Antiproliferative Effects of Cynara Cardunculus in Colorectal Cancer Cells Are Modulated by the Circadian Clock
Abstract
:1. Introduction
2. Results
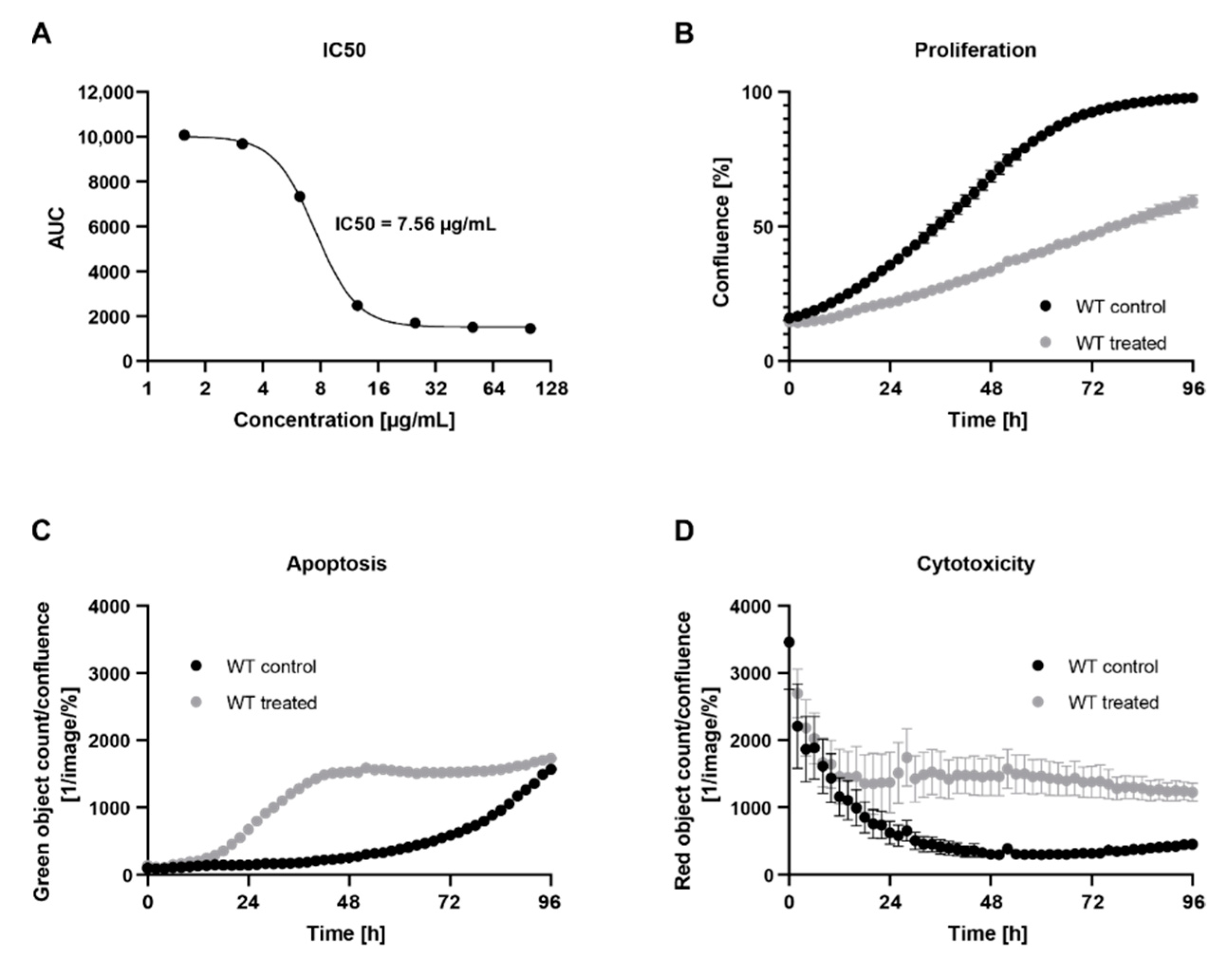
2.1. Treatment with C. cardunculus Leaf Extract Reduces Cell Proliferation and Induces Apoptosis and Cytotoxicity
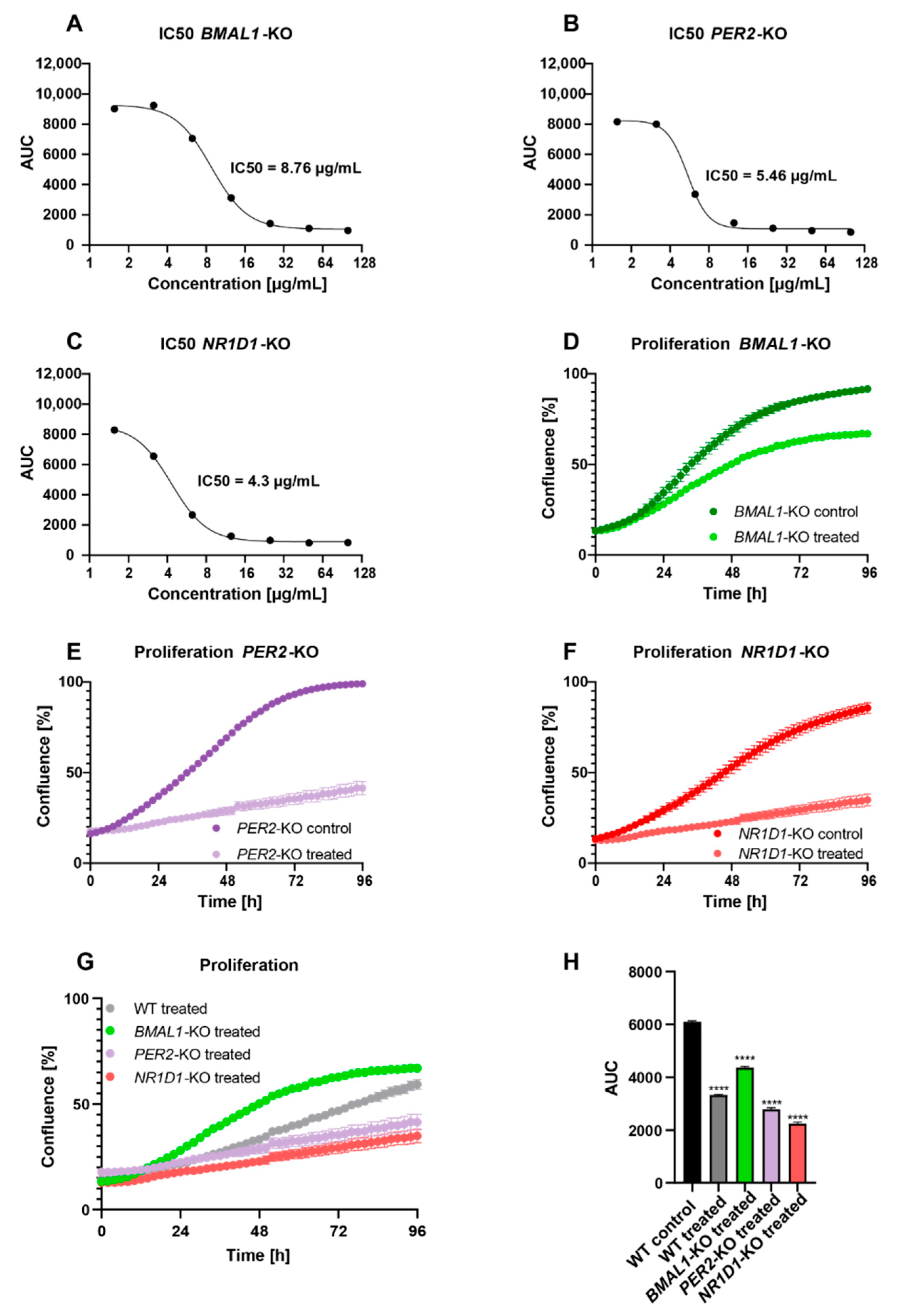
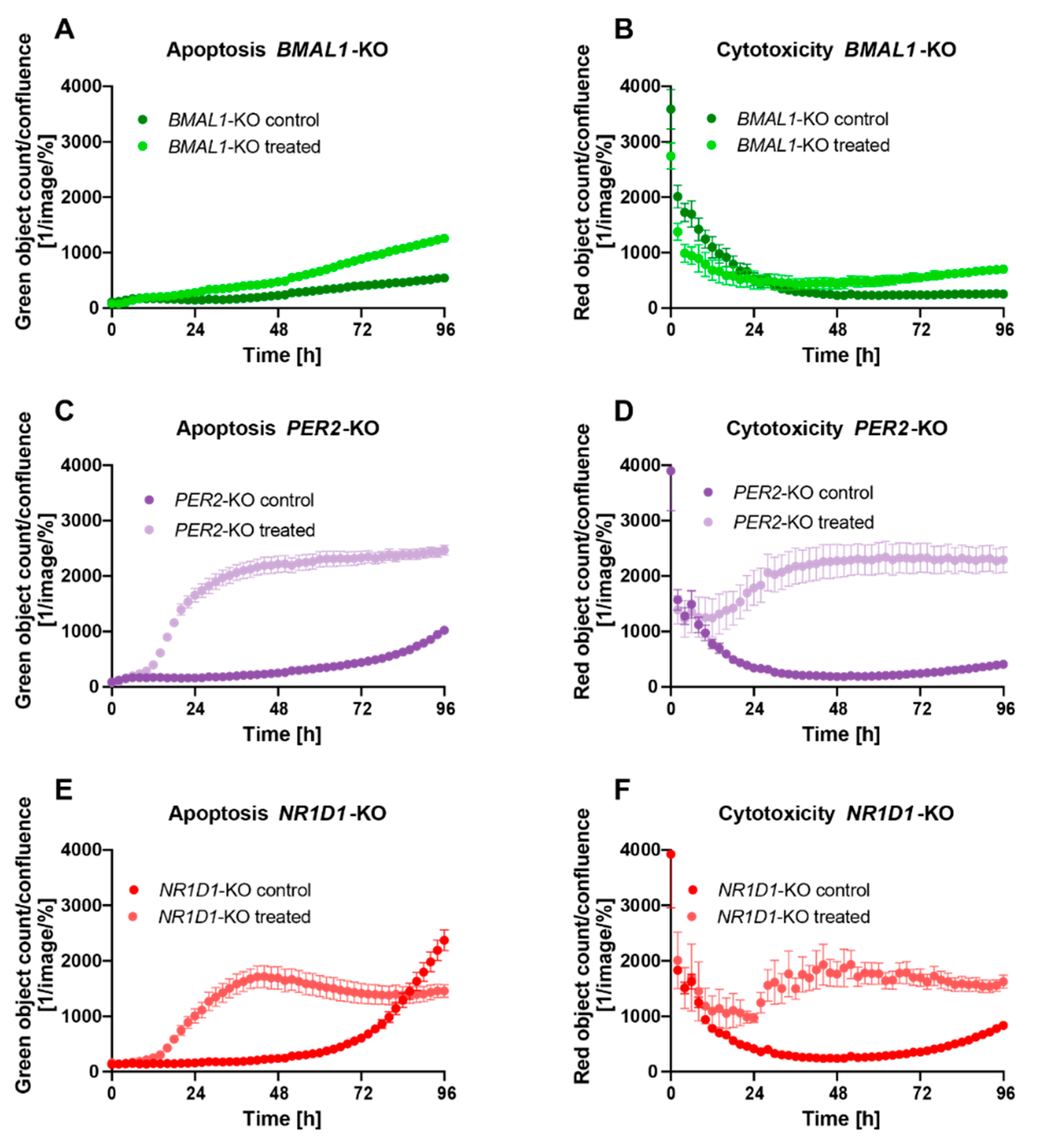
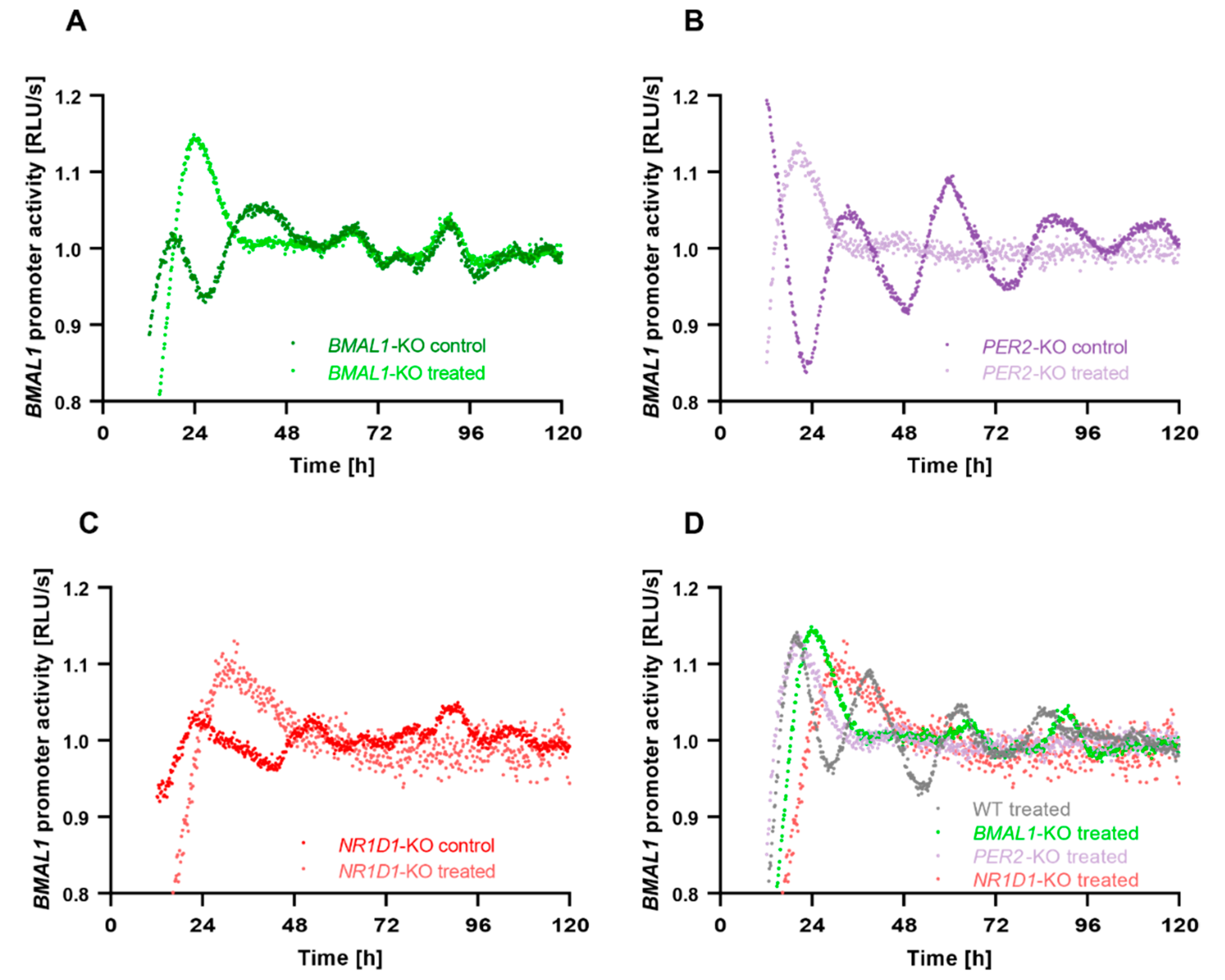
2.2. Core-Clock Disruption Influences Response to C. cardunculus Treatment
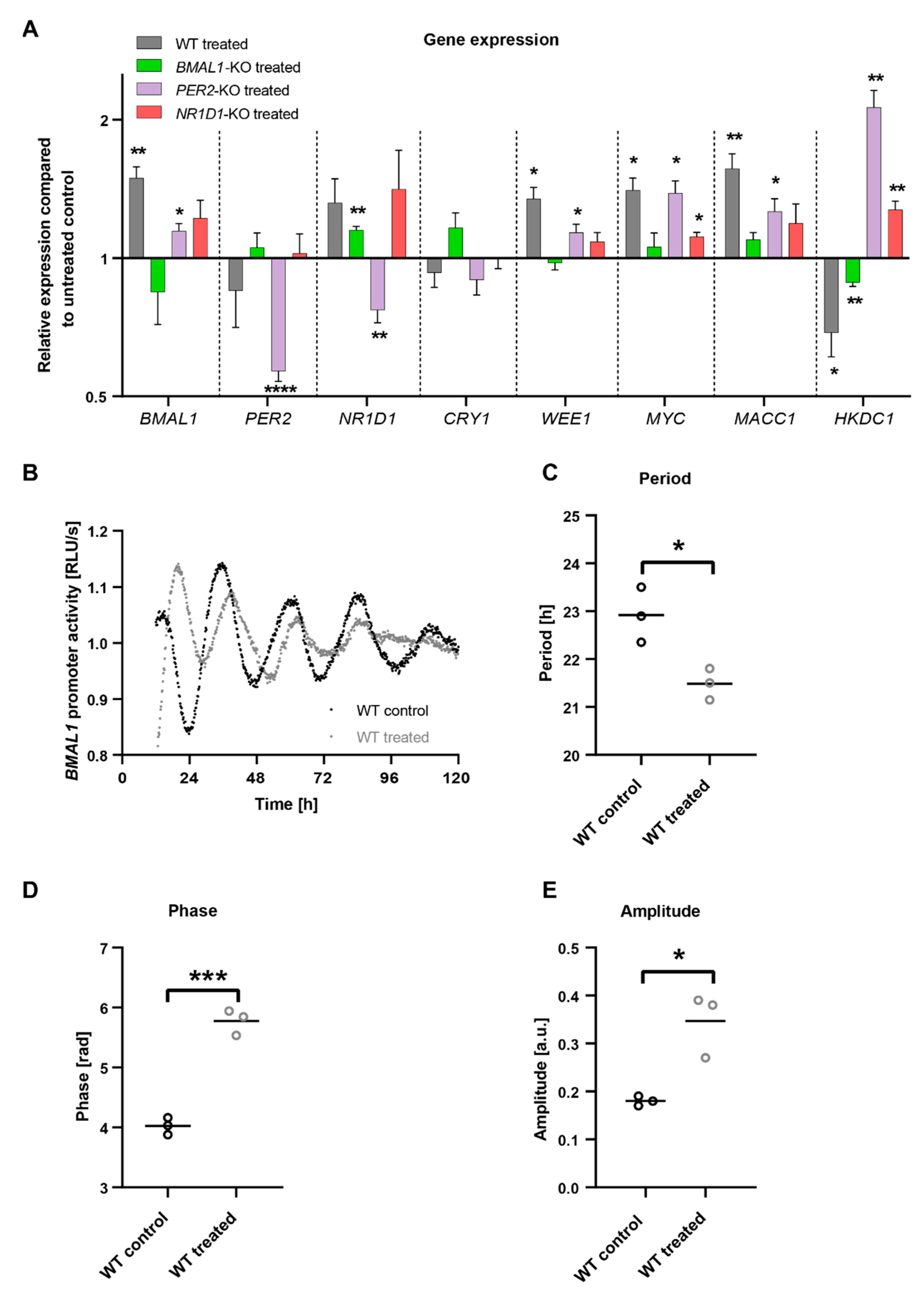
2.3. C. cardunculus Alters Circadian Properties in HCT116 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Determination of Treatment Concentration
4.3. Lentivirus Production
4.4. Transduction with Lentiviral Vectors
4.5. Bioluminescence Measurements
4.6. RNA Extraction, cDNA Synthesis (Reverse Transcription) and Quantitative Real-Time PCR (qPCR)
4.7. Proliferation, Apoptosis, and Cytotoxicity Measurements
4.8. Cynara cardunculus Plant Extracts
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Relógio, A.; Westermark, P.; Wallach, T.; Schellenberg, K.; Kramer, A.; Herzel, H. Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Comput. Biol. 2011, 7, e1002309. [Google Scholar] [CrossRef] [PubMed]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef]
- Sulli, G.; Manoogian, E.N.; Taub, P.R.; Panda, S. Training the Circadian Clock, Clocking the Drugs, and Drugging the Clock to Prevent, Manage, and Treat Chronic Diseases. Trends Pharmacol. Sci. 2018, 39, 812–827. [Google Scholar] [CrossRef]
- Fuhr, L.; Abreu, M.; Pett, P.; Relógio, A. Circadian systems biology: When time matters. Comput. Struct. Biotechnol. J. 2015, 13, 417–426. [Google Scholar] [CrossRef]
- Rijo-Ferreira, F.; Takahashi, J.S. Genomics of circadian rhythms in health and disease. Genome Med. 2019, 11, 82. [Google Scholar] [CrossRef]
- Kinouchi, K.; Sassone-Corsi, P. Metabolic rivalry: Circadian homeostasis and tumorigenesis. Nat. Rev. Cancer 2020, 20, 645–661. [Google Scholar] [CrossRef]
- Shafi, A.A.; Knudsen, K.E. Cancer and the Circadian Clock. Cancer Res. 2019, 79, 3806–3814. [Google Scholar] [CrossRef]
- Fuhr, L.; El-Athman, R.; Scrima, R.; Cela, O.; Carbone, A.; Knoop, H.; Li, Y.; Hoffmann, K.; Laukkanen, M.O.; Corcione, F.; et al. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. eBioMedicine 2018, 33, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Basti, A.; Fior, R.; Yalҫin, M.; Póvoa, V.; Astaburuaga, R.; Li, Y.; Naderi, J.; Ferreira, M.G.; Relógio, A. The Core-Clock Gene NR1D1 Impacts Cell Motility In Vitro and Invasiveness in A Zebrafish Xenograft Colon Cancer Model. Cancers 2020, 12, 853. [Google Scholar] [CrossRef]
- Ballesta, A.; Innominato, P.F.; Dallmann, R.; Rand, D.A.; Lévi, F.A. Systems Chronotherapeutics. Pharmacol. Rev. 2017, 69, 161–199. [Google Scholar] [CrossRef] [PubMed]
- Printezi, M.I.; Kilgallen, A.B.; Bond, M.J.G.; Štibler, U.; Putker, M.; Teske, A.J.; Cramer, M.J.; A Punt, C.J.; Sluijter, J.P.G.; Huitema, A.D.R.; et al. Toxicity and efficacy of chronomodulated chemotherapy: A systematic review. Lancet Oncol. 2022, 23, e129–e143. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. Thompson, The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Huang, M.; Lu, J.-J.; Ding, J. Natural Products in Cancer Therapy: Past, Present and Future. Nat. Prod. Bioprospecting 2021, 11, 5–13. [Google Scholar] [CrossRef]
- Lichota, A.; Gwozdzinski, K. Gwozdzinski, Anticancer Activity of Natural Compounds from Plant and Marine Environment. Int. J. Mol. Sci. 2018, 19, 3533. [Google Scholar] [CrossRef]
- Ullah, M.F.; Bhat, S.H.; Husain, E.; Abu-Duhier, F.; Hadi, S.M.; Sarkar, F.H.; Ahmad, A. Cancer chemopreventive pharmacology of phytochemicals derived from plants of dietary and non-dietary origin: Implication for alternative and complementary approaches. Phytochem. Rev. 2014, 13, 811–833. [Google Scholar] [CrossRef]
- Chadwick, M.; Trewin, H.; Gawthrop, F.; Wagstaff, C. Sesquiterpenoids lactones: Benefits to plants and people. Int. J. Mol. Sci. 2013, 14, 12780–12805. [Google Scholar] [CrossRef]
- Ramos, P.A.B.; Guerra, R.; Guerreiro, O.; Freire, C.S.R.; Silva, A.M.S.; Duarte, M.F.; Silvestre, A.J.D. Lipophilic extracts of Cynara cardunculus L. var. altilis (DC): A source of valuable bioactive terpenic compounds. J. Agric. Food Chem. 2013, 61, 8420–8429. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.A.; Guerra, R.; Guerreiro, O.; Santos, S.A.O.; Oliveira, H.; Freire, C.S.R.; Silvestre, A.J.D.; Duarte, M.F. Antiproliferative Effects of Cynara cardunculus L. var. altilis (DC) Lipophilic Extracts. Int. J. Mol. Sci. 2016, 18, 63. [Google Scholar] [CrossRef]
- Brás, T.; Rosa, D.; Gonçalves, A.C.; Gomes, A.C.; Brazinha, C.; Neves, L.A.; Duarte, M.F.; Crespo, J.G. Fractionation of Cynara cardunculus ethanolic extracts using diananofiltration. Sep. Purif. Technol. 2020, 256, 117856. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kim, A.R.; Jung, J.H.; Chun, T.; Rhee, M.H.; Yoo, E.S. Cytotoxic and pro-apoptotic activities of cynaropicrin, a sesquiterpene lactone, on the viability of leukocyte cancer cell lines. Eur. J. Pharmacol. 2004, 492, 85–94. [Google Scholar] [CrossRef]
- Geetha, B.S.; Nair, M.S.; Latha, P.G.; Remani, P. Sesquiterpene lactones isolated from Elephantopus scaber L. inhibits human lymphocyte proliferation and the growth of tumour cell lines and induces apoptosis in vitro. J. Biomed. Biotechnol. 2012, 2012, 721285. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Relógio, A.; Thomas, P.; Medina-Pérez, P.; Reischl, S.; Bervoets, S.; Gloc, E.; Riemer, P.; Mang-Fatehi, S.; Maier, B.; Schäfer, R.; et al. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014, 10, e1004338. [Google Scholar] [CrossRef]
- Ballesta, A.; Dulong, S.; Abbara, C.; Cohen, B.; Okyar, A.; Clairambault, J.; Levi, F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput. Biol. 2011, 7, e1002143. [Google Scholar] [CrossRef] [PubMed]
- Hesse, J.; Martinelli, J.; Aboumanify, O.; Ballesta, A.; Relógio, A. A mathematical model of the circadian clock and drug pharmacology to optimize irinotecan administration timing in colorectal cancer. Comput. Struct. Biotechnol. J. 2021, 19, 5170–5183. [Google Scholar] [CrossRef]
- El-Athman, R.; Fuhr, L.; Relógio, A. A Systems-Level Analysis Reveals Circadian Regulation of Splicing in Colorectal Cancer. eBioMedicine 2018, 33, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Cheng, B.; Xie, M.; Chen, Y.; Zhao, J.; Zhou, X.; Chen, L. Circadian Clock Gene Bmal1 Inhibits Tumorigenesis and Increases Paclitaxel Sensitivity in Tongue Squamous Cell Carcinoma. Cancer Res. 2017, 77, 532–544. [Google Scholar] [CrossRef]
- Zeng, Z.-L.; Luo, H.-Y.; Yang, J.; Wu, W.-J.; Chen, D.-L.; Huang, P.; Xu, R.-H. Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. 2014, 20, 1042–1052. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science 2021, 371, eabb0738. [Google Scholar] [CrossRef]
- Zhang, M.; Kobayashi, K.; Atsumi, H.; Katada, Y.; Nakane, Y.; Chen, J.; Nagano, R.; Kadofusa, N.; Nishiwaki-Ohkawa, T.; Kon, N.; et al. Modulation of circadian clock by crude drug extracts used in Japanese Kampo medicine. Sci. Rep. 2021, 11, 21038. [Google Scholar] [CrossRef]
- Irwin, D.M.; Tan, H. Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 96–107. [Google Scholar] [CrossRef]
- Li, G.H.; Huang, J.F. Inferring therapeutic targets from heterogeneous data: HKDC1 is a novel potential therapeutic target for cancer. Bioinformatics 2014, 30, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, S.; Wang, H.; Wu, J.; Chen, D.; Peng, B.; Zhou, Q. High expression of hexokinase domain containing 1 is associated with poor prognosis and aggressive phenotype in hepatocarcinoma. Biochem. Biophys. Res. Commun. 2016, 474, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, H.; Walther, W.; Zincke, F.; Kobelt, D.; Imbastari, F.; Erdem, M.; Kortüm, B.; Dahlmann, M.; Stein, U. MACC1-the first decade of a key metastasis molecule from gene discovery to clinical translation. Cancer Metastasis Rev. 2018, 37, 805–820. [Google Scholar] [CrossRef]
- Stein, U.; Walther, W.; Arlt, F.; Schwabe, H.; Smith, J.; Fichtner, I.; Birchmeier, W.; Schlag, P.M. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat. Med. 2009, 15, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Kobelt, D.; Perez-Hernandez, D.; Fleuter, C.; Dahlmann, M.; Zincke, F.; Smith, J.; Migotti, R.; Popp, O.; Burock, S.; Walther, W. The newly identified MEK1 tyrosine phosphorylation target MACC1 is druggable by approved MEK1 inhibitors to restrict colorectal cancer metastasis. Oncogene 2021, 40, 5286–5301. [Google Scholar] [CrossRef]
- Paço, A.; Brás, T.; Santos, J.O.; Sampaio, P.; Gomes, A.C.; Duarte, M.F. Anti-Inflammatory and Immunoregulatory Action of Sesquiterpene Lactones. Molecules 2022, 27, 1142. [Google Scholar] [CrossRef]
- Basti, A.; Malhan, D.; Dumbani, M.; Dahlmann, M.; Stein, U.; Relógio, A. Core-Clock Genes Regulate Proliferation and Invasion via a Reciprocal Interplay with MACC1 in Colorectal Cancer Cells. Cancers 2022, 14, 3458. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef]
- Khan, T.; Gurav, P. PhytoNanotechnology: Enhancing Delivery of Plant Based Anti-Cancer Drugs. Front. Pharmacol. 2018, 8, 1002. [Google Scholar] [CrossRef] [PubMed]
- Yalçin, M.; Malhan, D.; Basti, A.; Peralta, A.R.; Ferreira, J.J.; Relógio, A. A Computational Analysis in a Cohort of Parkinson’s Disease Patients and Clock-Modified Colorectal Cancer Cells Reveals Common Expression Alterations in Clock-Regulated Genes. Cancers 2021, 13, 5978. [Google Scholar] [CrossRef] [PubMed]
- Malhan, D.; Basti, A.; Relógio, A. Transcriptome analysis of clock disrupted cancer cells reveals differential alternative splicing of cancer hallmarks genes. npj Syst. Biol. Appl. 2022, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.; Fleury-Olela, F.; Nagoshi, E.; Hauser, C.; Juge, C.; A Meier, C.; Chicheportiche, R.; Dayer, J.-M.; Albrecht, U.; Schibler, U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005, 3, e338. [Google Scholar] [CrossRef]
- Spörl, F.; Schellenberg, K.; Blatt, T.; Wenck, H.; Wittern, K.-P.; Schrader, A.; Kramer, A. A circadian clock in HaCaT keratinocytes. J. Investig. Dermatol. 2011, 131, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Brás, T.; Paulino, A.F.C.; Neves, L.A.; Crespo, J.G.; Duarte, M.F. Ultrasound assisted extraction of cynaropicrin from Cynara cardunculus leaves: Optimization using the response surface methodology and the effect of pulse mode. Ind. Crops Prod. 2020, 150, 112395. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuhr, L.; Basti, A.; Brás, T.S.; Duarte, M.F.; Relógio, A. Antiproliferative Effects of Cynara Cardunculus in Colorectal Cancer Cells Are Modulated by the Circadian Clock. Int. J. Mol. Sci. 2022, 23, 9130. https://doi.org/10.3390/ijms23169130
Fuhr L, Basti A, Brás TS, Duarte MF, Relógio A. Antiproliferative Effects of Cynara Cardunculus in Colorectal Cancer Cells Are Modulated by the Circadian Clock. International Journal of Molecular Sciences. 2022; 23(16):9130. https://doi.org/10.3390/ijms23169130
Chicago/Turabian StyleFuhr, Luise, Alireza Basti, Teresa Silva Brás, Maria F. Duarte, and Angela Relógio. 2022. "Antiproliferative Effects of Cynara Cardunculus in Colorectal Cancer Cells Are Modulated by the Circadian Clock" International Journal of Molecular Sciences 23, no. 16: 9130. https://doi.org/10.3390/ijms23169130
APA StyleFuhr, L., Basti, A., Brás, T. S., Duarte, M. F., & Relógio, A. (2022). Antiproliferative Effects of Cynara Cardunculus in Colorectal Cancer Cells Are Modulated by the Circadian Clock. International Journal of Molecular Sciences, 23(16), 9130. https://doi.org/10.3390/ijms23169130






