Glycosylphosphatidylinositol Mannosyltransferase Ⅰ Protects Chinese Giant Salamander, Andrias davidianus, against Iridovirus
Abstract
1. Introduction
2. Results
2.1. Sequence Characterization of AdGPI-MT-I

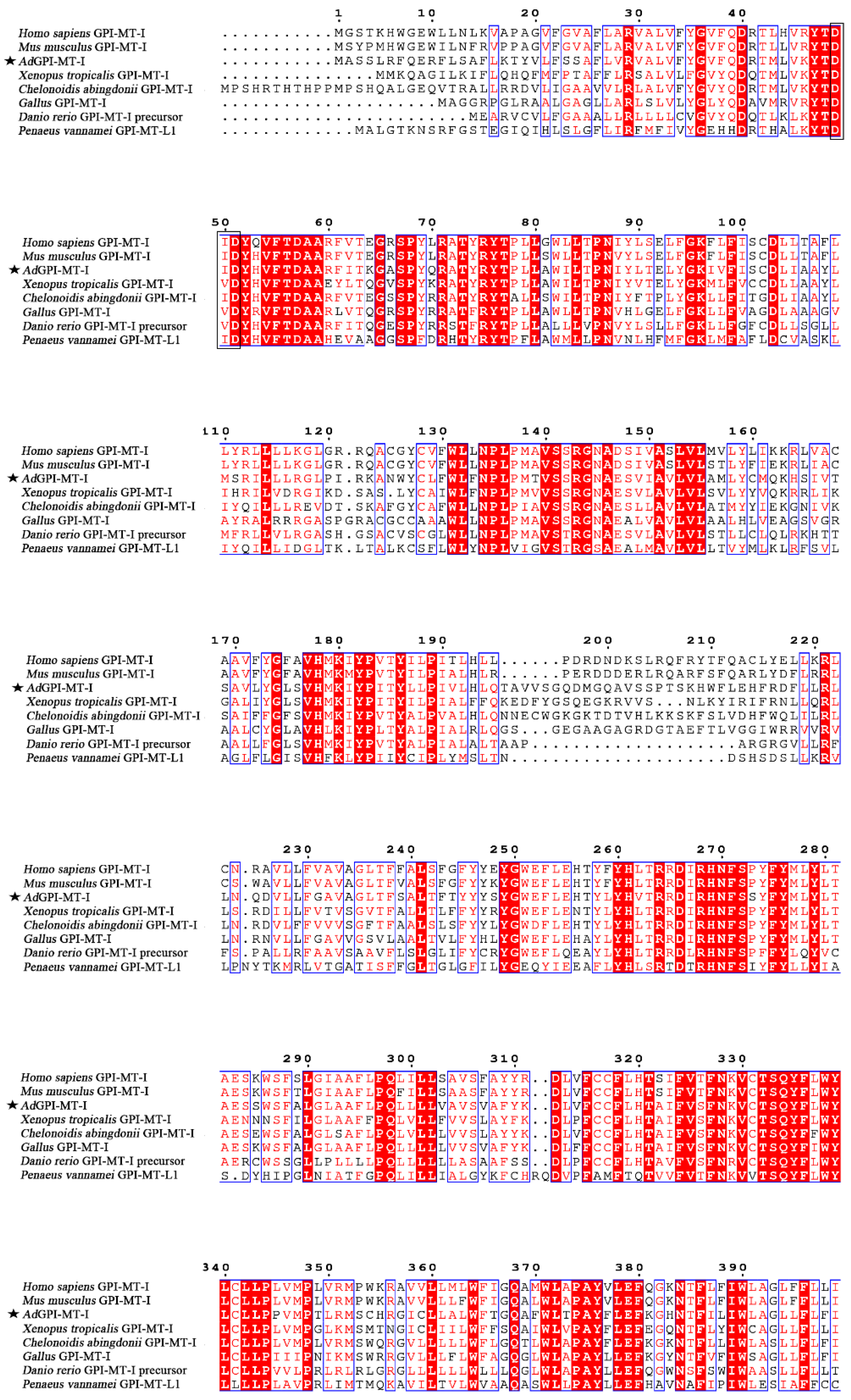
| Taxonomy | Species/Gene | Accession Number |
|---|---|---|
| Mammal | ||
| Homo sapiens GPI-MT-I | NP_660150.1 | |
| Pan troglodytes GPI-MT-I | NP_001233402.1 | |
| Hylobates moloch GPI-MT-I | XP_032009911.1 | |
| Rattus norvegicus GPI-MT-I | NP_077058.1 | |
| Mus musculus GPI-MT-I | NP_080510.1 | |
| Equus caballus GPI-MT-I | XP_005610027.1 | |
| Avian | ||
| Gallus gallus GPI-MT-I | NP_001026693.2 | |
| Anas platyrhynchos GPI-MT-I | XP_005023671.2 | |
| Reptile | ||
| Gopherus evgoodei GPI-MT-I | XP_030410133.1 | |
| Chelonoidis abingdonii GPI-MT-I | XP_032648289.1 | |
| Notechis scutatus GPI-MT-I | XP_026547789.1 | |
| Amphibian | ||
| Xenopus tropicalis GPI-MT-I | NP_001008120.1 | |
| Bufo bufo GPI-MT-I | XP_040289781.1 | |
| Fish | ||
| Polyodon spathula GPI-MT-I | XP_041101136.1 | |
| Danio rerio GPI-MT-I | NP_956684.1 | |
| Amphiprion ocellaris GPI-MT-I | XP_023121236.1 | |
| Poecilia latipinna GPI-MT-I | XP_014911736.1 | |
| Invertebrate | ||
| Octopus sinensis GPI-MT-I | XP_029633767.1 | |
| Exaiptasia diaphana GPI-MT-I | KXJ16443.1 | |
| Penaeus vannamei GPI-MT-I like | XP_027225082.1 | |
| Chionoecetes opilio GPI-MT-I | KAG0721514.1 |
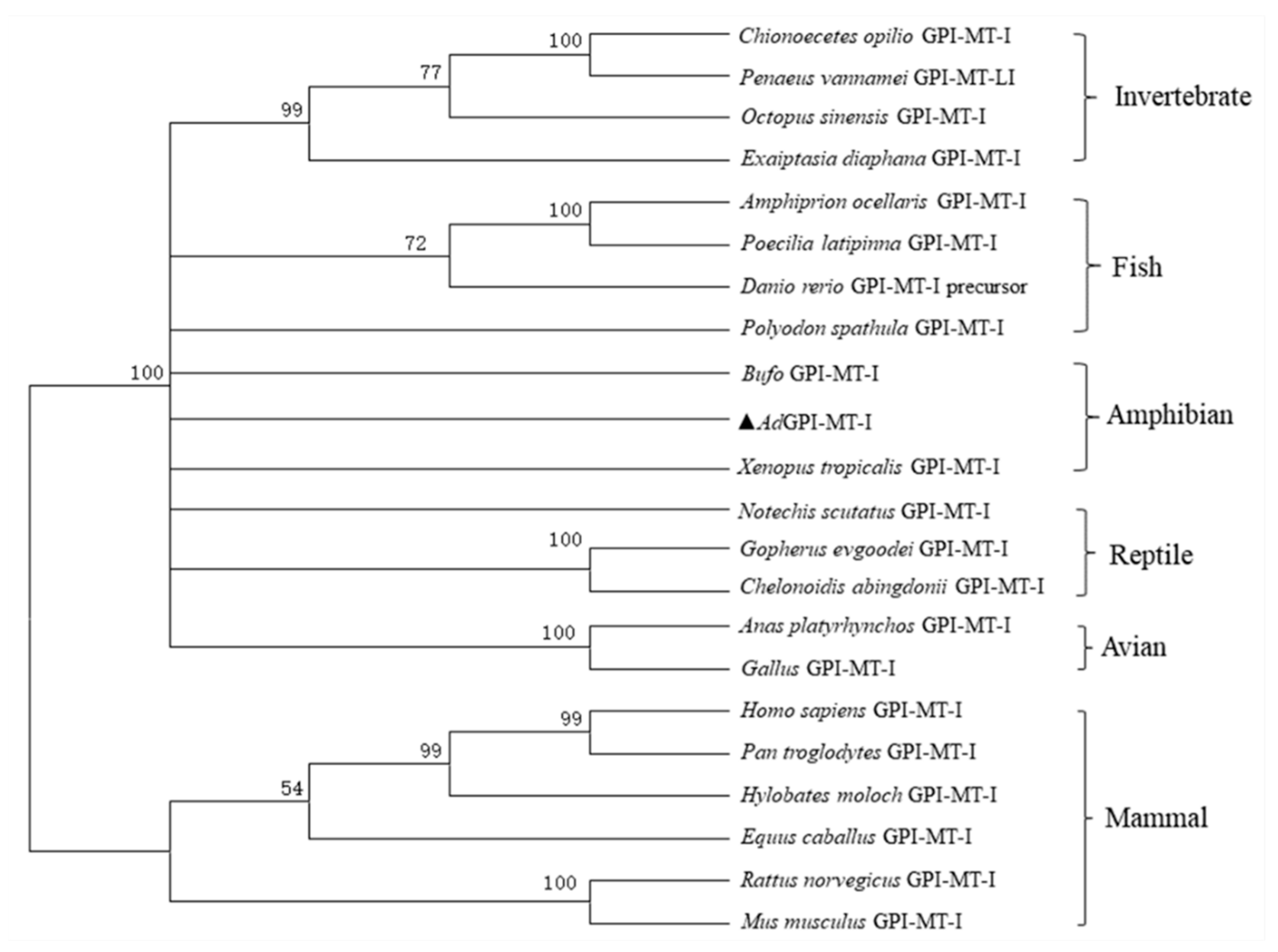
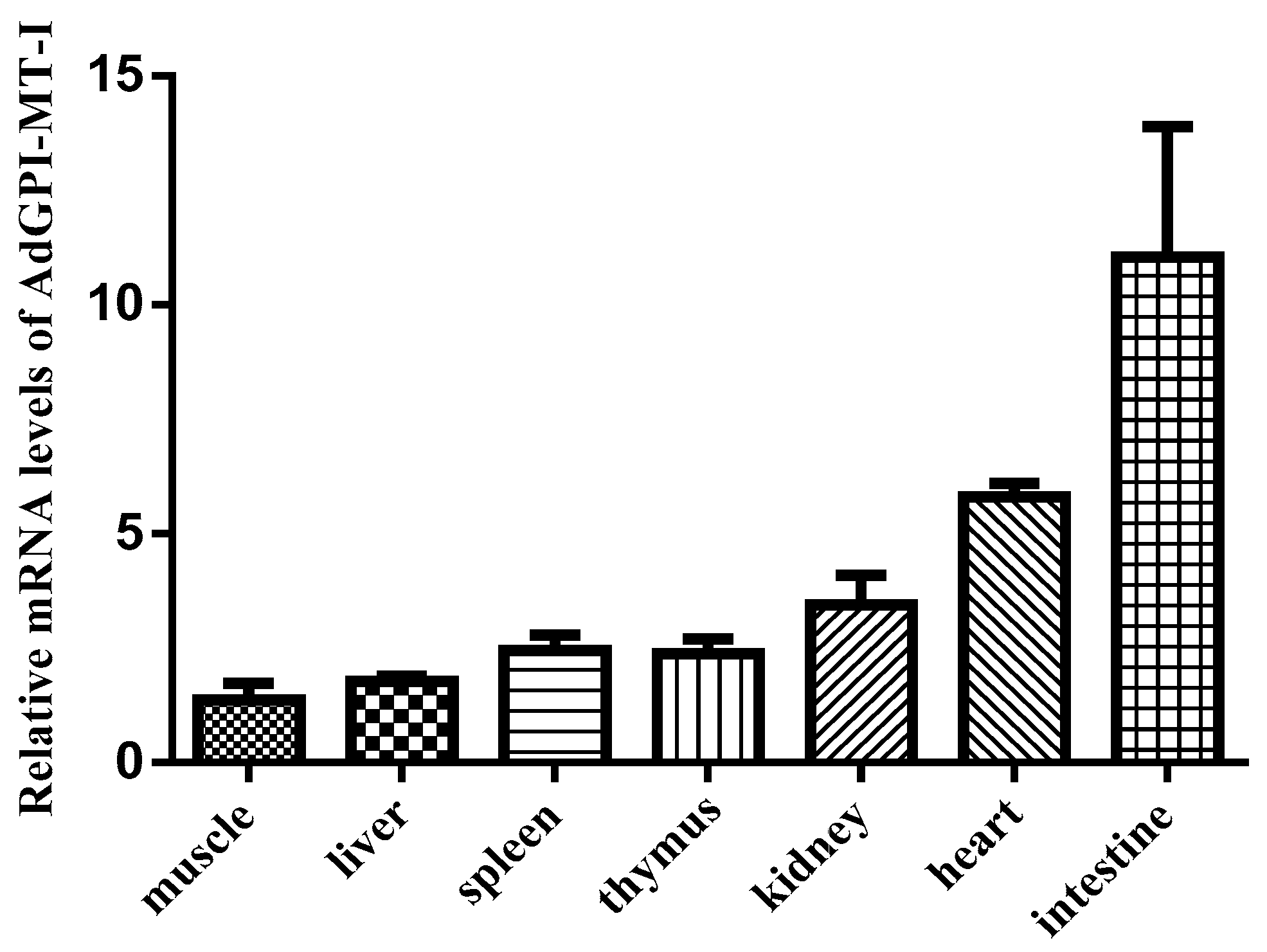
2.2. Expression Patterns of AdGPI-MT-I in Tissues and Cells
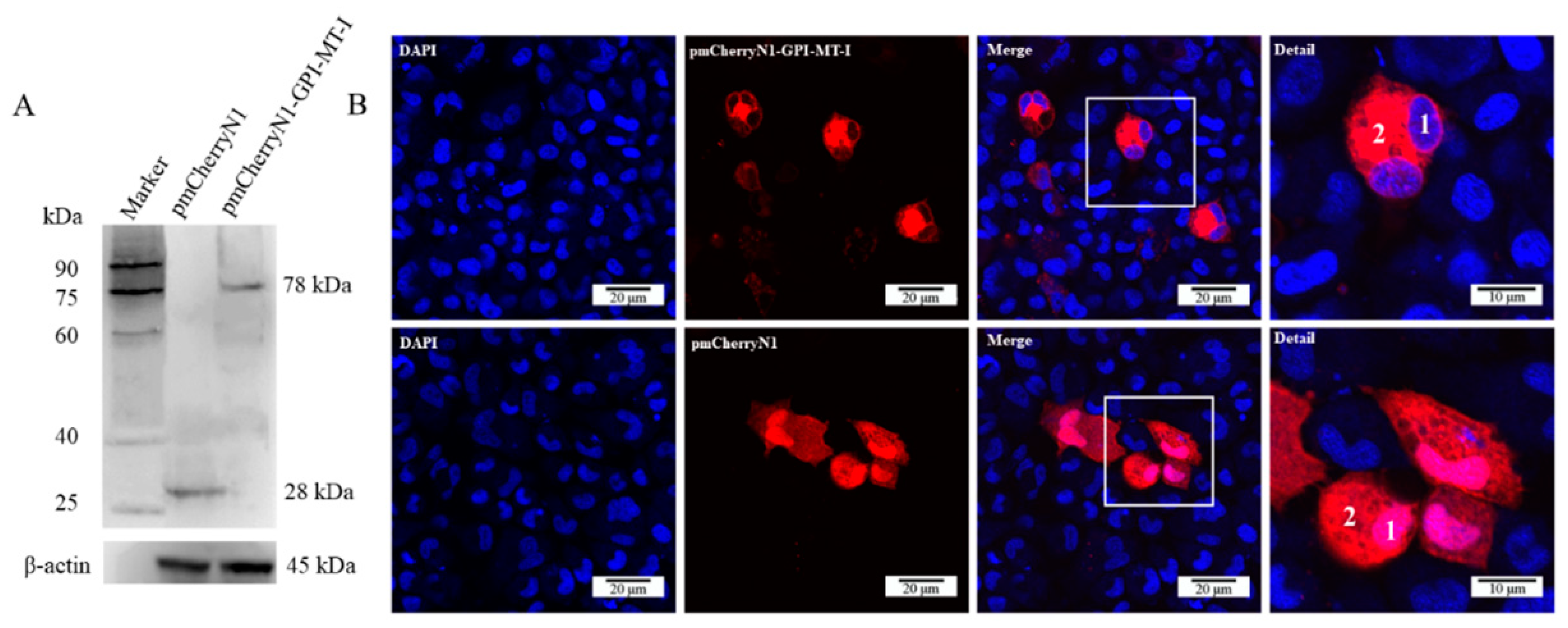
2.3. Subcellular Localization of AdGPI-MT-I
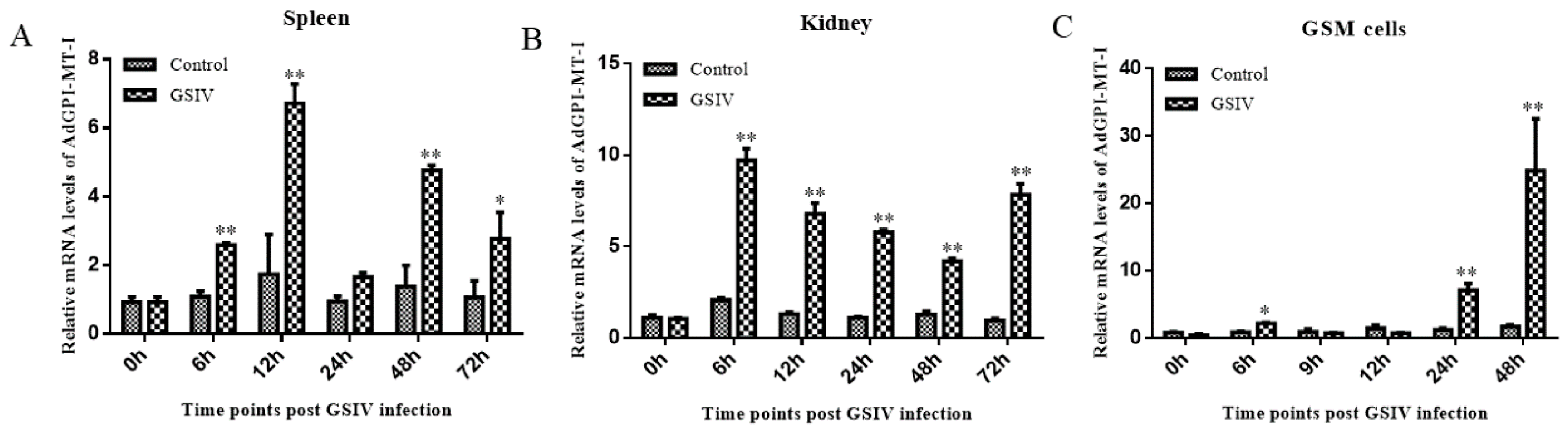
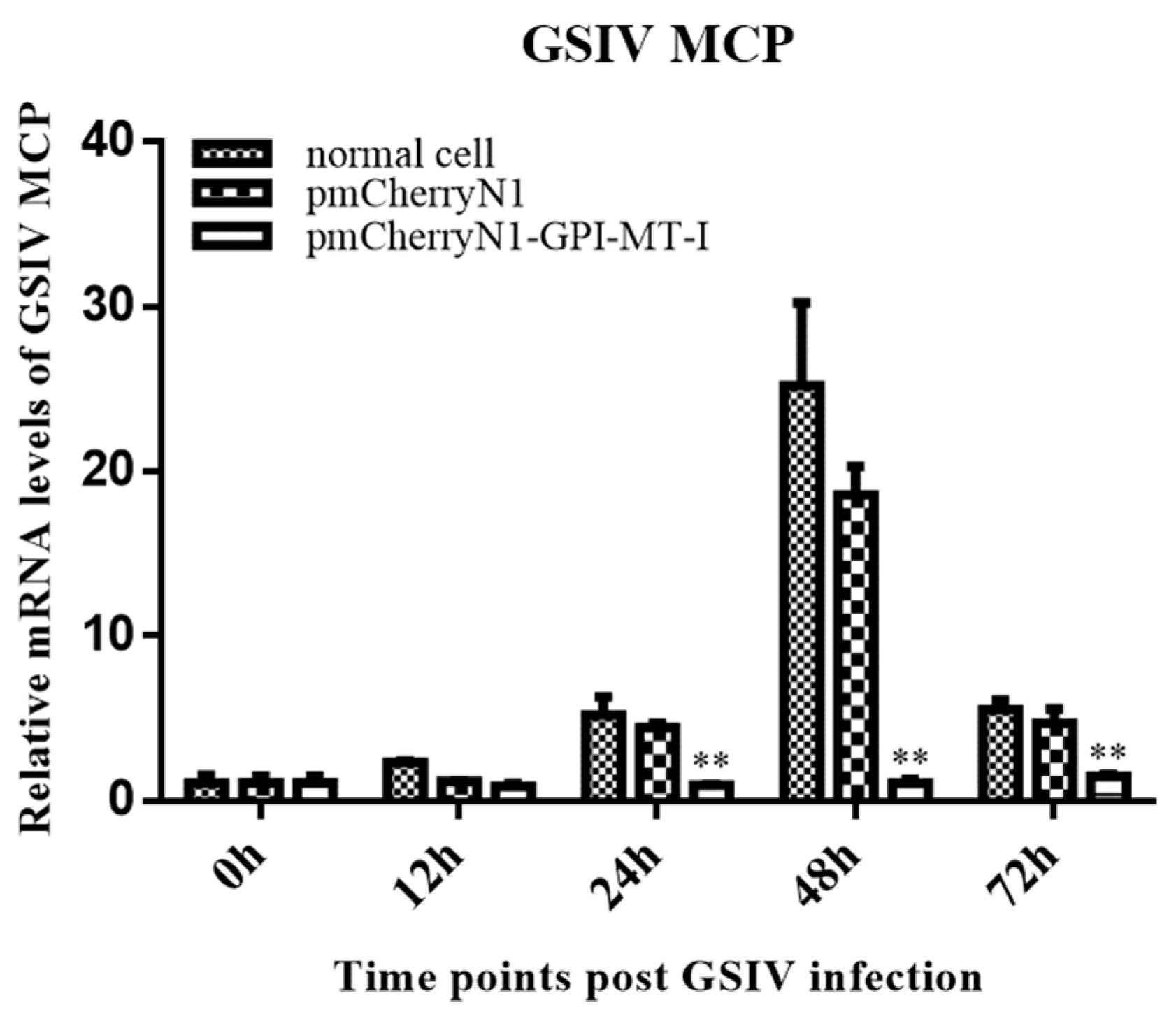
2.4. Antiviral Effect of AdGPI-MT-I in GSM Cells
3. Discussion
4. Materials and Methods
4.1. Animals, Cells, and Virus
4.2. DNA Extraction, RNA Extraction, and cDNA Preparation
4.3. cDNA Cloning of AdGPI-MT-I and Bioinformatic Analysis of Sequences
4.4. Detection of AdGPI-MT-I Distribution in Tissues by Quantitative Real-Time PCR (qRT-PCR)
4.5. The Effects of GSIV on the AdGPI-MT-I Expression in Tissues and GSM Cells
4.6. AdGPI-MT-I Expression Plasmid Construction, Subcellular Localization, and Western Blot Confirmation
4.7. Antiviral Activity of AdGPI-MT-I in GSM Cells
4.8. Digital Droplet PCR (ddPCR) Detection of Major Capsid Protein (MCP) Gene Copies of GSIV
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GPI-MT-I | Glycosylphosphatidylinositol mannosyltransferase I |
| GPI-APs | Glycosylphosphatidylinositol-anchor proteins |
| GSIV | Chinese giant salamander iridovirus |
| GSM cells | Chinese giant salamander muscle cell line |
| MCP | Chinese giant salamander iridovirus major capsid protein |
References
- Paulick, M.G.; Bertozzi, C.R. The glycosylphosphatidylinositol anchor: A complex membrane-anchoring structure for. Biochemistry 2008, 47, 6991–7000. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Murakami, Y.; Marmor, M.D.; Inoue, N.; Maeda, Y.; Hino, J.; Kangawa, K.; Julius, M.; Kinoshita, T. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 2000, 19, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Fujita, M.; Maeda, Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: Recent progress. J. Biochem. 2008, 144, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Leidich, S.D.; Drapp, D.A.; Orlean, P. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem. 1994, 269, 10193–10196. [Google Scholar] [CrossRef]
- Boutlis, C.S.; Riley, E.M.; Anstey, N.M.; de Souza, J.B. Glycosylphosphatidylinositols in malaria pathogenesis and immunity: Potential for therapeutic inhibition and vaccination. Immunol. Immunopathogenesis Malar. 2005, 297, 145–185. [Google Scholar]
- Debierre-Grockiego, F.; Schwarz, R.T. Immunological reactions in response to apicomplexan glycosylphosphatidylinositols. Glycobiology 2010, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.D.; Fernandez-Borja, M. Leukocyte adhesion and polarization: Role of glycosylphosphatidylinositol-anchored proteins. Bioarchitecture 2015, 5, 61–69. [Google Scholar] [CrossRef][Green Version]
- Huang, J.H.; Getty, R.R.; Chisari, F.V.; Fowler, P.; Greenspan, N.S.; Tykocinski, M.L. Protein transfer of preformed MHC-peptide complexes sensitizes target cells to T cell cytolysis. Immunity 1994, 1, 607–613. [Google Scholar] [CrossRef]
- Basagoudanavar, S.H.; Feng, X.; Krishnegowda, G.; Muthusamy, A.; Gowda, D.C. Plasmodium falciparum GPI mannosyltransferase-III has novel signature sequence and is functional. Biochem. Biophys. Res. Commun. 2007, 364, 748–754. [Google Scholar] [CrossRef]
- Ashida, H.; Hong, Y.; Murakami, Y.; Shishioh, N.; Sugimoto, N.; Kim, Y.U.; Maeda, Y.; Kinoshita, T. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell. 2005, 16, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.K.; Sharma, D.K.; Crossman, A.; Dix, A.; Brimacombe, J.S.; Ferguson, M.A. Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J. 1997, 16, 6667–6675. [Google Scholar] [CrossRef] [PubMed]
- Naderer, T.; Vince, J.E.; McConville, M.J. Surface determinants of Leishmania parasites and their role in infectivity in the mammalian host. Curr. Mol. Med. 2004, 4, 649–665. [Google Scholar] [CrossRef]
- Ribeiro, C.V.; Rocha, B.F.B.; Moreira, D.S.; Peruhype-Magalhães, V.; Murta, S.M.F. Mannosyltransferase (GPI-14) overexpression protects promastigote and amastigote forms of Leishmania braziliensis against trivalent antimony. Parasites Vectors 2019, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Mohan, T.; Berman, Z.; Luo, Y.; Wang, C.; Wang, S.; Compans, R.W.; Wang, B.Z. Chimeric virus-like particles containing influenza HA antigen and GPI-CCL28 induce long-lasting mucosal immunity against H3N2 viruses. Sci. Rep. 2017, 7, 40226. [Google Scholar] [CrossRef]
- Budirahardja, Y.; Doan, T.D.; Zaidel-Bar, R. Glycosyl phosphatidylinositol anchor biosynthesis is essential for maintaining epithelial integrity during Caenorhabditis elegans embryogenesis. PLoS Genet. 2015, 11, e1005082. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Zhang, K.; Wang, Z.; Ding, Y.; Wu, W.; Huang, S. The decline of the Chinese giant salamander Andrias davidianus and implications for its conservation. Oryx 2004, 38, 197–202. [Google Scholar] [CrossRef]
- Gao, K.Q.; Shubin, N.H. Earliest known crown-group salamanders. Nature 2003, 422, 424–428. [Google Scholar] [CrossRef]
- Robert, J.; Cohen, N. The genus Xenopus as a multispecies model for evolutionary and comparative immunobiology of the 21st century. Dev. Comp. Immunol. 2011, 35, 916–923. [Google Scholar] [CrossRef]
- Meng, Y.; Ma, J.; Jiang, N.; Zeng, L.B.; Xiao, H.B. Pathological and microbiological findings from mortality of the Chinese giant salamander (Andrias davidianus). Arch. Virol. 2014, 159, 1403–1412. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, Y.; Zhou, Y.; Liu, W.; Ma, J.; Meng, Y.; Xie, C.; Zeng, L. Characterization of Chinese giant salamander iridovirus tissue tropism and inflammatory response after infection. Dis. Aquat. Org. 2015, 114, 229–237. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, N.; Fan, Y.; Zhou, Y.; Liu, W.; Xue, M.; Meng, Y.; Zeng, L. Chinese Giant Salamander (Andrias davidianus) Iridovirus Infection Leads to Apoptotic Cell Death through Mitochondrial Damage, Caspases Activation, and Expression of Apoptotic-Related Genes. Int. J. Mol. Sci. 2019, 20, 6149. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Wang, K.; Zhou, Z.; Li, C.; Wang, J.; He, M.; Yin, Z.; Lai, W. First report of a ranavirus associated with morbidity and mortality in farmed Chinese giant salamanders (Andrias davidianus). J. Comp. Pathol. 2011, 145, 95–102. [Google Scholar] [CrossRef]
- Dong, W.; Zhang, X.; Yang, C.; An, J.; Qin, J.; Song, F.; Zeng, W. Iridovirus infection in Chinese giant salamanders, China, 2010. Emerg. Infect. Dis. 2011, 17, 2388–2389. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Watanabe, R.; Harris, C.L.; Hong, Y.; Ohishi, K.; Kinoshita, K.; Kinoshita, T. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001, 20, 250–261. [Google Scholar] [CrossRef]
- Orlean, P.; Menon, A.K. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar]
- Oriol, R.; Martinez-Duncker, I.; Chantret, I.; Mollicone, R.; Codogno, P. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol. Biol. Evol. 2002, 19, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mushegian, A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003, 12, 1418–1431. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Kinoshita, T.; Inoue, N. Dissecting and manipulating the pathway for glycosylphos-phatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000, 4, 632–638. [Google Scholar] [CrossRef]
- Gowda, D.C.; Gupta, P.; Davidson, E.A. Glycosylphosphatidylinositol anchors represent the major carbohydrate modification in proteins of intraerythrocytic stage Plasmodium falciparum. J. Biol. Chem. 1997, 272, 6428–6439. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.U.; Hong, Y. Functional analysis of the first mannosyltransferase (PIG-M) involved in glycosylphosphatidylinositol synthesis in Plasmodium falciparum. Mol. Cells 2007, 24, 294–300. [Google Scholar]
- McConville, M.J.; Menon, A.K. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol. 2000, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nosjean, O.; Briolay, A.; Roux, B. Mammalian GPI proteins: Sorting, membrane residence and functions. Biochim. Biophys. Acta 1997, 1331, 153–186. [Google Scholar] [CrossRef]
- Huang, L.; Fan, Y.; Zhou, Y.; Jiang, N.; Liu, W.; Meng, Y.; Zeng, L. Cloning, sequence analysis and expression profiles of Toll-like receptor 7 from Chinese giant salamander Andrias davidianus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2015, 184, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Zhou, Y.; Jiang, N.; Fan, Y.; Lin, G.; Zeng, L. Characterization, expression pattern and antiviral activities of oligoadenylate synthetase in Chinese Giant Salamander, Andrias davidianus. Dev. Comp. Immunol. 2022, 129, 104347. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Zhou, Y.; Jiang, N.; Fan, Y.; Zeng, L. Characterization, Expression Pattern and Antiviral Activities of Mx Gene in Chinese Giant Salamander, Andrias davidianus. Int. J. Mol. Sci. 2020, 21, 2246. [Google Scholar] [CrossRef]
- Chen, Q.; Ma, J.; Fan, Y.; Meng, Y.; Xu, J.; Zhou, Y.; Liu, W.; Zeng, X.; Zeng, L. Identification of type I IFN in Chinese giant salamander (Andrias davidianus) and the response to an iridovirus infection. Mol. Immunol. 2015, 65, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Bellai-Dussault, K.; Nguyen, T.T.M.; Baratang, N.V.; Jimenez-Cruz, D.A.; Campeau, P.M. Clinical variability in inherited glycosylphosphatidylinositol deficiency disorders. Clin. Genet. 2019, 95, 112–121. [Google Scholar] [CrossRef]
- Nebl, T.; De Veer, M.J.; Schofield, L. Stimulation of innate immune responses by malarial glycosylphosphatidylinositol via pattern recognition receptors. Parasitology 2005, 130 (Suppl. S1), S45–S62. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, M.A.; Brimacombe, J.S.; Brown, J.R.; Crossman, A.; Dix, A.; Field, R.A.; Güther, M.L.; Milne, K.G.; Sharma, D.K.; Smith, T.K. The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim. Biophys. Acta 1999, 1455, 327–340. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gallo-Oller, G.; Ordoñez, R.; Dotor, J. A new background subtraction method for Western blot densitometry band quantification through image analysis software. J. Immunol. Methods 2018, 457, 1–5. [Google Scholar] [CrossRef] [PubMed]

| Application | Primer Name | Nucleotide Sequence (5′-3′) |
|---|---|---|
| PCR | ||
| AdGPI-MT-I-ORF-F1 | ATGGCATCCTCGCTGAGATTTCA | |
| AdGPI-MT-I-ORF-R1 | CTAATGCAATTTAAGTTTTTCCACC | |
| AdGPI-MT-I-ORF-F2 | CCGGATCCATGGCATCCTCGCTGAGATTTCA | |
| AdGPI-MT-I-ORF-R2 | GCCTCGAGCTAATGCAATTTAAGTTTTTCCACC | |
| GSIV-F | GACTTGGCCACTTATGAC | |
| GSIV-R | GTCTCTGGAGAAGAAGAA | |
| qRT-PCR | ||
| AdGPI-MT-I-rqF | TTACTCCCCATTGTCCTCCA | |
| AdGPI-MT-I-rqR | GTGTTCCAAAAACTCCCAGC | |
| MCP-rqF | GCGGTTCTCACACGCAGTC | |
| MCP-rqR | ACGGGAGTGACGCAGGTGT | |
| β-actin-rqF | TGAACCCAAAAGCCAACCGAGAAAAGAT | |
| β-actin-rqR | TACGACCAGAGGCATACAGGGACAGGAC | |
| ddPCR | ||
| GSIV MCP-F | GCGGTTCTCACACGCAGTC | |
| GSIV MCP-R | ACGGGAGTGACGCAGGTGT | |
| GSIV probe | FAM + AGCCGACGGAAGGGTGTGTGAC + TAMARA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Dai, Y.; Fan, Y.; Jiang, N.; Zhou, Y.; Zeng, L.; Li, Y. Glycosylphosphatidylinositol Mannosyltransferase Ⅰ Protects Chinese Giant Salamander, Andrias davidianus, against Iridovirus. Int. J. Mol. Sci. 2022, 23, 9009. https://doi.org/10.3390/ijms23169009
Zhang J, Dai Y, Fan Y, Jiang N, Zhou Y, Zeng L, Li Y. Glycosylphosphatidylinositol Mannosyltransferase Ⅰ Protects Chinese Giant Salamander, Andrias davidianus, against Iridovirus. International Journal of Molecular Sciences. 2022; 23(16):9009. https://doi.org/10.3390/ijms23169009
Chicago/Turabian StyleZhang, Jingjing, Yanlin Dai, Yuding Fan, Nan Jiang, Yong Zhou, Lingbing Zeng, and Yiqun Li. 2022. "Glycosylphosphatidylinositol Mannosyltransferase Ⅰ Protects Chinese Giant Salamander, Andrias davidianus, against Iridovirus" International Journal of Molecular Sciences 23, no. 16: 9009. https://doi.org/10.3390/ijms23169009
APA StyleZhang, J., Dai, Y., Fan, Y., Jiang, N., Zhou, Y., Zeng, L., & Li, Y. (2022). Glycosylphosphatidylinositol Mannosyltransferase Ⅰ Protects Chinese Giant Salamander, Andrias davidianus, against Iridovirus. International Journal of Molecular Sciences, 23(16), 9009. https://doi.org/10.3390/ijms23169009







