Abstract
Despite its discovery in the early 1970s, m6A modification within mRNA molecules has only powerfully entered the oncology field in recent years. This chemical modification can control all aspects of the maturation of mRNAs, both in the nucleus and in the cytoplasm. Thus, the alteration in expression levels of writers, erasers, and readers may significantly contribute to the alteration of gene expression observed in cancer. In particular, the activation of oncogenic pathways can lead to an alteration of the global rate of mRNA translation or the selective translation of specific mRNAs. In both cases, m6A can play an important role. In this review, we highlight the role of m6A in the regulation of translation by focusing on regulatory mechanisms and cancer-related functions of this novel but still controversial field.
1. Introduction
Translational regulation plays a central role in the control of gene expression, and it is often dysregulated in cancer. In particular, oncogenic pathways may act by regulating the global rate of mRNA translation or the selective translation of specific mRNAs [1]. mRNA translation is a complex process divided into four steps: Initiation, elongation, termination, and ribosome recycling. Generally, mechanisms that regulate gene expression at the translational level act on translation initiation [2]. Eukaryotic mRNAs contain the cap structure at the 5′-end, which is constituted by an N7-methylguanosine (m7G) connected via a 5′ to 5′ triphosphate bond to the first transcribed nucleotide [3]. The cap structure plays a critical role in translation initiation and is recognized by the eukaryotic translation initiation factor (eIF) 4F, which contains the cap-binding protein eIF4E, the scaffold protein eIF4G, and the RNA helicase eIF4A. eIF4G also interacts with the poly(A)-binding protein (PABP), which promotes mRNA circularization and reinitiation efficiency of ribosomes after translation termination. The AUG start codon is recognized through a scanning mechanism by the 43S pre-initiation complex (PIC) that is recruited to the cap structure by eIF4F. The PIC is formed by the 40S small ribosomal subunit, different eukaryotic initiation factors (eIF1, eIF1A, eIF2, eIF3, eIF5), and the ternary complex (TC), composed of eIF2, initiator methionyl tRNA, and GTP [2]. AUG start codon recognition by PIC induces GTP hydrolysis in the ternary complex, the release of the initiation factors, and the joining of the large 60S ribosomal subunit to form the 80S initiation complex. The 80S complex is then ready to synthesize the peptide chain [2]. A single mRNA may contain multiple 80S ribosomes and it is referred to as a polyribosome or polysome. The higher the number of ribosomes, the higher the translation rate of the mRNA. The initiation of viral mRNAs and some cellular mRNAs can be cap-independent and rely on internal RNA structures, referred to as the internal ribosome entry site (IRES), which directly recruits the PIC on the AUG start codon [2] or an alternative cap-recognition mechanism mediated by eIF3d [4].
In addition to the m7G, mRNAs may also contain different internal chemical modifications. In mammalian cells, the first and second transcribed nucleotides are 2′-O-methylated on the ribose moiety [5]. Moreover, if the first nucleotide is A, it can also be methylated at the N6-position (m6Am) [6]. 2′-O ribose methylations do not affect mRNA expression but are important to discriminate between self and non-self RNA, while the role of m6Am in mRNA regulation and particularly in translation is still controversial. However, the most abundant internal modification is the N6-methyladenosine (m6A) [7]. m6A levels in mRNA are often deregulated in cancer and, more importantly, it is the only internal modification in mRNA with an established role in translation regulation.
In this review, we describe the expanding roles of translation regulation by m6A modification in cancer. In particular, we highlight regulatory mechanisms and cancer-related functions of this novel but still controversial field.
2. An Overview of m6A Regulators
The methyltransferase complex responsible for the vast majority of m6A modifications in mRNAs is composed of METTL3/METTL14 proteins, where METTL3 is the catalytic subunit and METTL14 is required for RNA binding. The complex recognizes and modifies A within the DRACH motif (D = A, G, U; R = A, G; H = A, C, U) during RNA transcription (reviewed in [7]). However, approximately 20% of DRACH motifs are methylated. m6A modifications are enriched in the terminal exon, near the STOP codon or in the 3′-untranslated region (3′-UTR). The U6 snRNA methyltransferase METTL16 is also involved in the modification of m6A sites in a small number of mRNAs and non-coding RNAs but it recognizes a different RNA sequence [8]. m6A modification can be removed by ALKBH5 (alkB homolog 5) and FTO (fat mass and obesity-associated protein) demethylases [7].
Even if m6A per se can impact the local RNA structure by altering the Hogsteen base-pairing, its effects on mRNA expression are generally mediated by specific protein readers. The YTH protein domain family are the only proteins that specifically recognize m6A modification, independently from the RNA sequence. In mammals, there are five YTH readers: YTHDC1, YTHDC2, and the paralogs YTHDF1, YTHDF2, and YTHDF3. YTHDC1 is the only nuclear reader, and it is involved in the regulation of nuclear processes such as transcription, splicing, and RNA export [9]. YTHDC2 is an RNA helicase that specifically acts during gametogenesis by degrading mRNAs. However, its function has been recently shown to be independent of the m6A binding domain [10,11]. Notably, several organisms have YTHDC2 orthologs in which the YTH is not present [12], thus indicating that YTHDC2 evolved to function in an m6A-independent manner. Cytoplasmic mRNA regulation by m6A is mainly controlled by the YTHDFs paralogs YTHDF1, YTHDF2, and YTHDF3. Although these proteins show high amino acid identity and equivalent binding sites in the transcriptome [9], different functions were initially ascribed to individual YTHDF proteins: YTHDF1 stimulates mRNA translation [13], YTHDF2 promotes mRNA degradation [14], and YTHDF3 has both functions [15]. However, recent studies reported that YTHDF proteins function together in a redundant manner only on the degradation of m6A-containing mRNAs [16,17]. At present, the action of YTHDF proteins is still controversial.
m6A regulators are often deregulated in cancer where they can play both oncogenic and oncosuppressive roles [18]. In the following sections, we focus on the specific role of m6A in translation regulation and its impact on cancer.
3. Regulation of mRNA Translation by m6A
The influence of m6A modifications on translation can be both positive and negative. There are currently many different, often controversial models. The position of m6A in mRNA influences the functional impact of its translation regulation. Indeed, proteins that bind within the coding region can be removed by ribosomes during translation elongation. Thus, m6A readers proteins bound on 5′- and 3′- untranslated regions (5′- and 3′- UTRs) will mediate cytoplasmic effects, while m6A modifications in coding regions will mainly affect the binding of tRNAs during the elongation process. Therein, the latter is generally independent of readers. In several cancer types, m6A modification was shown to be required for maintaining the high translational rate of oncogenic proteins [18].
3.1. Translational Regulation by m6A in 5′-UTRs
Although rare, m6A sites were identified in the 5′-UTR of different mRNAs. Furthermore, they were found to change in response to stress [19]. m6A in the 5′-UTR stimulates cap-independent translation by recruitment of the initiation factor eIF3 [20] (Figure 1a). This mechanism was initially demonstrated to be responsible for the induction of hsp70 (heat shock protein 70) upon heat shock [19,20]. In this condition, the YTHDF2 reader translocates in the nucleus protecting specific mRNAs from FTO-mediated demethylation, among which hsp70 mRNA, thereby enabling their cap-independent translation. However, the study did not clarify how these transcripts are selected by nuclear YTHDF2, which recognized only m6A without specificity of the sequence [9], as well as the mechanisms that translocate YTHDF2 in the nucleus without affecting the other two paralogs YTHDF1 and YTHDF3. A later study showed that the m6A-dependent translation of hsp70 requires the ABCF1 protein (also known as ABC50) [21]. ABCF1 belongs to the ATP-binding cassette (ABC) transporter family but lacks the transmembrane domains, which are characteristic of most ABC transporters and are regulators of translation initiation [22]. Conversely to the initial model, in which hsp70 translation was shown to depend on direct recognition of m6A by eIF3, ABCF1 stimulates cap-independent translation by interacting with the eIF4G and the TC component eIF2 in a stress-dependent manner [21] (Figure 1b). Notably, the m6A methyltransferase METTL3 was also found to be translated by ABCF1 in a positive feedback loop that could act when the cap-dependent translation is inhibited [21]. Surprisingly, ABCF1 also stimulates the translation of 5′-UTR modified mRNAs under normal growth conditions [21], thus indicating the existence of a cap-independent mechanism that relies on m6A modification even in the absence of stress. The same study showed that the YTHDF3 reader, but not YTHDF1 and YTHDF2, was required for cap-independent translation. However, it is not clear how ABCF1 is recruited to m6A modifications in the 5′-UTR region and whether YTHDF3 is involved in its recruitment. Further studies are needed to understand how specific m6A sites are maintained in the 5′-UTR of specific mRNAs.
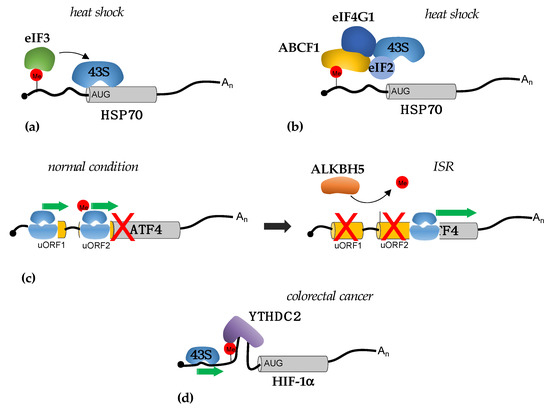
Figure 1.
Mechanisms of translational regulation mediated by m6A sites in 5′-UTRs. (a) Proposed mechanism for the cap-independent translational regulation of Hsp70 mRNA by eIF3 during heat shock (see main text for detail); (b) proposed mechanism for the cap-independent translational regulation of Hsp70 mRNA by ABCF1 during heat shock (see main text for detail); (c) during ISR, the decrease i TC levels and loss of removal of m6A from uORF2 of ATF4 mRNA by ALKBH5 induces ATF translation; (d) in colorectal cancer cells, m6A sites in the 5′-UTR of HIF-1α mRNA recruit the YTHDC2 helicase that removes secondary RNA structure facilitating 43S scanning.
M6A modification in the 5′-UTR was also shown to regulate the translation of ATF4 (Activating Transcription Factor 4) during the integrated stress response (ISR) [23] (Figure 1c). ISR is a prosurvival pathway that is initiated in response to different extrinsic and intrinsic factors [24]. ATF4 translation is required for the expression of stress-responsive genes [24]. ISR induction results in the phosphorylation of eIF2 with a consequent reduction in TC levels and a decrease in translation initiation efficiency. ATF4 mRNA contains two upstream open reading frames (uORFs) that precede the ATF4 coding region, uORF1 and uORF2. uORF2 overlaps with the ATF4 coding region but with a different reading frame. In normal conditions, with a high level of TC, initiation of the translation from uORFs is favored over ATF4 translation. Moreover, uORF2 start codon selection also depends on the presence of m6A modifications in the 5′-UTR of ATF mRNA. Upon stress, ATF4 translation is promoted by the concomitant decrease in TC and demethylation of m6A sites by ALKBH5. Global analysis of m6A levels upon nutrient deprivation showed that different transcripts lost m6A modifications in 5′-UTR, thus indicating that this can be a general mechanism for translation initiation by m6A from the non-canonical start codon in response to stress. However, the translational regulation by m6A, in this case, is cap-dependent and m6A in the 5′-UTR is strictly required for retaining initiating ribosomes in a transcript with multiple start sites. The mechanism responsible for this regulation has not yet been elucidated, but it has been suggested that the m6A can be read by specific RNA binding proteins that will, in turn, decrease the ribosome scanning efficiency [23]. Interestingly, ISR plays an important role in cancer, and its activation is required for tumor cell survival under stress and resistance to therapy. Therein, this indicates that the modulation of m6A levels can be utilized as a novel strategy to reduce ISR activation in tumors.
In tumors, the regulation of translation by m6A sites in the 5′-UTR was initially demonstrated in colorectal cancer [25]. Here, the YTHDC2 RNA helicase promotes the translation of the HIF-1α and Twist1 in hypoxia and facilitates the epithelial–mesenchymal transition (EMT), which plays a relevant role in cancer metastases. YTHDC2 binds to m6A sites and unwinds the RNA structures in the 5′-UTR, thus facilitating ribosome scanning during initiation [25] (Figure 1d).
The importance of m6A methylation within 5′-UTR in cancer was also shown in melanoma cells [26]. Melanoma cells that acquire resistance to BRAF (B-Raf Proto-Oncogene Serine/Threonine-Protein Kinase) and MEKs (Mitogen-Activated Protein Kinases) inhibitors, despite a general decrease in translation, showed increased translation of specific mRNAs that correlated with high m6A levels in their 5′-UTRs. These mRNAs encode for regulators of epigenetic modifications and signaling pathways that are connected to the presence of melanoma resistance cells. The effect of m6A modification on their translation is mediated by eIF4A helicase even if it is still not clear if it is directly recruited by m6A sites in the 5′-UTR [26]. This study suggests that the inhibition of METTL3 activity might be utilized as a novel strategy to overcome drug resistance in melanoma.
In breast cancer, the YTHDF3 reader was shown to be highly upregulated and bind to the m6A sites in 5′-UTR of its own transcript, stimulating translation in a positive feedback loop [27]. However, the molecular mechanism has not been clarified. High levels of YTHDF3 are then required for the translation of genes involved in brain metastases. A regulation depends on m6A sites that are not present in the 5′-UTR. Notably, the depletion of YTHDF3 is sufficient to inhibit brain metastasis and increases mice survival [27], thus indicating the lack of functional redundancy between YTHDF proteins in this context.
3.2. Translational Regulation by m6A in Coding Regions
The effect of m6A modifications within coding regions on mRNA translation, as well as their mechanisms of action, is still controversial. Translation elongation is conserved in all kingdoms of life, and initial work performed with bacteria showed that the presence of m6A within codons impairs tRNA accommodation, thus decreasing the translation elongation rate [28] (Figure 2a). Similar results were observed in HEK293T cells transfected with in vitro transcribed mRNAs containing different modifications [29]. This study showed that in human cells, m6A within coding regions also strongly inhibited translation, especially when it was present in the first codon position. A negative effect of m6A on elongation was also reported in the breast cancer cell line MCF7 in a study performed to correlate the transcription rate with translation efficiency by using reporter systems containing the luciferase gene under the control of different human promoters [30]. Additionally, in this case, the increase in m6A levels within the coding region of reporter transcripts produced a decrease in the translation rate. Moreover, the study also showed that the deposition of m6A is inversely proportional to the transcription rate. Notably, in different maize lines and during Xenopus laevis oogenesis, an inverse correlation between the m6A levels in the coding regions of mRNAs and their translational efficiency was confirmed [31,32]. The negative effect of m6A modification on translation elongation was also reported in mouse embryonic fibroblasts (MEF) [33], where the presence of m6A in highly structured regions produced ribosome pausing. Surprisingly, deleting m6A from these transcripts resulted in a further decrease in translation [33]. The authors of this study proposed a model in which the YTHDC2 reader, which also contains an RNA helicase domain, resolves the RNA structures containing m6A, therein promoting translation elongation by the ribosome (Figure 2b).
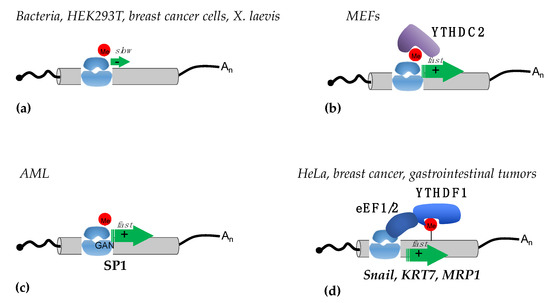
Figure 2.
Mechanisms of translational regulation mediated by m6A sites in coding regions. (a) In bacteria, different human cell lines, and in X. laevis, m6A sites in the coding regions were shown to slow down elongation (indicated by a green arrow); (b) in MEFs m6A in the coding regions are recognized by the YTHDC2 reader, which by removing secondary RNA structures facilitates elongation by the ribosome; (c) in AML the m6A modifications present in GAN codons of specific mRNAs, such as SP1, increase elongation by reducing ribosome stalling; (d) in different tumor cell lines, m6A sites in the coding regions of specific mRNAs (such as Snail, KRT, and MRP1) are bound by the YTHDF1 reader, which stimulates elongation by interacting with the eEF1 and eEF2 elongation factors.
The opposite results were reported in acute myeloid leukemia (AML) cells. In this blood cell cancer, METTL3 plays an oncogenic role and is required for sustaining AML cell proliferation [34]. In this case, METTL3 is recruited to specific promoters, independently from METTL14, to install m6A modification within the coding region of oncogenic mRNAs during transcription. However, later crystallographic studies demonstrated that METTL14 is strictly required for METTL3 methyltransferase activity [7]. Therein, it is not clear how METTL3 can act independently from METTL14 to methylate specific mRNAs. However, methylation by METTL3 in their coding region results in an increased translation rate. The proposed mechanism relies on the reduction of ribosome stalling on m6A methylated GAN codons, which results in faster elongation speed [34] (Figure 2c). This peculiar mechanism has only been described in AML to date.
The importance of translation elongation regulation by m6A in coding regions was also demonstrated in the regulation of EMT [35]. In this case, m6A sites in the coding region of the Snail transcript, which encodes for a transcription factor that regulates EMT, promote Snail translation by recruiting the eukaryotic translation elongation factor (eEF) 2 through the YTHDF1 reader [35] (Figure 2d). Interestingly, the study also reported that Snail mRNA also contains several m6A sites in the 3′-UTR that are not required for Snail regulation. Moreover, by using cell-line-derived xenograft mice, the authors showed that the overexpression of Snail can also stimulate lung colonization by HeLa cells in the absence of the methyltransferase METTL3 [35]. Similar results were reported in breast cancer and gastrointestinal stromal tumors [36,37]. In the first case, the m6A deposition in keratin 7 (KRT7), an important mediator of cancer metastases, stimulates its translation via the YTHDF1/eEF1 axis [36]. In the latter, the YTHDF1/eEF1 interaction stimulates the translation of the multidrug transporter MRP1 mRNA, also known as ABCC1 (ATP Binding Cassette Subfamily C Member 1), which is involved in drug resistance [37].
3.3. Translational Regulation by m6A in 3′-UTRs
3′-UTRs contain cis-regulatory elements that are recognized by RNAs and proteins to regulate translation. This regulation mainly takes place at the level of the translation initiation phase. Indeed, the interaction between eIF4F and PABP brings 3′-UTR elements close to the translation initiation complex. m6A sites in the 3′-UTR were initially shown to stimulate mRNA circularization via the interaction between the YTHDF1 reader and the initiation factors eIF3A and eIF3B [14] (Figure 3a). Moreover, the tethering of YTHDF1 on reporter constructs was sufficient to stimulate its translation. However, it was reported that eIF3 can also directly recognize m6A in the 5′-UTR to stimulate the cap-independent translation (see above), so it is not clear how eIF3 would not bind to m6A in the 3′-UTR, which is close to the 5′-end of mRNA, independently from YTHDF1. Even if the specific effect of YTHDF paralogue proteins is currently under debate [16,17], many studies reported the importance of YTHDF1 in the regulation of translation in cancer [38,39,40,41,42,43,44,45,46,47,48,49,50,51]. Notably, a strong antitumor response was described in YTHDF1-deficient mice [39]. In dendritic cells, YTHDF1 stimulates the translation of mRNA encoding for proteases that results in efficient antigen degradation. The lack of YTHDF1 increases the presentation of tumor antigens and the activation of CD8+ T cells, which are required for containing tumor infiltration [39]. Therein, these results suggest that inhibition of YTHDF1 activity might be used in combination with immune checkpoint inhibitors to enhance the killing of cancer cells by the immune system.
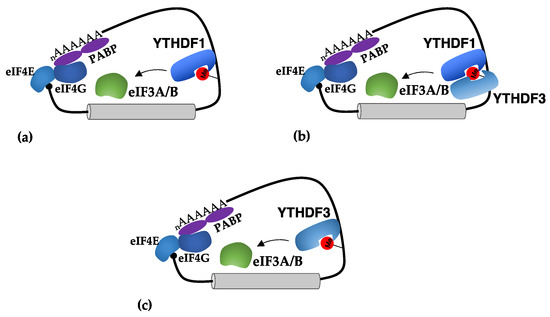
Figure 3.
Mechanisms of translational regulation mediated by m6A sites in 3′-UTRs. (a) The binding of YTHDF1 in m6A sites present in the 3′-UTR stimulates translation initiation by recruiting eIF3A and eIF3B; (b) the YTHDF3 reader was shown to act in cooperation with YTHDF1 in stimulation of translation; (c) in some tumors, YTHDF3 can act independently from YTHDF1 in stimulating translation.
Similarly, YTHDF3 was also shown to stimulate the translation of mRNAs containing m6A sites in the 3′-UTR. Initial studies indicated that YTHDF3 acts in cooperation with YTHDF1 [15,52] (Figure 3b). However, further studies indicated that YTHDF3 can also function independently from YTHDF1 [26] (Figure 3c). Interestingly, it was shown that the MYC oncogene, which is a general activator of ribosome biogenesis and translation in cancer, inhibits the translation of specific transcripts by downregulating their m6A levels through the transcriptional induction of the demethylase ALKBH5 [53]. Furthermore, in this case, YTHDF3 was required for their translational activation, and this effect was lost upon MYC activation [53].
3.4. Direct Translational Regulation by m6A Methyltransferases
Notably, in cancer cells, METTL3 and METTL16 methyltransferases can translocate to the cytoplasm and act as positive regulators for the translation of oncogenic m6A-modified mRNAs. This mechanism was initially described for METTL3 in lung cancer [54,55] and chronic myeloid leukemia (CML) [56,57]. The binding of METTL3 in the 3′-UTR of these transcripts promotes translation via interaction with the eIF3 component eIF3h [55] (Figure 4a). The tethering of METTL3 in the 3′-UTR of reported genes is sufficient to stimulate translation. Notably, the expression of a METTL3 derivative without the eIF3h interacting region is not able to induce tumors in lung cancer cell line xenograft models; thus, indicating the important role of cytoplasmic regulation of translation by METTL3 in tumorigenesis [55]. Importantly, the cytoplasmic activity of METTL3 does not require its catalytic domain. However, m6A installation is apparently required for METTL3 binding to mRNA in the cytoplasm. It has been proposed that cytoplasmic METTL3 does not require any readers for the recognition of m6A sites in the 3′-UTR. However, it is not clear how METTL3 would recognize and bind to modified transcripts in the cytoplasm because, in the nucleus, it strictly requires METTL14 for binding to consensus DRACH sequences, which do not contain m6A-modified adenines [7].
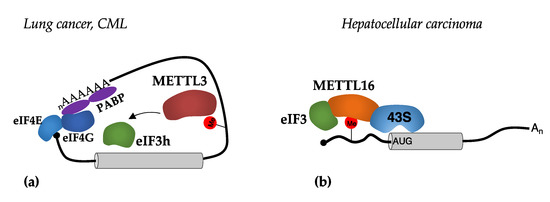
Figure 4.
Mechanisms of translational regulation mediated by METTL3 and METTL16 methyltransferases. (a) The binding of METTL3 in m6A sites present in the 3′-UTR stimulates translation initiation by recruiting eIF3h; (b) the binding of METTL16 in m6A sites present in the 5′-UTR stimulates translation initiation by facilitating the interaction between the 43S PIC and the eIF3 translation initiation factor.
More recently, a similar mechanism was described for METTL16 in hepatocellular carcinoma [58]. The authors reported that METTL16 binds to m6A sites close to the AUG start codon and promotes translation by facilitating the interaction between eIF3a and eIF3b, and the 18 rRNA component of the 40S small subunit in the PIC [57] (Figure 4b). Conversely to METTL3, the methyltransferase domain was shown to be strictly required for the activity of METTL16 as a translational regulator and interaction with the eIF3 components. Therein, this suggests that the use of a catalytic inhibitor would inhibit both catalytic-dependent and -independent activities of METTL16. Furthermore, in hepatocellular carcinoma cells, the depletion of METTL16 produced a strong decrease in translation and impaired cell survival. Notably, METTL16 can bind more than a thousand transcripts, the m6A modification of which does not, however, depend on METTL16 [58].
For both METTL3 and METTL16, further studies are needed to understand the mechanism that promotes their cytoplasmic translocation and how they can discriminate and bind to specific m6A-modified transcripts.
3.5. Translational Regulation by m6A in Circular RNAs (circRNAs)
circRNAs are covalently closed RNA molecules produced by the back-splicing of coding transcripts (review in [59]). As many circRNAs are derived from coding exons, they may still have coding potential but without the cap structure and poly-A tail required for cap-dependent translation. Notably, many circRNAs contain m6A close to the AUG start codon, and a single m6A site is sufficient to promote circRNA translation [60,61]. m6A sites within circRNAs are recognized by the YTHDF3 reader that recruits the non-canonical eIF4G2 (also known as Dap5 and Nat1) translation initiation factor to stimulate cap-independent translation [60,61]. In hepatocellular carcinoma, an additional mechanism has been described for circMAP3K4, derived from the MAP3K4 gene (Mitogen-Activated Protein Kinase 4). This circRNA was found upregulated and translated into a peptide of 455 amino acids that protects cancer cells from apoptosis induced by chemotherapeutic agents [62]. In this case, circMAP3K4 translation depends on m6A sites that are recognized by the IGF2BP1 protein (Insulin-Like Growth Factor 2 MRNA Binding Protein 1). This is a peculiarity of circRNAs because, in linear m6A-modified transcripts, the binding of IGF2BP1 increases their stability [7]. Several translated circRNAs have been discovered (reviewed in [63]), and many of the produced proteins play important roles in different types of cancer [63,64,65,66,67]. Nevertheless, the mechanism of cap-independent translation has not yet been identified for most of them. Therein, the role of m6A in the translation of circRNAs warrants further investigation.
4. Conclusions
Translational regulation is crucial for cancer development [1]. Highly proliferating cells demand elevated levels of ribosomes and intense translational rates. Moreover, cancer cells need to adapt their translation under stress conditions, such as hypoxia, derived from their growth environment. Thus, they require mechanisms that integrate different steps of gene expression, from transcription to translation, to produce a general increase in protein synthesis or to enhance the translation of specific mRNAs. The greatly expanding field of RNA modifications fits precisely in this context. Among the hundreds of different modifications that RNA can undergo, the one that is most studied in the tumor field is undoubtedly m6A within mRNA molecules. Inhibitors against writers and erasers of m6A have been recently developed and have shown promising results in cancer cells and cancer mouse models [68,69,70].
However, a major problem in the field is the many different mechanisms identified for m6A in mRNA metabolism and translation regulation, often with opposite effects, and the lack of reproducible results between different studies. This could be due, at least partially, to the lack of specific and quantitative methods for m6A detection, context dependency, and the complexity of gene expression regulation by m6A and, eventually, its indirect effect on translation. We urgently need mechanistic studies to determine the contribution of m6A to translation. In this context, targeted m6A editors might be used to install or remove m6A at desired sites in cancer-related mRNAs to address the specific positional effects of m6A on translation. Moreover, another critical point to clarify is the redundancy between cytoplasmic readers of the YTH family and their individual contributions to translation regulation.
Nevertheless, the identification of specific translational regulations mediated by m6A in cancer cells might bring the development of novel therapeutic strategies. One of these might be the interference with the cytoplasmic function of METTL3 and METTL16 as positive translational regulators of oncogenes. Understanding the mechanisms and signaling pathways that drive their translocation to the cytoplasm can be useful in the design of combination therapy with drugs already used in the clinic.
The m6A field is relatively new and it is hoped that, in the future, methodological advancement and scientific rigor can produce definitive results on its contribution to gene expression regulation, including translation. This will surely benefit cancer studies.
Author Contributions
Writing—original draft preparation, A.F.; writing—review and editing, G.F.R. and B.C. All authors have read and agreed to the published version of the manuscript.
Funding
The authors received funding from H2020-MSCA-ITN-2018, Project number 813091; and “Progetti Ateneo” Sapienza University of Rome, Project number RP1201729D714976.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Silvera, D.; Formenti, S.C.; Schneider, R.J. Translational control in cancer. Nat. Rev. Cancer 2010, 10, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H.D. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Alvin Chew, B.L.; Lai, Y.; Dong, H.; Xu, L.; Balamkundu, S.; Cai, W.M.; Cui, L.; Liu, C.F.; Fu, X.-Y.; et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019, 47, e130. [Google Scholar] [CrossRef] [PubMed]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II–Associated methyltransferase. Science 2019, 363, eaav0080. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m6A in the Transcriptome: m6A-Binding Proteins. Trends Cell Biol. 2017, 28, 113–127. [Google Scholar] [CrossRef]
- Li, L.; Krasnykov, K.; Homolka, D.; Gos, P.; Mendel, M.; Fish, R.J.; Pandey, R.R.; Pillai, R.S. The XRN1-regulated RNA helicase activity of YTHDC2 ensures mouse fertility independently of m6A recognition. Mol. Cell 2022, 82, 1678–1690.e12. [Google Scholar] [CrossRef]
- Saito, Y.; Hawley, B.R.; Puno, M.R.; Sarathy, S.N.; Lima, C.D.; Jaffrey, S.R.; Darnell, R.B.; Keeney, S.; Jain, D. YTHDC2 control of gametogenesis requires helicase activity but not m6A binding. Genes Dev. 2022, 36, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Puno, M.R.; Meydan, C.; Lailler, N.; Mason, C.E.; Lima, C.D.; Anderson, K.V.; Keeney, S. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 2018, 7, e30919. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Lasman, L.; Krupalnik, V.; Viukov, S.; Mor, N.; Aguilera-Castrejon, A.; Schneir, D.; Bayerl, J.; Mizrahi, O.; Peles, S.; Tawil, S.; et al. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev. 2020, 34, 1373–1391. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Coots, R.A.; Liu, X.M.; Mao, Y.; Dong, L.; Zhou, J.; Wan, J.; Zhang, X.; Qian, S.B. m6A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell 2017, 68, 504–514.e7. [Google Scholar] [CrossRef]
- Paytubi, S.; Wang, X.; Lam, Y.W.; Izquierdo, L.; Hunter, M.J.; Jan, E.; Hundal, H.S.; Proud, C.G. ABC50 Promotes Translation Initiation in Mammalian Cells. J. Biol. Chem. 2009, 284, 24061–24073. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Shu, X.E.; Mao, Y.; Liu, X.-M.; Yuan, X.; Zhang, X.; Hess, M.E.; Brüning, J.C.; Qian, S.-B. N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell 2018, 69, 636–647.e7. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Faouzi, S.; Bastide, A.; Martineau, S.; Malka-Mahieu, H.; Fu, Y.; Sun, X.; Mateus, C.; Routier, E.; Roy, S.; et al. An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat. Commun. 2019, 10, 5713–5714. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871.e7. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Ieong, K.-W.; Demirci, H.; Chen, J.; Petrov, A.; Prabhakar, A.; O’Leary, S.E.; Dominissini, D.; Rechavi, G.; Soltis, S.M.; et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016, 23, 110–115. [Google Scholar] [CrossRef]
- Hoernes, T.P.; Heimdörfer, D.; Köstner, D.; Faserl, K.; Nußbaumer, F.; Plangger, R.; Kreutz, C.; Lindner, H.; Erlacher, M.D. Eukaryotic Translation Elongation is Modulated by Single Natural Nucleotide Derivatives in the Coding Sequences of mRNAs. Genes 2019, 10, 84. [Google Scholar] [CrossRef]
- Slobodin, B.; Han, R.; Calderone, V.; Vrielink, J.A.O.; Loayza-Puch, F.; Elkon, R.; Agami, R. Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell 2017, 169, 326–337.e12. [Google Scholar] [CrossRef]
- Qi, S.T.; Ma, J.-Y.; Wang, Z.-B.; Guo, L.; Hou, Y.; Sun, Q.-Y. N6-Methyladenosine Sequencing Highlights the Involvement of mRNA Methylation in Oocyte Meiotic Maturation and Embryo Development by Regulating Translation in Xenopus laevis. J. Biol. Chem. 2016, 291, 23020–23026. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.-H.; Wang, Y.; Wang, M.; Zhang, L.-Y.; Peng, H.-R.; Zhou, Y.-Y.; Jia, G.-F.; He, Y. Natural Variation in RNA m6A Methylation and Its Relationship with Translational Status. Plant Physiol. 2020, 182, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Dong, L.; Liu, X.-M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.-B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chai, G.; Wu, Y.; Li, J.; Chen, F.; Liu, J.; Luo, G.; Tauler, J.; Du, J.; Lin, S.; et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019, 10, 2065. [Google Scholar] [CrossRef]
- Chen, F.; Chen, Z.; Guan, T.; Zhou, Y.; Ge, L.; Zhang, H.; Wu, Y.; Jiang, G.M.; He, W.; Li, J.; et al. N6-Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res. 2021, 81, 2847–2860. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, Q.; Chen, M.; Li, B.; Wang, N.; Li, C.; Gao, Z.; Zhang, D.; Yang, L.; Xu, Z.; et al. N6-methyladenosine modification regulates imatinib resistance of gastrointestinal stromal tumor by enhancing the expression of multidrug transporter MRP1. Cancer Lett. 2022, 530, 85–99. [Google Scholar] [CrossRef]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef]
- Pi, J.; Wang, W.; Ji, M.; Wang, X.; Wei, X.; Jin, J.; Liu, T.; Qiang, J.; Qi, Z.; Li, F.; et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 2021, 81, 2651–2665. [Google Scholar] [CrossRef]
- Li, Q.; Ni, Y.; Zhang, L.; Jiang, R.; Xu, J.; Yang, H.; Hu, Y.; Qiu, J.; Pu, L.; Tang, J.; et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 2021, 6, 76. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, X.; Yang, P.; Zhang, X.; Peng, Y.; Li, D.; Yu, Y.; Wu, Y.; Wang, Y.; Zhang, J.; et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 2021, 12, 1394. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Liang, R.; Yi, Y.-C.; Fan, H.-N.; Chen, M.; Zhang, J.; Zhu, J.-S. The m6A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m6A-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 647702. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, Q.; Kang, J.; Wei, Q.; Yang, Y.; Yang, D.; Liu, X.; Liu, T.; Yi, P. YTHDF1 Aggravates the Progression of Cervical Cancer Through m6A-Mediated Up-Regulation of RANBP2. Front. Oncol. 2021, 11, 650383. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Ning, J.; Liu, W.; Li, K.; Qian, B.; Xu, D.; Wu, Y.; Zhang, D.; Cui, W. YTHDF1 Promotes Cyclin B1 Translation through m6A Modulation and Contributes to the Poor Prognosis of Lung Adenocarcinoma with KRAS/TP53 Co-Mutation. Cells 2021, 10, 1669. [Google Scholar] [CrossRef]
- Wang, S.; Gao, S.; Zeng, Y.; Zhu, L.; Mo, Y.; Wong, C.C.; Bao, Y.; Su, P.; Zhai, J.; Wang, L.; et al. N6-Methyladenosine Reader YTHDF1 Promotes ARHGEF2 Translation and RhoA Signaling in Colorectal Cancer. Gastroenterology 2022, 162, 1183–1196. [Google Scholar] [CrossRef]
- Chen, H.; Yu, Y.; Yang, M.; Huang, H.; Ma, S.; Hu, J.; Xi, Z.; Guo, H.; Yao, G.; Yang, L.; et al. YTHDF1 promotes breast cancer progression by facilitating FOXM1 translation in an m6A-dependent manner. Cell Biosci. 2022, 12, 19. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, L.; Wang, L.; Huang, W.; Tan, L.; Liu, H.; Huo, J.; Su, T.; Zhang, M.; Kuang, M.; et al. YTHDF1 promotes intrahepatic cholangiocarcinoma progression via regulating EGFR mRNA translation. J. Gastroenterol. Hepatol. 2022, 37, 1156–1168. [Google Scholar] [CrossRef]
- Yao, X.; Li, W.; Li, L.; Li, M.; Zhao, Y.; Fang, D.; Zeng, X.; Luo, Z. YTHDF1 upregulation mediates hypoxia-dependent breast cancer growth and metastasis through regulating PKM2 to affect glycolysis. Cell Death Dis. 2022, 13, 258. [Google Scholar] [CrossRef]
- Xiong, J.; He, J.; Zhu, J.; Pan, J.; Liao, W.; Ye, H.; Wang, H.; Song, Y.; Du, Y.; Cui, B.; et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 2022, 82, 1660–1677.e10. [Google Scholar] [CrossRef]
- Zhu, Y.; Peng, X.; Zhou, Q.; Tan, L.; Zhang, C.; Lin, S.; Long, M. METTL3-mediated m6A modification of STEAP2 mRNA inhibits papillary thyroid cancer progress by blocking the Hedgehog signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. 2022, 13, 358. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Suo, C.; Yang, Y.; Shen, S.; Sun, L.; Li, S.-T.; Zhou, Y.; Yang, D.; Wang, Y.; Cai, Y.; et al. MYC promotes cancer progression by modulating m6A modifications to suppress target gene translation. EMBO Rep. 2021, 22, e51519. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramírez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Sorci, M.; Ianniello, Z.; Cruciani, S.; Larivera, S.; Ginistrelli, L.C.; Capuano, E.; Marchioni, M.; Fazi, F.; Fatica, A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018, 9, 796. [Google Scholar] [CrossRef]
- Ianniello, Z.; Sorci, M.; Ceci Ginistrelli, L.; Iaiza, A.; Marchioni, M.; Tito, C.; Capuano, E.; Masciarelli, S.; Ottone, T.; Attrotto, C.; et al. New insight into the catalytic -dependent and -independent roles of METTL3 in sustaining aberrant translation in chronic myeloid leukemia. Cell Death Dis. 2021, 12, 870. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; He, P.C.; Liu, W.; Wei, J.; Zhao, Z.; Gao, L.; Han, L.; et al. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 2022, 24, 205–216. [Google Scholar] [CrossRef]
- Liu, C.-X.; Chen, L.-L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.-L.; Chen, W.; Xie, J.-J.; Zhang, M.-L.; Nie, R.-C.; Liang, H.; Mei, J.; Han, K.; Xiang, Z.-C.; Wang, F.-W.; et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 2022, 21, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, C.; Du, Y.; Li, Z.; Li, M.; Hou, P.; Shen, Z.; Chu, S.; Zheng, J.; Bai, J. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer 2022, 21, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef]
- Jiang, T.; Xia, Y.; Lv, J.; Li, B.; Li, Y.; Wang, S.; Xuan, Z.; Xie, L.; Qiu, S.; He, Z.; et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 2021, 20, 66. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, K.; Xu, X.; Yang, Y.; Yan, S.; Wei, P.; Liu, H.; Xu, J.; Xiao, F.; Zhou, H.; et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018, 9, 4475. [Google Scholar] [CrossRef]
- Liang, Z.-X.; Liu, H.-S.; Xiong, L.; Yang, X.; Wang, F.-W.; Zeng, Z.-W.; He, X.-W.; Wu, X.-R.; Lan, P. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 2021, 20, 103. [Google Scholar] [CrossRef]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691.e10. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96.e11. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).