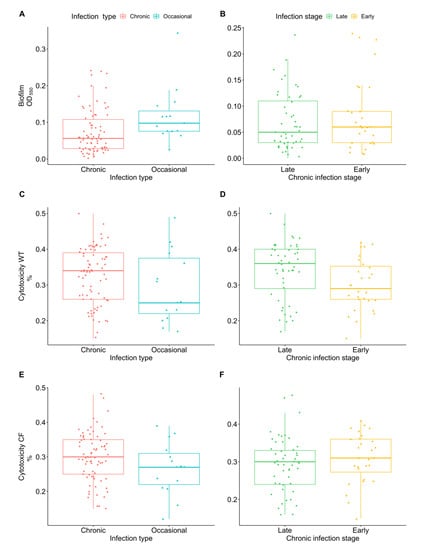
Figure 2.
Achromobacter spp. biofilm formation and cytotoxicity on WT and CF bronchial epithelial cells. Biofilm formation in occasional and chronic isolates (A) and in early and late chronic isolates (B) measured by crystal violet staining (OD550). Cytotoxicity of occasional and chronic isolates and of early and late chronic isolates on WT cells (C,D) and CF cells (E,F) expressed as percentage of LDH release compared to the maximum value (positive control).
The ability to cause cytotoxicity could play an important role in tissue inflammation and degeneration. Cytotoxicity was assessed on both WT and CF bronchial epithelial cells. Although no statistically significant difference was found, we observed that chronic infection isolates showed a trend for greater cytotoxicity than occasional isolates in both cell types (Figure 2C,E). Moreover, when comparing early and late chronic isolates we observed an increase (Wilcoxon Mann–Whitney test p-value = 0.05 after 10,000 permutations) of cytotoxicity from early to late isolates in WT cells (Figure 2D).
Virulence, cytotoxicity and biofilm formation results per isolate are shown in Figure S2.
2.2. Antimicrobial Susceptibility
An important factor for the survival of infectious bacteria is their resistance to administered antibiotics, making infections hard to eradicate. We evaluated six antibiotics that have been reported to show variable susceptibility in chronic strains []: SXT, TGC, SSS, IPM, TZP and MEM.
The majority of strains were resistant to SXT and TGC and sensitive to IPM and TZP, while different susceptibility between occasional and chronic isolates was observed for SSS and MEM (Figure 3): when compared to occasional isolates, strains from chronically-infected patients were significantly more resistant to SSS (Fisher’s exact test p-value = 0.04 after 10,000 permutations; CI = 0.042–0.62; odds ratio = 0.17) and MEM (Fisher’s exact test p-value = 0.01 after 10,000 permutations; CI = 0–0.34; odds ratio = 0). No significant difference in resistance was observed when comparing early and late chronic isolates (Figure 4). Antimicrobial susceptibility results per isolate are shown in Figure S3.
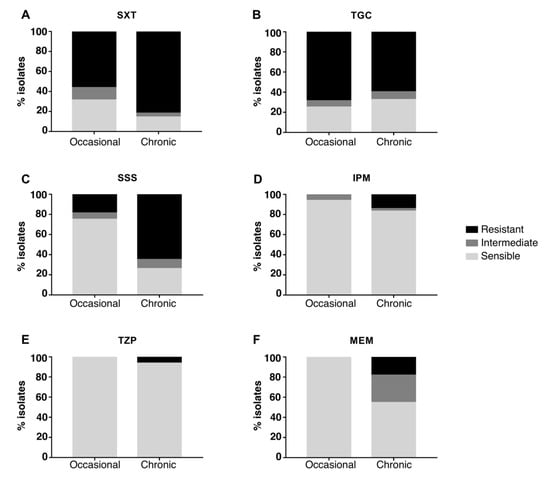
Figure 3.
Antimicrobial susceptibility testing of chronic and occasional isolates. The percentage of resistant, intermediate and sensible isolates from chronic and occasional infection are represented for each antibiotic tested. SXT = trimethoprim–sulfamethoxazole (A), TGC = tigecycline (B), SSS = sulfonamides (C), IPM = imipenem (D), TZP = piperacillin-tazobactam (E), MEM = meropenem (F).
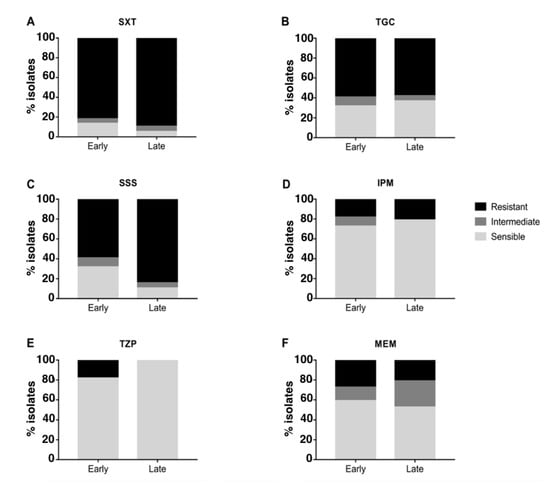
Figure 4.
Antimicrobial susceptibility testing of early and late chronic isolates. The percentage of resistant, intermediate and sensible isolates from chronic and occasional infection are represented for each antibiotic tested. SXT = trimethoprim–sulfamethoxazole (A), TGC = tigecycline (B), SSS = sulfonamides (C), IPM = imipenem (D), TZP = piperacillin-tazobactam (E), MEM = meropenem (F).
2.3. Biomarkers of Antimicrobial Resistance
Genome analysis of the 54 Achromobacter spp. isolates from the Verona collection was performed to identify genetic components significantly associated with virulence, biofilm, cytotoxicity and antimicrobial resistance. As regards the former three features, no genetic loci were significantly associated to the phenotypic traits. Within the tested antibiotics, we found various candidate biomarkers of resistance or sensitivity linked to transmembrane transporters and efflux pumps (e.g., secretory components, ABC transporters), transcriptional regulators (e.g., AraC) and metabolic enzymes (Table 1 and Table S1 for details). In addition, some hypothetical proteins were associated with sensitivity to SSS and resistance to MEM. Some node sequences showed statistically significant correlation (q-value ≤ 0.05) with both IPM and MEM, and with both SSS and SXT (Table S1).

Table 1.
Genetic loci significantly associated with antimicrobial resistance. The most significant results of association analysis are reported. IPM = imipenem, SXT = trimethoprim–sulfamethoxazole, SSS = sulfonamides (sulphadiazine, sulphathiazole), MEM = meropenem, q-value = p-value adjusted for the False Discovery Rate.
3. Discussion
To identify phenotypic features related to the ability of Achromobacter spp. to establish chronic or occasional colonization in CF airways, we evaluated virulence, biofilm formation, cytotoxicity, and antimicrobial susceptibility of 95 clinical isolates and compared results between chronic and occasional isolates and between early and late chronic ones.
Isolates were collected at the CF Center of Verona (Italy) and Bambino Gesù Hospital in Rome (Italy). While the Verona collection included a larger number of occasional isolates, the Rome one comprised many longitudinally-isolated chronic strains encompassing a long period of time. The two collections showed similar phenotypic characteristics (Figures S2 and S3); so, their combination allowed the analysis of a more homogeneous group of strains than using them separately; however, the final collection still included a lower number of occasional (n = 16) than chronic isolates (n = 79, 30 early and 49 late chronic isolates).
To evaluate the virulence of Achromobacter spp. isolates, we used the well-characterized G. mellonella larvae model [,]. Occasional infection isolates caused higher mortality than strains from chronic infection, indicating that Achromobacter spp. exhibit higher virulence during occasional infection. This observation suggests that virulence attenuation could be a key factor during the establishment of chronic infection. Differences in virulence between early and late chronic strains of another CF pathogen, Pseudomonas aeruginosa, were also previously highlighted [], leading to the hypothesis that multiple mutations could be responsible for virulence attenuation during late infection. Although we did not observe a significant difference in virulence between early and late chronic isolates of Achromobacter spp., a similar mechanism of adaptation could be proposed for occasional and chronic isolates, where selection of strains with an increasing ability to persist may occur.
As for biofilm formation, the majority of our isolates showed a low-moderate production of biofilm, confirming the poor adhesion ability of Achromobacter spp. on surfaces [,]. Although no significant difference in biofilm production was observed between chronic and occasional isolates nor between early and late chronic isolates, the great majority of strains unable to form biofilm were isolated from chronic infection. This could suggest a mechanism of within-host adaptation in the CF lung; e.g., acquisition of mutations in genes with a role in surface adhesion could lead to decreased biofilm production or to formation of unattached aggregates [,,].
To investigate whether a reduced virulence in the chronic isolates coincided with lower cytotoxicity, we compared the cytotoxic potential of chronic and occasional strains in WT and F508del human bronchial epithelial cultured cells, but no significant difference was found. Although no statistically significant difference was found, we observed that chronic infection isolates induced slightly greater cytotoxicity than occasional isolates—an opposite trend compared to the results of biofilm and virulence testing. We observed an increased cytotoxicity from early to late isolates in cells expressing WT CFTR. No significant difference was observed in CF cells; this could indicate an underlying adaptation of late chronic strains to the CF lung environment leading to a more indolent colonization.
Biomarkers analysis of virulence traits (biofilm, cytotoxicity and virulence in G. mellonella larvae) showed no associated genetic loci. This is probably due to the fact these aspects are known to be mainly regulated through RNA modulation [,] rather than through the accumulation of genomic mutations.
Achromobacter spp. are reported to increasingly develop resistance to various antibiotics. We tested susceptibility of all isolates to 6 antibiotics that have been reported to show variable susceptibility in chronic strains []. Achromobacter spp. strains generally displayed resistance to TGC and susceptibility to TZP and IPM, confirming their previously-reported innate resistance to tetracyclines [] and susceptibility to beta-lactams and carbapenems []. Interestingly, an increased resistance to MEM was observed in chronic isolates. Moreover, even though IPM and MEM belong to the same class of antimicrobials, we observed both imipenem resistant but meropenem susceptible (IRMS) and meropenem resistant but imipenem susceptible (MRIS) phenotypes. These phenotypes were previously reported for P. aeruginosa and various members of the Enterobacteriaceae family [] as well as for Achromobacter spp. clinical isolates [,]. In addition, we observed significantly higher resistance to SSS in chronic infection isolates than in occasional ones, in concordance with previous studies [,]. Resistance of chronic infection isolates to SSS could be associated with the use of SXT to eradicate CF pathogens. The majority of both occasional and chronic isolates was resistant also to SXT, in contrast with previous investigations; e.g., a recent one found that the majority of Achromobacter spp. strains from Danish CF patients were sensitive to SXT [], considered as one of the most active agents against Achromobacter spp. infections []. This suggests that the acquisition of resistance to SXT might have occurred among isolates in our collections, e.g., through the spread of mobile genetic elements carrying resistance genes [] or due to the different antibiotic treatment regimens used.
Candidate biomarkers were identified for SXT, IPM, MEM and SSS by analyzing sequenced isolates. Some of them are known to be involved in antibiotic resistance, such as ABC transporters [], while the role of other candidates—associated with antibiotic resistance or with sensitivity—should be further investigated. Of particular interest are hypothetical proteins, which could provide additional information on Achromobacter spp. resistance mechanisms upon further characterization. These candidate biomarkers could help in the identification of strains that are becoming persistent and support their eradication before chronic infection is fully developed. Finally, in order to assess whether biomarkers presence could be linked to an adaptation mechanism such as clonal expansion or horizontal gene transfer, we further performed genome analysis but neither of these hypotheses was confirmed. In conclusion, our results show that Achromobacter spp. isolates from chronic and occasional lung infection exhibit different virulence and antibiotic resistance characteristics, some of which might be linked to persistence in CF lungs. We identified potential predictive markers of persistence such as decreased virulence, higher cytotoxicity, resistance to antibiotics, as well as genetic biomarkers [], that could be translated into the clinical setting either to help preventing the development of chronic infections or to support therapeutic treatments aimed at eradicating Achromobacter spp.
4. Materials and Methods
4.1. Samples Collection
Ninety-five Achromobacter spp. isolates were collected from the sputum samples of 38 patients followed at the CF Center of Verona and Bambino Gesù Hospital in Rome (Italy): 54 isolates were recovered from 26 patients in Verona [,], while 41 isolates were recovered from 12 patients in Rome. Patients were classified as occasionally- and chronically-infected with Achromobacter spp. according to the European Consensus Criteria or Leeds criteria. In the Verona collection, 43 longitudinal isolates were collected from 17 patients with chronic infections while 11 strains were collected from 9 patients with occasional infection. The Rome collection comprised 36 strains collected over time from 7 patients with chronic infection and 5 strains collected from 5 patients with occasional infections. Isolates from chronically-infected patients were further classified as early (<1 year from 1st colonization event) and late isolates (>1 year from 1st colonization). Relatedness of strains was verified to confirm that subsequent isolates from one patient actually represented a single strain. An overview of the isolates included in each collection is reported in Table 2. Informed consent was obtained according to projects CRCFC-CEPPO026 and CRCFC-CEPPO031, approved by the Ethical Committee. All the isolates included in this study were identified as Achromobacter spp. by MALDI-TOF-MS (bioMerieux Marcy-l’Étoile, France). Strains were stored in Microbank (Pro-Lab Diagnostics, Neston, UK) at −80 °C. Detailed information regarding the collections is reported in Table S2.

Table 2.
Achromobacter spp. collections summary. The number of isolates included in each collection is reported in the table; the number of patients from which the strains were collected is indicated in parenthesis. E = early chronic infection isolates; L = late chronic infection isolates.
4.2. Virulence Testing
Virulence was assessed in Galleria mellonella larvae. Ten larvae were inoculated with a 1 × 106 CFU bacterial suspension of each clinical isolate through the last proleg into the haemocoel using a 0.3 mL syringe and incubated in Petri dishes, on filter paper, at 37 °C, in the dark. In the control group, larvae were injected with sterile saline solution. Larvae were monitored daily up to 72 h and death was assessed by lack of movement after stimulation and blackening.
4.3. Biofilm Formation Assay
Bacterial strains were plated onto LB agar and grown at 37 °C for 24–48 h. A single colony was inoculated in BHI medium and grown for 16 h at 37 °C with shaking. OD600 was measured, cultures were diluted to 0.1 OD/mL and 200 µL/well were incubated in a 96-well plate for 24 h at 37 °C. Wells were washed with saline solution and stained with 0.1% crystal violet solution for 15 min, then rinsed, washed with water and air dried. After 30 min of incubation with 30% acetic acid at 37 °C, absorbance at 550 nm was measured.
4.4. Cytotoxicity Testing
Human CF bronchial epithelial cell lines CFBE14o- 4.7 WT-CFTR (WT cells) and DeltaF508-CFTR (CF cells) (Merck, Darmstadt, Germany), overexpressing WT and F508del CFTR cDNA, respectively, were cultured in 200 µL EMEM supplemented with 1% Fetal Bovine Serum, 0.5–2 µg/mL Puromycin and 2 mM L-Glutamine into Fibronectin/Collagen/BSA-coated 96-well plates incubated at 37 °C and 5% CO2. At 80% confluency, 50 µL/well of 2 OD600/mL bacterial suspension were added to cell cultures and incubated at 37 °C for 4 h. CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) measuring LDH release was used according to manufacturer’s instructions. Briefly, 50 µL of cells suspension were added with 50 µL of CytoTox 96 Reagent and incubated for 30 min in the dark. After adding 50 µL of Stop Solution, absorbance at 450 nm was recorded. Cytotoxicity was calculated by dividing for the absorbance of the positive control (treated with Lysis Solution).
4.5. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was determined by disk diffusion assay. Bacterial suspension at 0.5 McFarland was streaked onto Mueller–Hinton agar plates, antibiotic-containing disks (Oxoid) were placed onto the agar surface, plates were incubated at 37 °C for 48 h and the diameter of the zone-of-inhibition was measured. Disks contained 1.25/23.75 µg trimethoprim–sulfamethoxazole (SXT), 15 µg tigecycline (TGC), 300 µg sulfonamides (sulphadiazine, sulphathiazole) (SSS), 10 µg imipenem (IPM), 100/10 µg piperacillin-tazobactam (TZP) or 10 µg meropenem (MEM). Since no EUCAST or CLSI breakpoint standard is available for Achromobacter spp., susceptibility profiles were interpreted as resistant (R), intermediately resistant (I) or sensible (S) based on breakpoints proposed in previous literature [,,].
4.6. Statistical Analysis
Statistical analysis was carried out to compare chronic and occasional isolates and early and late chronic isolates. Virulence results were tested by the Kaplan–Meier method using the log-rank test to compare the overall survival of larvae over an observation period of 72 h. Hazard ratios were computed with a Cox regression model. Cytotoxicity and biofilm formation results were tested using a Wilcoxon Mann–Whitney test. Outlier values were observed in biofilm formation results (n = 2, isolates 7-3 and 12-2) and were excluded from statistical analysis. Fisher’s exact test was used to ascertain the significance of antibiotic susceptibility results. Since observations per isolate were not independent due to a longitudinal collection strategy, for each test, p-values were adjusted performing 10,000 permutations of the infection type (chronic, occasional) or infection stage (early chronic, late chronic) stratified by collection (Verona, Rome). R version 4.0.4 (R Foundation for Statistical Computing, Vienna, Austria) [] was used for statistical analysis and for results visualization. Boxplots were generated using ggpubr v0.4.0, survival curves using survminer v0.4.9, and heatmaps using pheatmap v1.0.8 R libraries.
4.7. Identification of Genetic Loci Associated with Virulence, Biofilm, Cytotoxicity and Antimicrobial Resistance
The DBGWAS 0.5.4 software (bioMerieux, Lyon, France) [] was used to identify genetic components significantly associated with virulence, biofilm, cytotoxicity and antimicrobial resistance. In previous work [,] we sequenced the whole genome of the 54 Achromobacter spp. isolates from the Verona collection. A phylogenetic analysis and a comparison of virulence and resistance genes, genetic variants and mutations, and hypermutability mechanisms between chronic and occasional isolates was also performed. The de novo assembled contigs of these genomes were used as input for the association analysis. The following phenotypic cut-offs were used as parameters when running the analysis: biofilm production of 0.115 (absorbance at 550 nm), cytotoxicity of 0.36 (% cytotoxicity vs. positive control), virulence causing 5 dead larvae in 48 h; while the cut-offs for antimicrobial resistance were defined as described in Section 4.5. In particular, DBGWAS accepts continuous phenotypes, so we translated the categorical variables “resistant (R)”, “intermediately resistant (I)” and “sensible (S)” to a dummy variable such that S = 0, I = 1 and R = 2. All available annotations of Achromobacter genes from the UniProt database (www.uniprot.org, ref. 320,589 genes; accessed on 1 March 2022) were used in the annotation step of the virulence, biofilm and cytotoxicity association analysis, whereas all known bacterial resistance genes from the UniProt database (www.uniprot.org, ref. 36,658 genes; accessed on 1 March 2022) were used for annotation in the antimicrobial resistance association analysis.
Sensitivity and specificity of candidate components having the same order of magnitude of the lowest q-value—i.e., p-value adjusted for the False Discovery Rate—of each analysis were calculated using the following formulas: for nodes positively associated to the phenotype sensitivity = Pheno1Count/Pheno1TotalCount and specificity = (Pheno0TotCount-Pheno0Count)/Pheno0TotCount; for nodes negatively associated to the phenotype sensitivity = Pheno0Count/Pheno0TotCount and specificity = (Pheno1TotCount-Pheno1Count)/Pheno1TotCount. In particular, Pheno1Count = isolates displaying the phenotype and carrying the allele, Pheno1TotalCount = isolates displaying the phenotype, Pheno0Count = isolates not displaying the phenotype and carrying the allele, Pheno0TotCount = isolates not displaying the phenotype. All candidate components having the highest sensitivity and specificity, down to a threshold of 80%, and lowest q-value were further analyzed (tests were considered to be statistically significant if q-value < 0.05). Every k-mer present in only one isolate was discarded.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169265/s1.
Author Contributions
Conceptualization: A.S.; Methodology: A.S., L.V. and R.P.M.; Validation: A.S., L.V., R.P.M. and M.C.; Formal analysis: A.S. and L.V.; Investigation: A.S., L.V., R.P.M., G.M.S., G.B., S.P. and C.P.; Resources: C.S. (Claudio Sorio), P.M., E.V.F., G.M., C.S. (Caterina Signoretto), M.B. and M.M.L.; Data curation: A.S. and L.V.; Writing—Original Draft Preparation: A.S., L.V. and G.M.S.; Writing—Review & Editing: R.P.M., M.C., C.S. (Claudio Sorio), P.M., E.V.F., A.L.M., C.P., C.S. (Caterina Signoretto), M.B., M.M.L. and G.M.; Visualization: A.S. and L.V.; Supervision: M.B., M.M.L. and G.M.; Project Administration: A.S., M.B. and M.M.L.; Funding Acquisition: M.M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fondazione per la Ricerca sulla Fibrosi Cistica (grant number FFC#18/2019 adopted by Delegazione FFC di Novara, Messina, Lodi, Foggia, Olbia and by Gruppo di sostegno FFC di Seregno and San Giovanni Rotondo).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Azienda Ospedaliera Universitaria di Verona (protocol code CRCFC-CEPPO026 and CRCFC-CEPPO031).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Sequences of the 54 Achromobacter spp. isolates from the Verona collection have been deposited at the NCBI SRA database under project n. PRJEB40979.
Acknowledgments
We thank Marta Donini (Department of Medicine, University of Verona) for technical support and the Technological Platform Center of the University of Verona for providing the Genomic Platform used for genomic sequences generation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Firmida, M.C.; Pereira, R.H.V.; Silva, E.A.S.R.; Marques, E.A.; Lopes, A.J. Clinical impact of Achromobacter xylosoxidans colonization/infection in patients with cystic fibrosis. Braz. J. Med. Biol. Res. 2016, 49, e5097. [Google Scholar] [CrossRef] [PubMed]
- Swenson, C.E.; Sadikot, R.T. Achromobacter Respiratory Infections. Annals ATS 2015, 12, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Veschetti, L.; Sandri, A.; Patuzzo, C.; Melotti, P.; Malerba, G.; Lleo, M.M. Genomic characterization of Achromobacter species isolates from chronic and occasional lung infection in cystic fibrosis patients. Microbial. Genomics. 2021, 7, 000606. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; Gong, J.; Zhang, L.; Wang, G. Comparative genome characterization of Achromobacter members reveals potential genetic determinants facilitating the adaptation to a pathogenic lifestyle. Appl. Microbiol. Biotechnol. 2013, 97, 6413–6425. [Google Scholar] [CrossRef]
- Mantovani, R.P.; Levy, C.E.; Yano, T. A heat-stable cytotoxic factor produced by Achromobacter xylosoxidans isolated from Brazilian patients with CF is associated with in vitro increased proinflammatory cytokines. J. Cyst. Fibros. 2012, 11, 305–311. [Google Scholar] [CrossRef][Green Version]
- Jeukens, J.; Freschi, L.; Vincent, A.T.; Emond-Rheault, J.-G.; Kukavica-Ibrulj, I.; Charette, S.J.; Levesque, R.C. A Pan-Genomic Approach to Understand the Basis of Host Adaptation in Achromobacter. Genome Biol. Evol. 2017, 9, 1030–1046. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Hansen, M.A.; Jensen, P.Ø.; Hansen, L.; Riber, L.; Cockburn, A.; Kolpen, M.; Rønne Hansen, C.; Ridderberg, W.; Eickhardt, S.; et al. Complete Genome Sequence of the Cystic Fibrosis Pathogen Achromobacter xylosoxidans NH44784-1996 Complies with Important Pathogenic Phenotypes. PLoS ONE 2013, 8, e68484. [Google Scholar] [CrossRef]
- Fiester, S.E.; Arivett, B.A.; Schmidt, R.E.; Beckett, A.C.; Ticak, T.; Carrier, M.V.; Ghosh, R.; Ohneck, E.J.; Metz, M.L.; Sellin Jeffries, M.K.; et al. Iron-Regulated Phospholipase C Activity Contributes to the Cytolytic Activity and Virulence of Acinetobacter baumannii. PLoS ONE 2016, 11, e0167068. [Google Scholar] [CrossRef]
- Veschetti, L.; Sandri, A.; Krogh Johansen, H.; Lleò, M.M.; Malerba, G. Hypermutation as an Evolutionary Mechanism for Achromobacter xylosoxidans in Cystic Fibrosis Lung Infection. Pathogens 2020, 9, 72. [Google Scholar] [CrossRef]
- Nielsen, S.M.; Nørskov-Lauritsen, N.; Bjarnsholt, T.; Meyer, R.L. Achromobacter Species Isolated from Cystic Fibrosis Patients Reveal Distinctly Different Biofilm Morphotypes. Microorganisms 2016, 4, 33. [Google Scholar] [CrossRef]
- Dantam, J.; Subbaraman, L.N.; Jones, L. Adhesion of Pseudomonas aeruginosa, Achromobacter xylosoxidans, Delftia acidovorans, Stenotrophomonas maltophilia to contact lenses under the influence of an artificial tear solution. Biofouling 2020, 36, 32–43. [Google Scholar] [CrossRef]
- Vijay, A.K.; Willcox, M.D.P. Adhesion of Stenotrophomonas maltophilia, Delftia acidovorans, and Achromobacter xylosoxidans to Contact Lenses. Eye Contact Lens 2018, 44 (Suppl. S2), S120–S126. [Google Scholar] [CrossRef]
- Ridderberg, W.; Nielsen, S.M.; Nørskov-Lauritsen, N. Genetic Adaptation of Achromobacter sp. during Persistence in the Lungs of Cystic Fibrosis Patients. PLoS ONE 2015, 10, e0136790. [Google Scholar] [CrossRef]
- Trancassini, M.; Iebba, V.; Citerà, N.; Tuccio, V.; Magni, A.; Varesi, P.; De Biase, R.V.; Totino, V.; Santangelo, F.; Gagliardi, A.; et al. Outbreak of Achromobacter xylosoxidans in an Italian Cystic fibrosis center: Genome variability, biofilm production, antibiotic resistance, and motility in isolated strains. Front. Microbiol. 2014, 5, 138. [Google Scholar] [CrossRef]
- Almuzara, M.; Limansky, A.; Ballerini, V.; Galanternik, L.; Famiglietti, A.; Vay, C. In vitro susceptibility of Achromobacter spp. isolates: Comparison of disk diffusion, Etest and agar dilution methods. Int. J. Antimicrob. Agents 2010, 35, 68–71. [Google Scholar] [CrossRef]
- Gabrielaite, M.; Bartell, J.A.; Nørskov-Lauritsen, N.; Pressler, T.; Nielsen, F.C.; Johansen, H.K.; Marvig, R.L. Transmission and antibiotic resistance of Achromobacter in cystic fibrosis. J. Clin. Microbiol. 2021, 59, e02911-20. [Google Scholar] [CrossRef]
- Whiley, R.A.; Sheikh, N.P.; Mushtaq, N.; Hagi-Pavli, E.; Personne, Y.; Javaid, D.; Waite, R.D. Differential Potentiation of the Virulence of the Pseudomonas aeruginosa Cystic Fibrosis Liverpool Epidemic Strain by Oral Commensal Streptococci. J. Infect. Dis. 2014, 209, 769–780. [Google Scholar] [CrossRef]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef]
- Sandri, A.; Haagensen, J.A.J.; Veschetti, L.; Johansen, H.K.; Molin, S.; Malerba, G.; Signoretto, C.; Boaretti, M.; Lleo, M.M. Adaptive Interactions of Achromobacter spp. with Pseudomonas aeruginosa in Cystic Fibrosis Chronic Lung Co-Infection. Pathogens 2021, 10, 978. [Google Scholar] [CrossRef]
- Quereda, J.J.; Cossart, P. Regulating Bacterial Virulence with RNA. Annu. Rev. Microbiol. 2017, 71, 263–280. [Google Scholar] [CrossRef]
- Millar, J.A.; Raghavan, R. Modulation of bacterial fitness and virulence through antisense RNAs. Front. Cell. Infect. Microbiol. 2021, 10, 596277. [Google Scholar] [CrossRef] [PubMed]
- Kayama, S.; Koba, Y.; Shigemoto, N.; Kuwahara, R.; Kakuhama, T.; Kimura, K.; Hisatsune, J.; Onodera, M.; Yokozaki, M.; Ohge, H.; et al. Imipenem-Susceptible, Meropenem-Resistant Klebsiella pneumoniae Producing OXA-181 in Japan. Antimicrob. Agents Chemother. 2015, 59, 1379–1380. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isler, B.; Kidd, T.J.; Stewart, A.G.; Harris, P.; Paterson, D.L. Achromobacter Infections and Treatment Options. Antimicrob. Agents Chemother. 2020, 64, e01025-20. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Mushtaq, S.; Warner, M.; Woodford, N. Comparative in vitro activity of sulfametrole/trimethoprim and sulfamethoxazole/trimethoprim and other agents against multiresistant Gram-negative bacteria. J. Antimicrob. Chemother. 2014, 69, 1050–1056. [Google Scholar] [CrossRef][Green Version]
- Abbott, I.; Peleg, A. Stenotrophomonas, Achromobacter, and Nonmelioid Burkholderia Species: Antimicrobial Resistance and Therapeutic Strategies. Semin. Respir. Crit. Care Med. 2015, 36, 099–110. [Google Scholar] [CrossRef]
- Veschetti, L.; Sandri, A.; Patuzzo, C.; Melotti, P.; Malerba, G.; Lleò, M.M. Mobilome Analysis of Achromobacter spp. Isolates from Chronic and Occasional Lung Infection in Cystic Fibrosis Patients. Microorganisms 2021, 9, 130. [Google Scholar] [CrossRef]
- Choi, C.H. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int. 2005, 5, 1–13. [Google Scholar] [CrossRef][Green Version]
- Dupont, C.; Jumas-Bilak, E.; Michon, A.-L.; Chiron, R.; Marchandin, H. Impact of High Diversity of Achromobacter Populations within Cystic Fibrosis Sputum Samples on Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 2017, 55, 206–215. [Google Scholar] [CrossRef]
- R Core Team. R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Jaillard, M.; Lima, L.; Tournoud, M.; Mahé, P.; van Belkum, A.; Lacroix, V.; Jacob, L. A fast and agnostic method for bacterial genome-wide association studies: Bridging the gap between k-mers and genetic events. PLoS Genet. 2018, 14, e1007758. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).