Recombinant Adeno-Associated Viral Vector-Mediated Gene Transfer of hTBX18 Generates Pacemaker Cells from Ventricular Cardiomyocytes
Abstract
:1. Introduction
2. Results
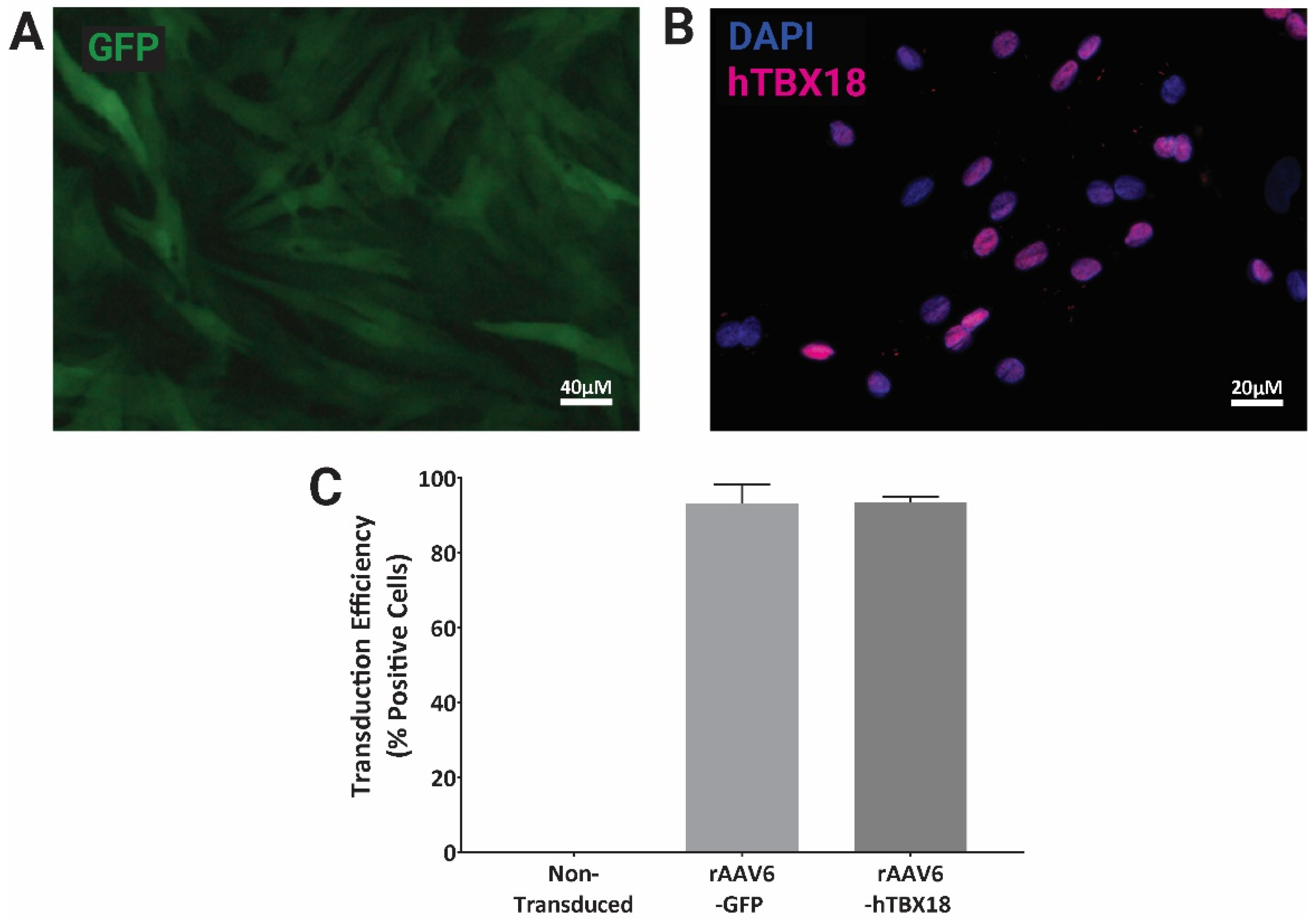
2.1. rAAV Vectors Expressing GFP or hTBX18 Showed Highly Efficient Transduction in NRVMs
2.2. NRVMs Transduced with rAAV6-hTBX18 Faithfully Reproduced the Distinctive Molecular and Physiological Characteristics of Pacemaker Cells
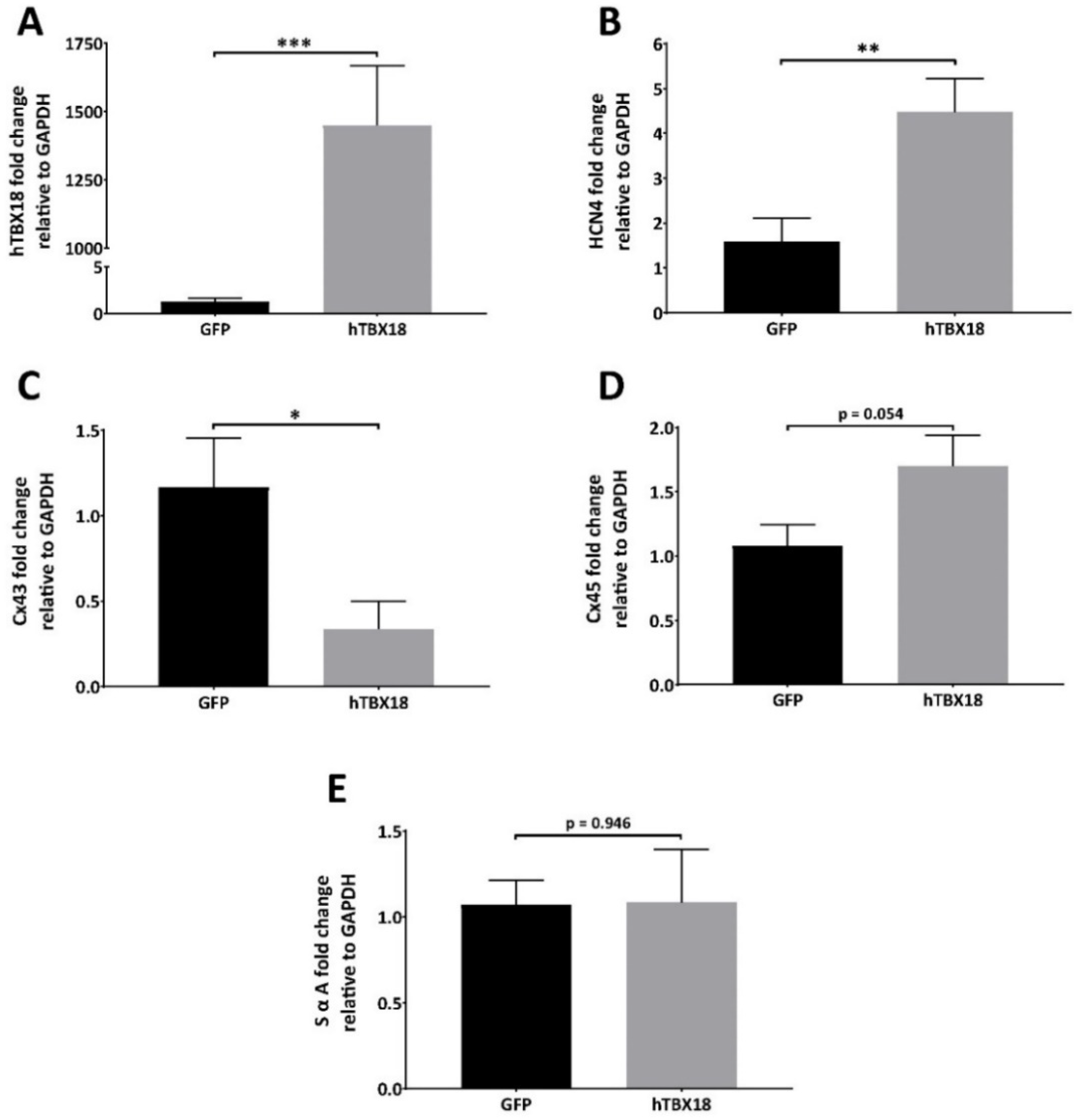
2.2.1. Analysis of mRNA Expression Showed Changes Characteristic of Pacemaker Cells
2.2.2. Analysis of Protein Expression Showed Changes Characteristic of Pacemaker Cells
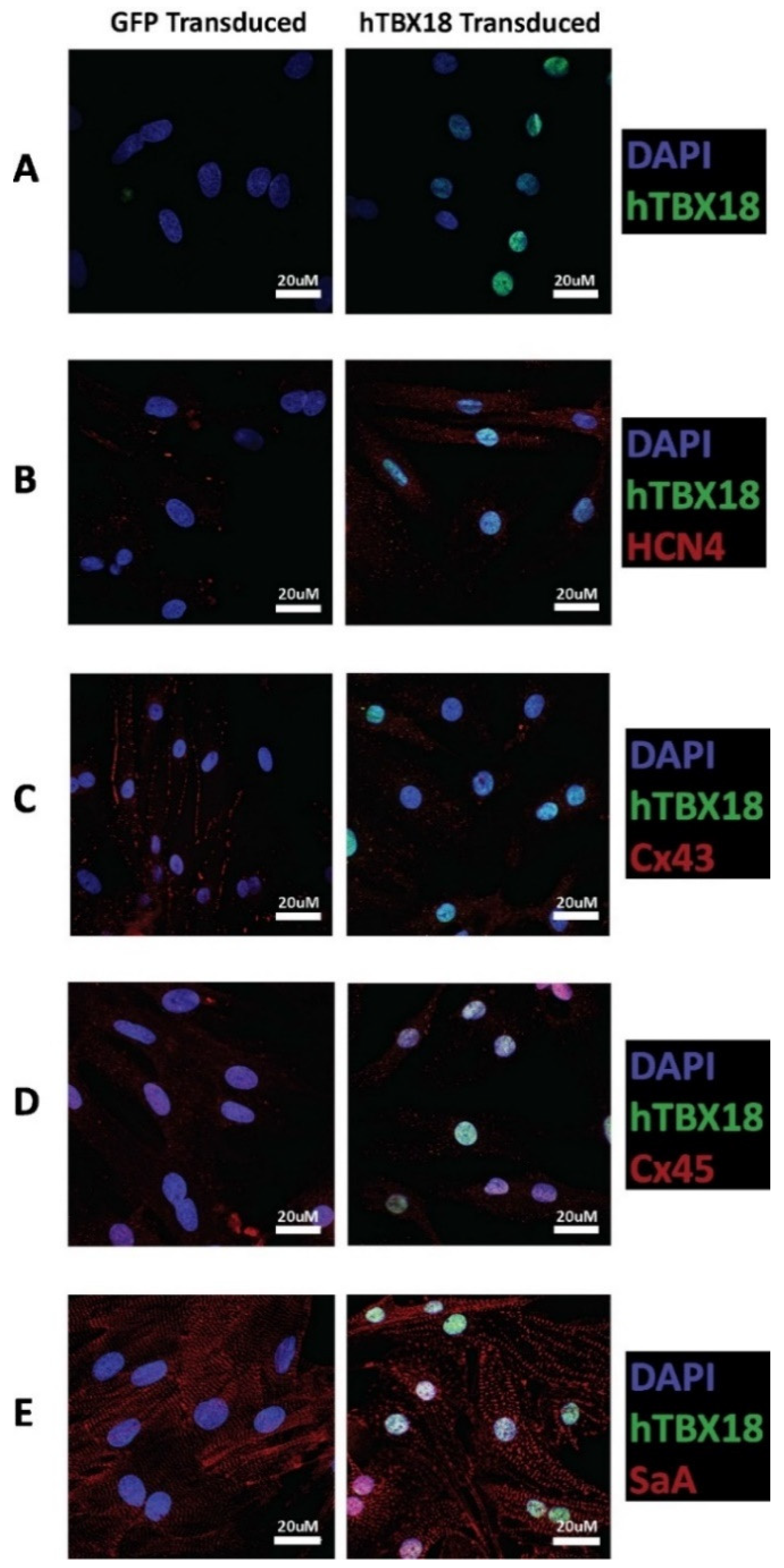
2.2.3. Immunocytochemical Analysis of Cells Transduced with rAAV6-hTBX18 Displayed Distinctive Characteristics of Pacemaker Cells
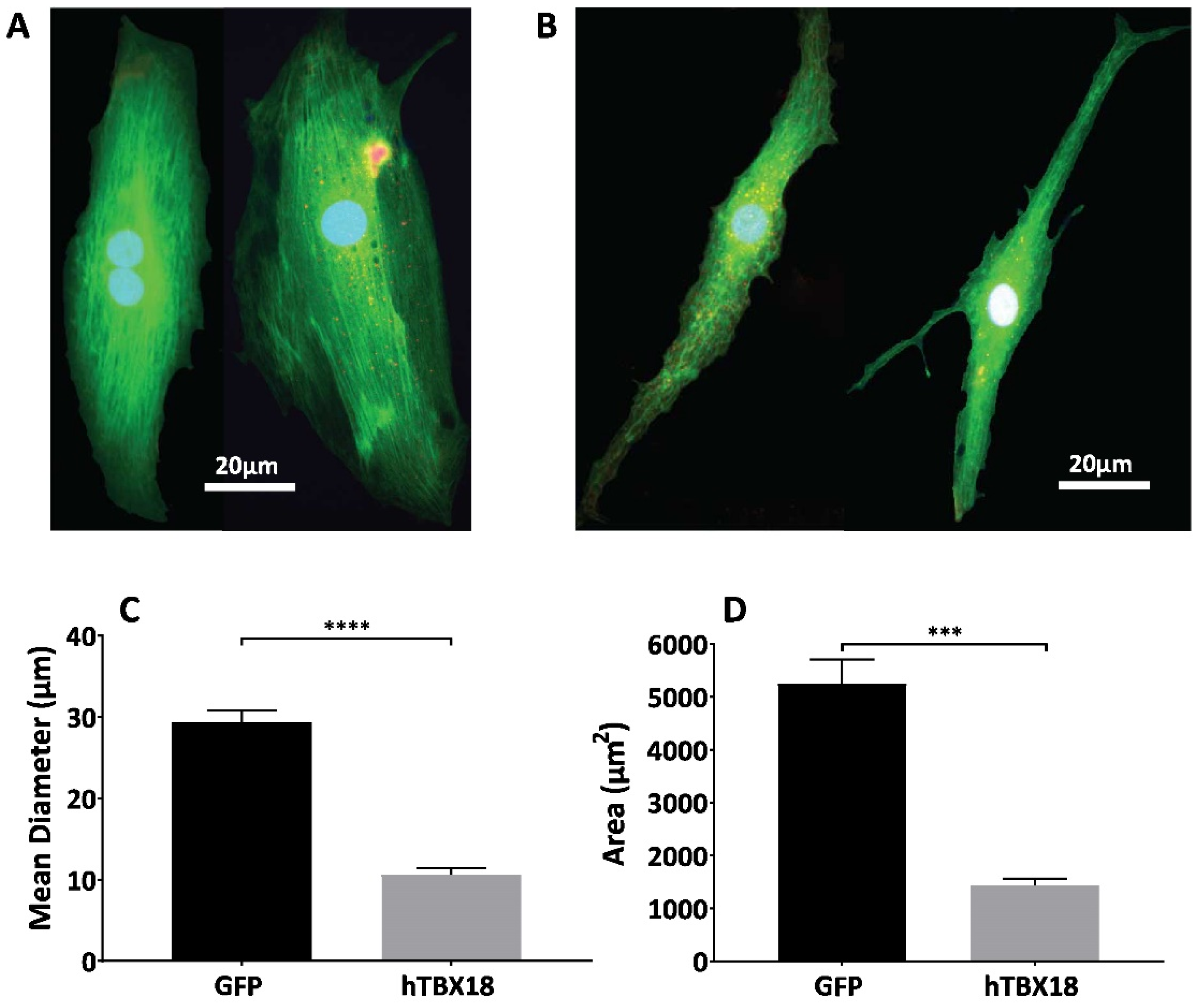
2.3. NRVMs Transduced with rAAV6-hTBX18 Faithfully Recapitulate the Morphological Features of Pacemaker Cells
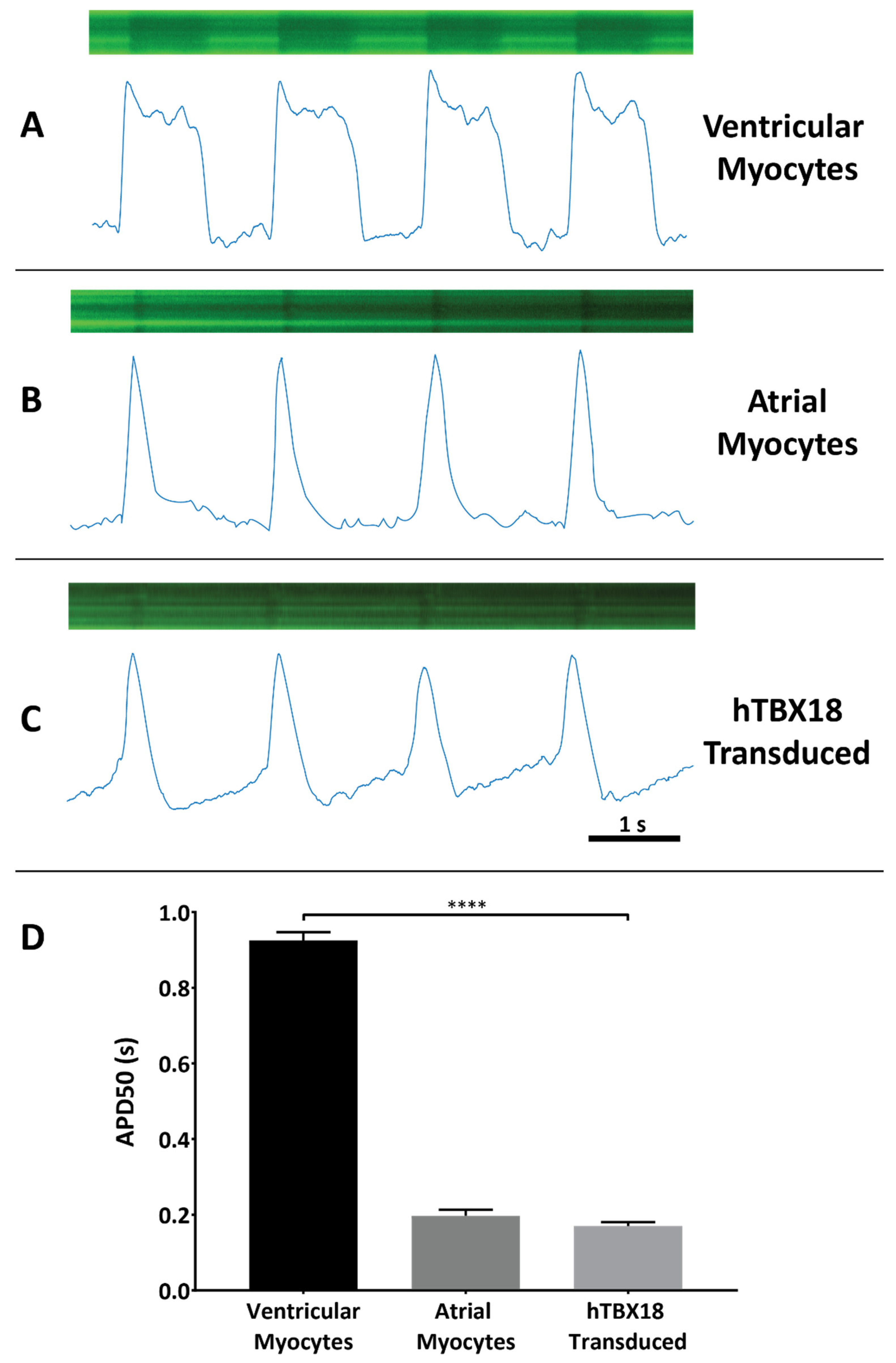
2.4. NRVMs Co-Transduced with rAAV6-hTBX18 and LV.Arclight Generated Pacemaker-like Action Potentials
3. Discussion
4. Materials and Methods
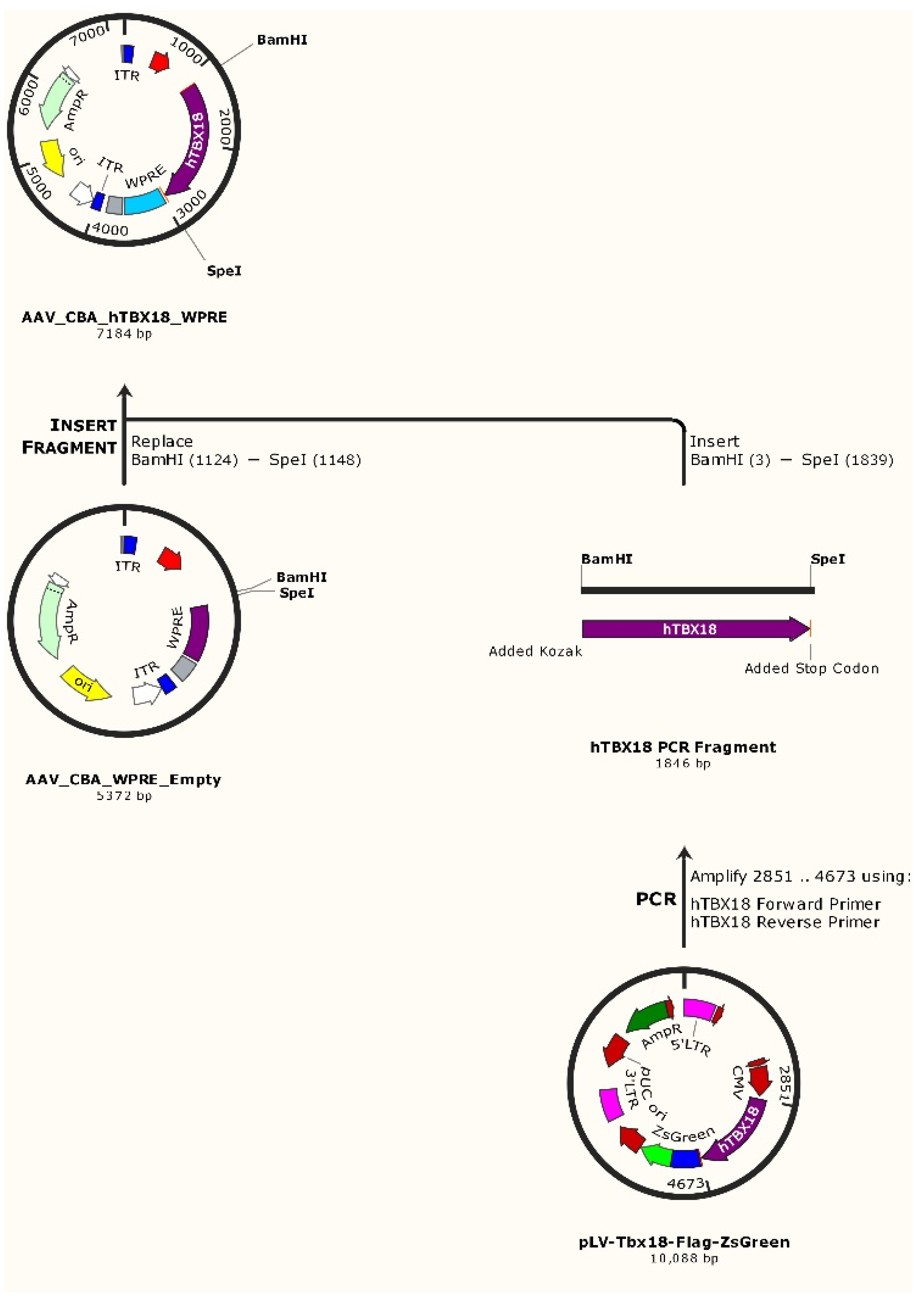
4.1. Molecular Cloning of hTBX18 Gene into a rAAV Expression Vector
4.2. rAAV6 Production and Purification
4.3. Neonatal Rat Ventricular Myocyte Isolation, Culture and rAAV Transduction
4.4. Messenger RNA Extraction and Complementary DNA Synthesis
4.5. Real-Time PCR Analysis
4.6. Protein Extraction and Immunoblotting
4.7. Immunocytochemistry and Microscopy
4.8. Flow Cytometry Analysis
4.9. Lentiviral Arclight Production, Titration and Transduction
4.10. Arclight Confocal Line Scanning
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapoor, N.; Liang, W.; Marbán, E.; Cho, H.C. Transcription factor-driven conversion of quiescent cardiomyocytes to pacemaker cells. Nat. Biotechnol. 2013, 31, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, W.K.; Mackaay, A.J.; Masson-Pévet, M.; Bouman, L.N.; Becker, A.E. Functional and morphological organization of the rabbit sinus node. Circ. Res. 1980, 46, 11–22. [Google Scholar] [CrossRef]
- Jensen, P.N.; Gronroos, N.N.; Chen, L.Y.; Folsom, A.R.; de Filippi, C.; Heckbert, S.R.; Alonso, A. Incidence of and risk factors for sick sinus syndrome in the general population. J. Am. Coll. Cardiol. 2014, 64, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Dakkak, W.; Doukky, R. Sick Sinus Syndrome. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Adan, V.; Crown, L.A. Diagnosis and treatment of sick sinus syndrome. Am. Fam. Physician 2003, 67, 1725–1732. [Google Scholar] [PubMed]
- Semelka, M.; Gera, J.; Usman, S. Sick sinus syndrome: A review. Am. Fam. Physician 2013, 87, 691–696. [Google Scholar] [PubMed]
- Bradshaw, P.J.; Stobie, P.; Knuiman, M.W.; Briffa, T.G.; Hobbs, M.S. Trends in the incidence and prevalence of cardiac pacemaker insertions in an ageing population. Open Heart 2014, 1, e000177. [Google Scholar] [CrossRef]
- John, R.M.; Kumar, S. Sinus Node and Atrial Arrhythmias. Circulation 2016, 133, 1892–1900. [Google Scholar] [CrossRef]
- Keller, K.B.; Lemberg, L. The Sick Sinus Syndrome. Am. J. Crit. Care 2006, 15, 226–229. [Google Scholar] [CrossRef]
- Cronin, B.; Essandoh, M.K. Update on Cardiovascular Implantable Electronic Devices for Anesthesiologists. J. Cardiothorac. Vasc. Anesth. 2017, 4, 1871–1884. [Google Scholar] [CrossRef]
- Madhavan, M.; Mulpuru, S.K.; McLeod, C.J.; Cha, Y.-M.; Friedman, P.A. Advances and Future Directions in Cardiac Pacemakers: Part 2 of a 2-Part Series. J. Am. Coll. Cardiol. 2017, 69, 211–235. [Google Scholar] [CrossRef]
- Banaszewski, M.; Stępińska, J. Right heart perforation by pacemaker leads. Arch. Med. Sci. 2012, 8, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.N.; Joseph, G.; Khaykin, Y.; Ziada, K.M.; Wilkoff, B.L. Delayed Lead Perforation: A Disturbing Trend. Pacing Clin. Electrophysiol. 2005, 28, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, E.; Goldhaber, J.I.; Marban, E. Next-generation pacemakers: From small devices to biological pacemakers. Nat. Rev. Cardiol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Chien, K.R.; Domian, I.J.; Parker, K.K. Cardiogenesis and the Complex Biology of Regenerative Cardiovascular Medicine. Science 2008, 322, 1494–1497. [Google Scholar] [CrossRef]
- Irisawa, H. Comparative physiology of the cardiac pacemaker mechanism. Physiol. Rev. 1978, 58, 461–498. [Google Scholar] [CrossRef]
- Christoffels, V.M.; Smits, G.J.; Kispert, A.; Moorman, A.F.M. Development of the Pacemaker Tissues of the Heart. Circ. Res. 2010, 106, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, R.J.; Hahurij, N.D.; Kuijper, S.; Just, S.; Wisse, L.J.; Deissler, K.; Maxelon, T.; Anastassiadis, K.; Spitzer, J.; Hardt, S.E.; et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 2007, 115, 1830–1838. [Google Scholar] [CrossRef]
- Jay, P.Y.; Berul, C.I.; Tanaka, M.; Ishii, M.; Kurachi, Y.; Izumo, S. Cardiac conduction and arrhythmia: Insights from Nkx2.5 mutations in mouse and humans. Novartis Found. Symp. 2003, 250, 227–238. [Google Scholar]
- Jay, P.Y.; Harris, B.S.; Maguire, C.T.; Buerger, A.; Wakimoto, H.; Tanaka, M.; Kupershmidt, S.; Roden, D.M.; Schultheiss, T.M.; O’Brien, T.X.; et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Investig. 2004, 113, 1130–1137. [Google Scholar] [CrossRef]
- Liu, H.; Espinoza-Lewis, R.A.; Chen, C.; Hu, X.; Zhang, Y.; Chen, Y. The role of Shox2 in SAN development and function. Pediatric Cardiol. 2012, 33, 882–889. [Google Scholar] [CrossRef]
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Hoogaars, W.M.; Engel, A.; Brons, J.F.; Verkerk, A.O.; de Lange, F.J.; Wong, L.Y.; Bakker, M.L.; Clout, D.E.; Wakker, V.; Barnett, P.; et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007, 21, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Mori, A.D.; Zhu, Y.; Vahora, I.; Nieman, B.; Koshiba-Takeuchi, K.; Davidson, L.; Pizard, A.; Seidman, J.G.; Seidman, C.E.; Chen, X.J.; et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006, 297, 566–586. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.M.; Svensson, E.C. Setting the pace: Tbx3 and Tbx18 in cardiac conduction system development. Circ. Res. 2009, 104, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Wiese, C.; Grieskamp, T.; Airik, R.; Mommersteeg, M.T.; Gardiwal, A.; de Gier-de Vries, C.; Schuster-Gossler, K.; Moorman, A.F.; Kispert, A.; Christoffels, V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009, 104, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.F.; Dawkins, J.F.; Cho, H.C.; Marban, E.; Cingolani, E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 2014, 6, 245–294. [Google Scholar] [CrossRef]
- Liu, Q.; Zaiss, A.K.; Colarusso, P.; Patel, K.; Haljan, G.; Wickham, T.J.; Muruve, D.A. The role of capsid-endothelial interactions in the innate immune response to adenovirus vectors. Hum. Gene Ther. 2003, 14, 627–643. [Google Scholar] [CrossRef]
- Muruve, D.A. The innate immune response to adenovirus vectors. Hum. Gene Ther. 2004, 15, 1157–1166. [Google Scholar] [CrossRef]
- Lusky, M.; Christ, M.; Rittner, K.; Dieterle, A.; Dreyer, D.; Mourot, B.; Schultz, H.; Stoeckel, F.; Pavirani, A.; Mehtali, M. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 1998, 72, 2022–2032. [Google Scholar] [CrossRef]
- Gorziglia, M.I.; Kadan, M.J.; Yei, S.; Lim, J.; Lee, G.M.; Luthra, R.; Trapnell, B.C. Elimination of both E1 and E2 from adenovirus vectors further improves prospects for in vivo human gene therapy. J. Virol. 1996, 70, 4173–4178. [Google Scholar] [CrossRef]
- Gorabi, A.M.; Hajighasemi, S.; Khori, V.; Soleimani, M.; Rajaei, M.; Rabbani, S.; Atashi, A.; Ghiaseddin, A.; Saeid, A.K.; Ahmadi Tafti, H.; et al. Functional biological pacemaker generation by T-Box18 protein expression via stem cell and viral delivery approaches in a murine model of complete heart block. Pharm. Res. 2019, 141, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Gorabi, A.M.; Hajighasemi, S.; Tafti, H.A.; Atashi, A.; Soleimani, M.; Aghdami, N.; Saeid, A.K.; Khori, V.; Panahi, Y.; Sahebkar, A. TBX18 transcription factor overexpression in human-induced pluripotent stem cells increases their differentiation into pacemaker-like cells. J. Cell. Physiol. 2019, 234, 1534–1546. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, N.; Liu, L.; Zhang, H.; Xue, X.; Shao, X.; Zhang, Y.; Lang, X. Genetically Modified Porcine Mesenchymal Stem Cells by Lentiviral Tbx18 Create a Biological Pacemaker. Stem Cells Int. 2019, 2019, 3621314. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.A.; Darche, F.F.; Ullrich, N.D.; Geschwill, P.; Greber, B.; Rivinius, R.; Seyler, C.; Müller-Decker, K.; Draguhn, A.; Utikal, J.; et al. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res. 2017, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, G.G.; Wang, T.; Wang, X.; Tang, Y.H.; Huang, H.; Barajas-Martinez, H.; Hu, D.; Huang, C.X. TBX18 gene induces adipose-derived stem cells to differentiate into pacemaker-like cells in the myocardial microenvironment. Int. J. Mol. Med. 2016, 38, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, H.; Quan, D.; Tang, Y.; Wang, X.; Huang, C. Tbx18 promoted the conversion of human-induced pluripotent stem cell-derived cardiomyocytes into sinoatrial node-like pacemaker cells. Cell Biol. Int. 2022, 46, 403–414. [Google Scholar] [CrossRef]
- Ranzani, M.; Cesana, D.; Bartholomae, C.C.; Sanvito, F.; Pala, M.; Benedicenti, F.; Gallina, P.; Sergi, L.S.; Merella, S.; Bulfone, A.; et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat. Methods 2013, 10, 155–161. [Google Scholar] [CrossRef]
- Bokhoven, M.; Stephen, S.L.; Knight, S.; Gevers, E.F.; Robinson, I.C.; Takeuchi, Y.; Collins, M.K. Insertional Gene Activation by Lentiviral and Gammaretroviral Vectors. J. Virol. 2009, 83, 283–294. [Google Scholar] [CrossRef]
- Chamberlain, K.; Riyad, J.M.; Weber, T. Cardiac gene therapy with adeno-associated virus-based vectors. Curr. Opin. Cardiol. 2017, 32, 275–282. [Google Scholar] [CrossRef]
- Zacchigna, S.; Zentilin, L.; Giacca, M. Adeno-associated virus vectors as therapeutic and investigational tools in the cardiovascular system. Circ. Res. 2014, 114, 1827–1846. [Google Scholar] [CrossRef] [PubMed]
- Pacak, C.A.; Byrne, B.J. AAV vectors for cardiac gene transfer: Experimental tools and clinical opportunities. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Asokan, A.; Samulski, R.J. Adeno-associated virus serotypes: Vector toolkit for human gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2006, 14, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Chauveau, S.; Anyukhovsky, E.P.; Ben-Ari, M.; Naor, S.; Jiang, Y.P.; Danilo, P., Jr.; Rahim, T.; Burke, S.; Qiu, X.; Potapova, I.A.; et al. Induced Pluripotent Stem Cell-Derived Cardiomyocytes Provide In Vivo Biological Pacemaker Function. Circ. Arrhythmia Electrophysiol. 2017, 10, e004508. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.J.; Husse, B.; Rimmbach, C.; Krebs, S.; Stieber, J.; Steinhoff, G.; Dendorfer, A.; Franz, W.M.; David, R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Rep. 2014, 2, 592–605. [Google Scholar] [CrossRef]
- Protze, S.I.; Liu, J.; Nussinovitch, U.; Ohana, L.; Backx, P.H.; Gepstein, L.; Keller, G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017, 35, 56–68. [Google Scholar] [CrossRef]
- Saito, Y.; Nakamura, K.; Ito, H. Cell-based Biological Pacemakers: Progress and Problems. Acta Med. Okayama 2018, 72, 1–7. [Google Scholar] [CrossRef]
- Cai, J.; Yi, F.F.; Li, Y.H.; Yang, X.C.; Song, J.; Jiang, X.J.; Jiang, H.; Lin, G.S.; Wang, W. Adenoviral gene transfer of HCN4 creates a genetic pacemaker in pigs with complete atrioventricular block. Life Sci. 2007, 80, 1746–1753. [Google Scholar] [CrossRef]
- Cingolani, E.; Yee, K.; Shehata, M.; Chugh, S.S.; Marban, E.; Cho, H.C. Biological pacemaker created by percutaneous gene delivery via venous catheters in a porcine model of complete heart block. Heart Rhythm 2012, 9, 1310–1318. [Google Scholar] [CrossRef]
- Manno, C.S.; Chew, A.J.; Hutchison, S.; Larson, P.J.; Herzog, R.W.; Arruda, V.R.; Tai, S.J.; Ragni, M.V.; Thompson, A.; Ozelo, M.; et al. AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B. Blood 2003, 101, 2963–2972. [Google Scholar] [CrossRef]
- Ginn, S.L.; Alexander, I.E.; Edelstein, M.L.; Abedi, M.R.; Wixon, J. Gene therapy clinical trials worldwide to 2012—An update. J. Gene Med. 2013, 15, 65–77. [Google Scholar] [CrossRef]
- Aitken, M.L.; Moss, R.B.; Waltz, D.A.; Dovey, M.E.; Tonelli, M.R.; McNamara, S.C.; Gibson, R.L.; Ramsey, B.W.; Carter, B.J.; Reynolds, T.C. A phase I study of aerosolized administration of tgAAVCF to cystic fibrosis subjects with mild lung disease. Hum. Gene Ther. 2001, 12, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, L.; Johnson, J.S.; Zhijian, W.; Grieger, J.C.; Ping-Jie, X.; Drouin, L.M.; Agbandje-McKenna, M.; Pickles, R.J.; Samulski, R.J. Generation of novel AAV variants by directed evolution for improved CFTR delivery to human ciliated airway epithelium. Mol. Ther. J. Am. Soc. Gene Ther. 2009, 17, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, J.; Xiao, X. Directed evolution of adeno-associated virus (AAV) as vector for muscle gene therapy. Methods Mol. Biol. (Clifton N.J.) 2011, 709, 127–139. [Google Scholar] [CrossRef]
- Zincarelli, C.; Soltys, S.; Rengo, G.; Rabinowitz, J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1073–1080. [Google Scholar] [CrossRef]
- Larsson, H.P. How is the heart rate regulated in the sinoatrial node? Another piece to the puzzle. J. Gen. Physiol. 2010, 136, 237–241. [Google Scholar] [CrossRef]
- Stieber, J.; Hofmann, F.; Ludwig, A. Pacemaker channels and sinus node arrhythmia. Trends Cardiovasc. Med. 2004, 14, 23–28. [Google Scholar] [CrossRef]
- DiFrancesco, D. The role of the funny current in pacemaker activity. Circ. Res. 2010, 106, 434–446. [Google Scholar] [CrossRef]
- Ueda, K.; Nakamura, K.; Hayashi, T.; Inagaki, N.; Takahashi, M.; Arimura, T.; Morita, H.; Higashiuesato, Y.; Hirano, Y.; Yasunami, M.; et al. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J. Biol. Chem. 2004, 279, 27194–27198. [Google Scholar] [CrossRef]
- Shi, W.; Wymore, R.; Yu, H.; Wu, J.; Wymore, R.T.; Pan, Z.; Robinson, R.B.; Dixon, J.E.; McKinnon, D.; Cohen, I.S. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ. Res. 1999, 85, e1–e6. [Google Scholar] [CrossRef]
- Chandler, N.J.; Greener, I.D.; Tellez, J.O.; Inada, S.; Musa, H.; Molenaar, P.; Difrancesco, D.; Baruscotti, M.; Longhi, R.; Anderson, R.H.; et al. Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation 2009, 119, 1562–1575. [Google Scholar] [CrossRef]
- Vozzi, C.; Dupont, E.; Coppen, S.R.; Yeh, H.I.; Severs, N.J. Chamber-related differences in connexin expression in the human heart. J. Mol. Cell. Cardiol. 1999, 31, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.M.; Rodefeld, M.E.; Green, K.; Beyer, E.C.; Saffitz, J.E. Gap junction protein phenotypes of the human heart and conduction system. J. Cardiovasc. Electrophysiol. 1995, 6, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Kodama, I.; Takagishi, Y.; Honjo, H.; Boyett, M.R.; Severs, N.J.; Coppen, S.R.; Opthof, T. Heterogeneous expression of connexins in rabbit sinoatrial node cells: Correlation between connexin isotype and cell size. Cardiovasc. Res. 2002, 53, 89–96. [Google Scholar] [CrossRef]
- Desplantez, T. Cardiac Cx43, Cx40 and Cx45 co-assembling: Involvement of connexins epitopes in formation of hemichannels and Gap junction channels. BMC Cell Biol. 2017, 18, 3–17. [Google Scholar] [CrossRef]
- Kapoor, N.; Galang, G.; Marban, E.; Cho, H.C. Transcriptional suppression of connexin43 by TBX18 undermines cell-cell electrical coupling in postnatal cardiomyocytes. J. Biol. Chem. 2011, 286, 14073–14079. [Google Scholar] [CrossRef]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the Muscle Cytoskeleton. Compr. Physiol. 2017, 7, 891–944. [Google Scholar] [CrossRef]
- Honjo, H.; Boyett, M.R.; Kodama, I.; Toyama, J. Correlation between electrical activity and the size of rabbit sino-atrial node cells. J. Physiol. 1996, 496, 795–808. [Google Scholar] [CrossRef]
- Trautwein, W.; Uchizono, K. Electron Microscopic and Electrophysiologic Study of the Pacemaker in the Sino-atrial Node of the Rabbit Heart. Z. Fur Zellforsch. Und Mikrosk. Anatomie (Vienna Austria 1948) 1963, 61, 96–109. [Google Scholar] [CrossRef]
- Barnett, L.; Platisa, J.; Popovic, M.; Pieribone, V.A.; Hughes, T. A Fluorescent, Genetically-Encoded Voltage Probe Capable of Resolving Action Potentials. PLoS ONE 2012, 7, e43454. [Google Scholar] [CrossRef]
- Shinnawi, R.; Huber, I.; Maizels, L.; Shaheen, N.; Gepstein, A.; Arbel, G.; Tijsen, A.J.; Gepstein, L. Monitoring Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes with Genetically Encoded Calcium and Voltage Fluorescent Reporters. Stem Cell Rep. 2015, 5, 582–596. [Google Scholar] [CrossRef]
- Murata, Y.; Iwasaki, H.; Sasaki, M.; Inaba, K.; Okamura, Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 2005, 435, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Leyton-Mange, J.S.; Mills, R.W.; Macri, V.S.; Jang, M.Y.; Butte, F.N.; Ellinor, P.T.; Milan, D.J. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep. 2014, 2, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Guo, L.; Fiene, S.J.; Anson, B.D.; Thomson, J.A.; Kamp, T.J.; Kolaja, K.L.; Swanson, B.J.; January, C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. Am. J. Physiol.-Heart Circ. Physiol. 2011, 301, 2006–2017. [Google Scholar] [CrossRef]
- Snyder, R.O. Adeno-associated virus-mediated gene delivery. J. Gene Med. 1999, 1, 166–175. [Google Scholar] [CrossRef]
- Choi, V.W.; Asokan, A.; Haberman, R.A.; Samulski, R.J. Production of Recombinant Adeno-Associated Viral Vectors. In Current Protocols in Human Genetics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Gregorevic, P.; Blankinship, M.J.; Allen, J.M.; Crawford, R.W.; Meuse, L.; Miller, D.G.; Russell, D.W.; Chamberlain, J.S. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat. Med. 2004, 10, 828–834. [Google Scholar] [CrossRef]
- Strobel, B.; Miller, F.D.; Rist, W.; Lamla, T. Comparative Analysis of Cesium Chloride- and Iodixanol-Based Purification of Recombinant Adeno-Associated Viral Vectors for Preclinical Applications. Hum. Gene Ther. Methods 2015, 26, 147–157. [Google Scholar] [CrossRef]
- Iravanian, S.; Nabutovsky, Y.; Kong, C.R.; Saha, S.; Bursac, N.; Tung, L. Functional reentry in cultured monolayers of neonatal rat cardiac cells. Am. J. Physiol.-Heart Circ. Physiol. 2003, 285, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Bursac, N.; Papadaki, M.; Cohen, R.J.; Schoen, F.J.; Eisenberg, S.R.; Carrier, R.; Vunjak-Novakovic, G.; Freed, L.E. Cardiac muscle tissue engineering: Toward an in vitro model for electrophysiological studies. Am. J. Physiol.-Heart Circ. Physiol. 1999, 277, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Toraason, M.; Luken, M.E.; Breitenstein, M.; Krueger, J.A.; Biagini, R.E. Comparative toxicity of allylamine and acrolein in cultured myocytes and fibroblasts from neonatal rat heart. Toxicology 1989, 56, 107–117. [Google Scholar] [CrossRef]
- Kizana, E.; Ginn, S.L.; Allen, D.G.; Ross, D.L.; Alexander, I.E. Fibroblasts can be genetically modified to produce excitable cells capable of electrical coupling. Circulation 2005, 111, 394–398. [Google Scholar] [CrossRef]

| Primer | Direction | Sequence |
|---|---|---|
| Human TBX18 | Forward (5′-3′) | TTCTGGCGACCATCACTACG |
| Human TBX18 | Reverse (5′-3′) | ACGCCATTCCCAGTACCTTG |
| Rat HCN4 | Forward (5′-3′) | CGCATCCACGACTACTACGAAC |
| Rat HCN4 | Reverse (5′-3′) | GGTCTGCATTGGCGAACAG |
| Rat Cx43 | Forward (5′-3′) | AGCCTGAACTCTCATTTTTCCTT |
| Rat Cx43 | Reverse (5′-3′) | CCATGTCTGGGCACCTCT |
| Rat Cx45 | Forward (5′-3′) | TGCCTACAAGCAGAACAAAGC |
| Rat Cx45 | Reverse (5′-3′) | TCCTCGTGGCTGCCATAC |
| Rat Actn2 | Forward (5′-3′) | CTATTGGGGCTGAAGAAATCGTC |
| Rat Actn2 | Reverse (5′-3′) | CTGAGATGTCCTGAATGGCG |
| Rat GAPDH | Forward (5′-3′) | GCATCACCCCATTTGATGTT |
| Rat GAPDH | Reverse (5′-3′) | TGGGAAGCTGGTCATCAAC |
| Primary Antibody | Source | Catalogue Number | Dilution |
|---|---|---|---|
| Goat anti-TBX18 | Santa Cruz | sc-17869 | 1:200 |
| Mouse anti-HCN4 | Abcam | ab85023 (S114-10) | 1:250 |
| Rabbit anti-Cx43 | Merck Millipore | Ab1727 | 1:500 |
| Rabbit anti-Cx45 | Koval | Donation | 1:500 |
| Mouse anti-SαA | Sigma | A7811 | 1:3000 |
| Rabbit anti-β-Actin | Abcam | Ab8227 | 1:5000 |
| Secondary Antibody | Source | Catalogue Number | Dilution |
| Anti-rabbit HRP | Thermofisher | 31460 | 1:10,000 |
| Anti-mouse HRP | Dako | P0447 | 1:10,000 |
| Anti-goat HRP | Dako | P0449 | 1:10,000 |
| Primary Antibody | Source | Catalogue Number | Dilution |
|---|---|---|---|
| Goat anti-TBX18 | Santa Cruz | sc-17869 | 1:150 |
| Mouse anti-HCN4 | Abcam | ab85023 (S114-10) | 1:150 |
| Rabbit anti-Cx43 | Merck Millipore | MAB3068 | 1:250 |
| Rabbit anti-Cx45 | Merck Millipore | MAB3100 | 1:150 |
| Mouse anti-SαA | Sigma | A7811 | 1:1000 |
| Secondary Antibody | Source | Catalogue Number | Dilution |
| Donkey anti-Goat Alexa fluor 594 | Life Technologies | A-11058 | 1:1000 |
| Donkey anti-Goat Alexa fluor 647 | Invitrogen | A-21447 | 1:1000 |
| Rabbit anti-Mouse Alexa fluor 594 | Molecular Probes | A-11067 | 1:1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farraha, M.; Rao, R.; Igoor, S.; Le, T.Y.L.; Barry, M.A.; Davey, C.; Kok, C.; Chong, J.J.H.; Kizana, E. Recombinant Adeno-Associated Viral Vector-Mediated Gene Transfer of hTBX18 Generates Pacemaker Cells from Ventricular Cardiomyocytes. Int. J. Mol. Sci. 2022, 23, 9230. https://doi.org/10.3390/ijms23169230
Farraha M, Rao R, Igoor S, Le TYL, Barry MA, Davey C, Kok C, Chong JJH, Kizana E. Recombinant Adeno-Associated Viral Vector-Mediated Gene Transfer of hTBX18 Generates Pacemaker Cells from Ventricular Cardiomyocytes. International Journal of Molecular Sciences. 2022; 23(16):9230. https://doi.org/10.3390/ijms23169230
Chicago/Turabian StyleFarraha, Melad, Renuka Rao, Sindhu Igoor, Thi Y. L. Le, Michael A. Barry, Christopher Davey, Cindy Kok, James J.H. Chong, and Eddy Kizana. 2022. "Recombinant Adeno-Associated Viral Vector-Mediated Gene Transfer of hTBX18 Generates Pacemaker Cells from Ventricular Cardiomyocytes" International Journal of Molecular Sciences 23, no. 16: 9230. https://doi.org/10.3390/ijms23169230
APA StyleFarraha, M., Rao, R., Igoor, S., Le, T. Y. L., Barry, M. A., Davey, C., Kok, C., Chong, J. J. H., & Kizana, E. (2022). Recombinant Adeno-Associated Viral Vector-Mediated Gene Transfer of hTBX18 Generates Pacemaker Cells from Ventricular Cardiomyocytes. International Journal of Molecular Sciences, 23(16), 9230. https://doi.org/10.3390/ijms23169230







