(-)-Epicatechin Is a Biased Ligand of Apelin Receptor
Abstract
:1. Introduction
2. Results
2.1. APLNR Isolation in Mouse Skeletal Muscle
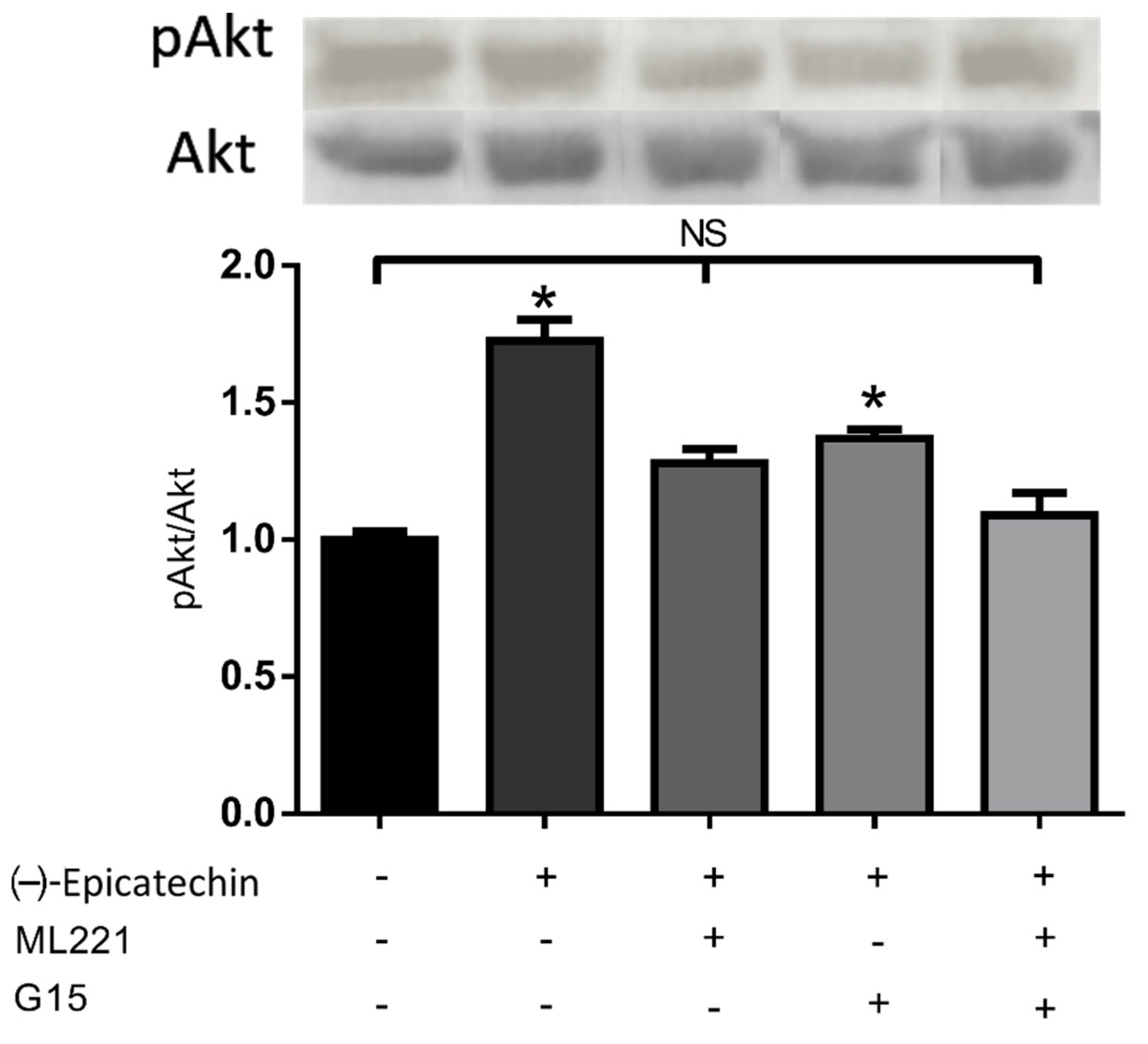
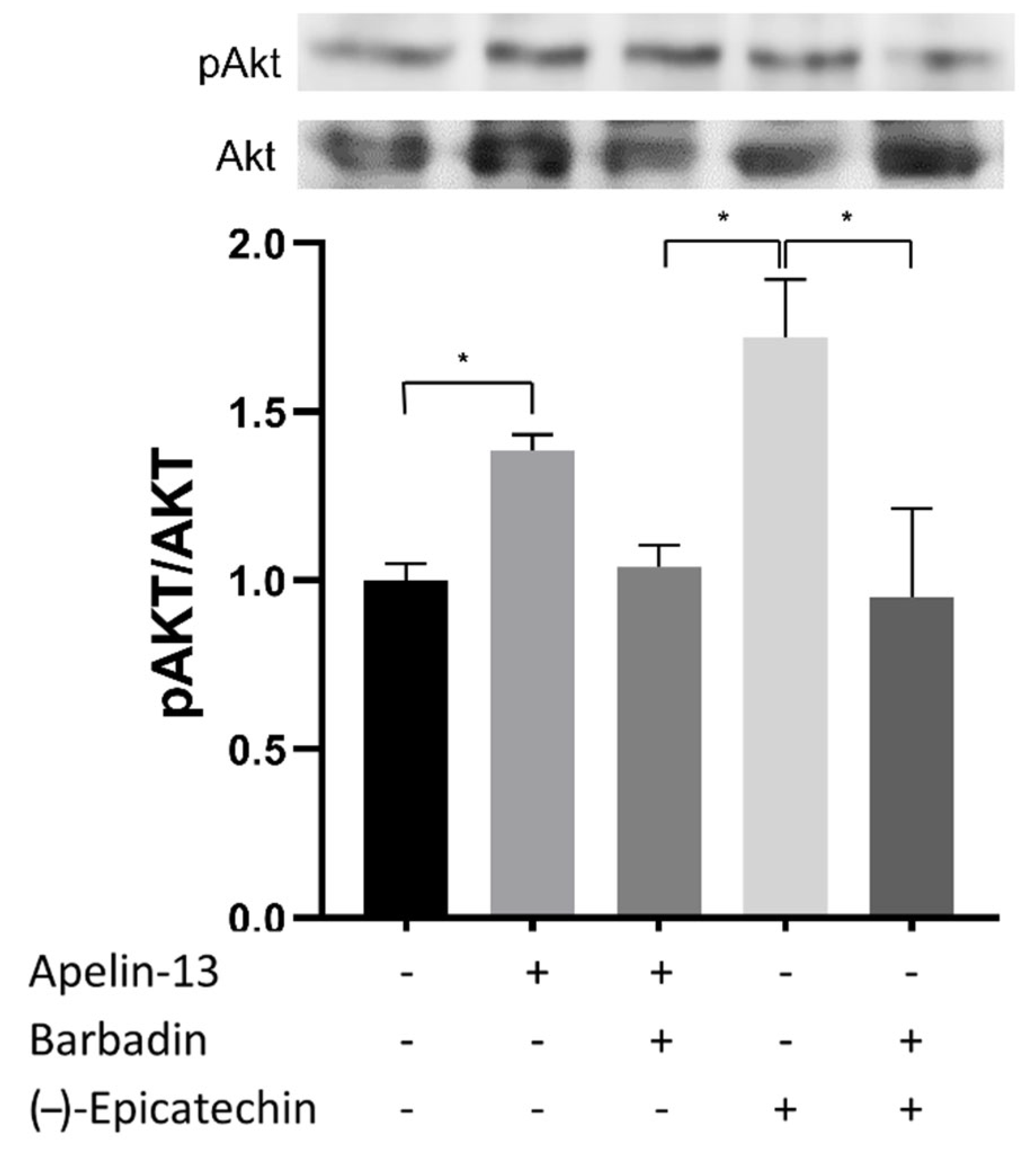
2.2. Akt Phosphorylation in C2C12 Assay
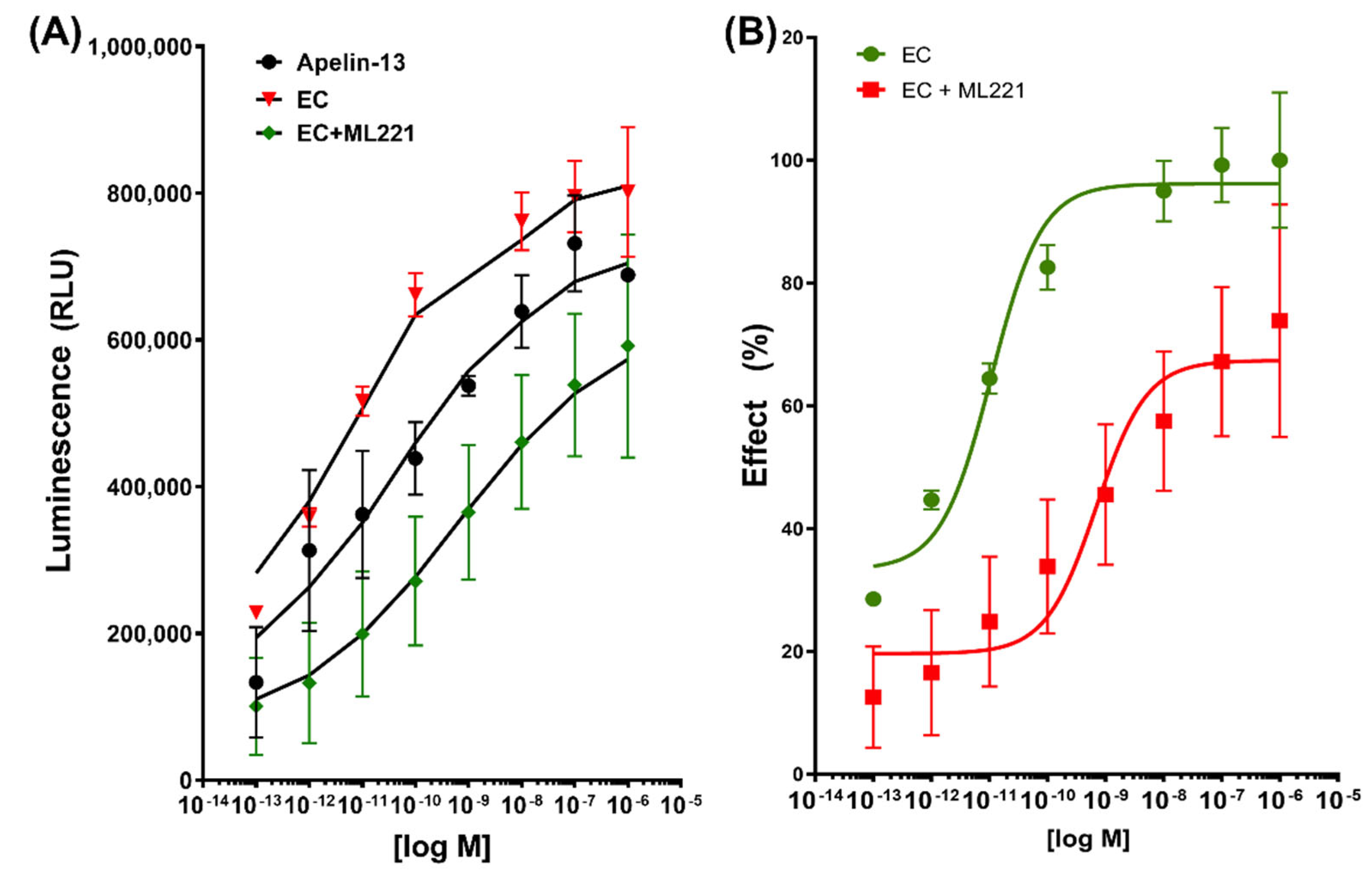
2.3. β-Arrestin Recruitment Assay
2.4. G Protein Pathways in EPI-Induced Activation of APLNR
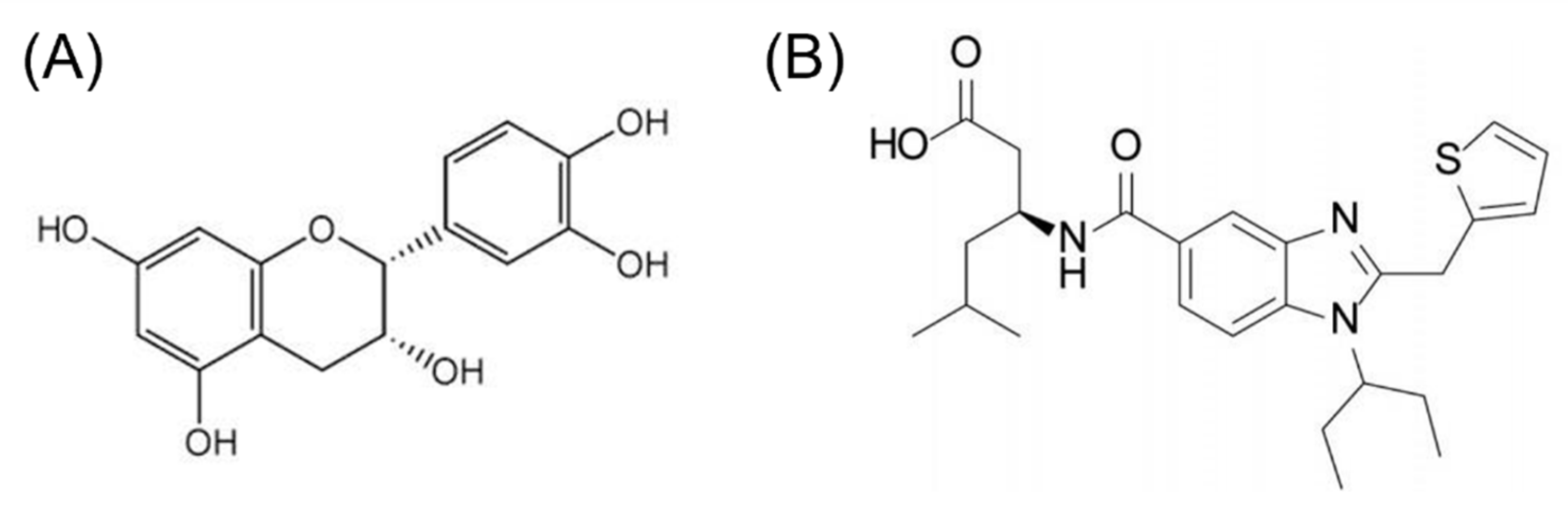
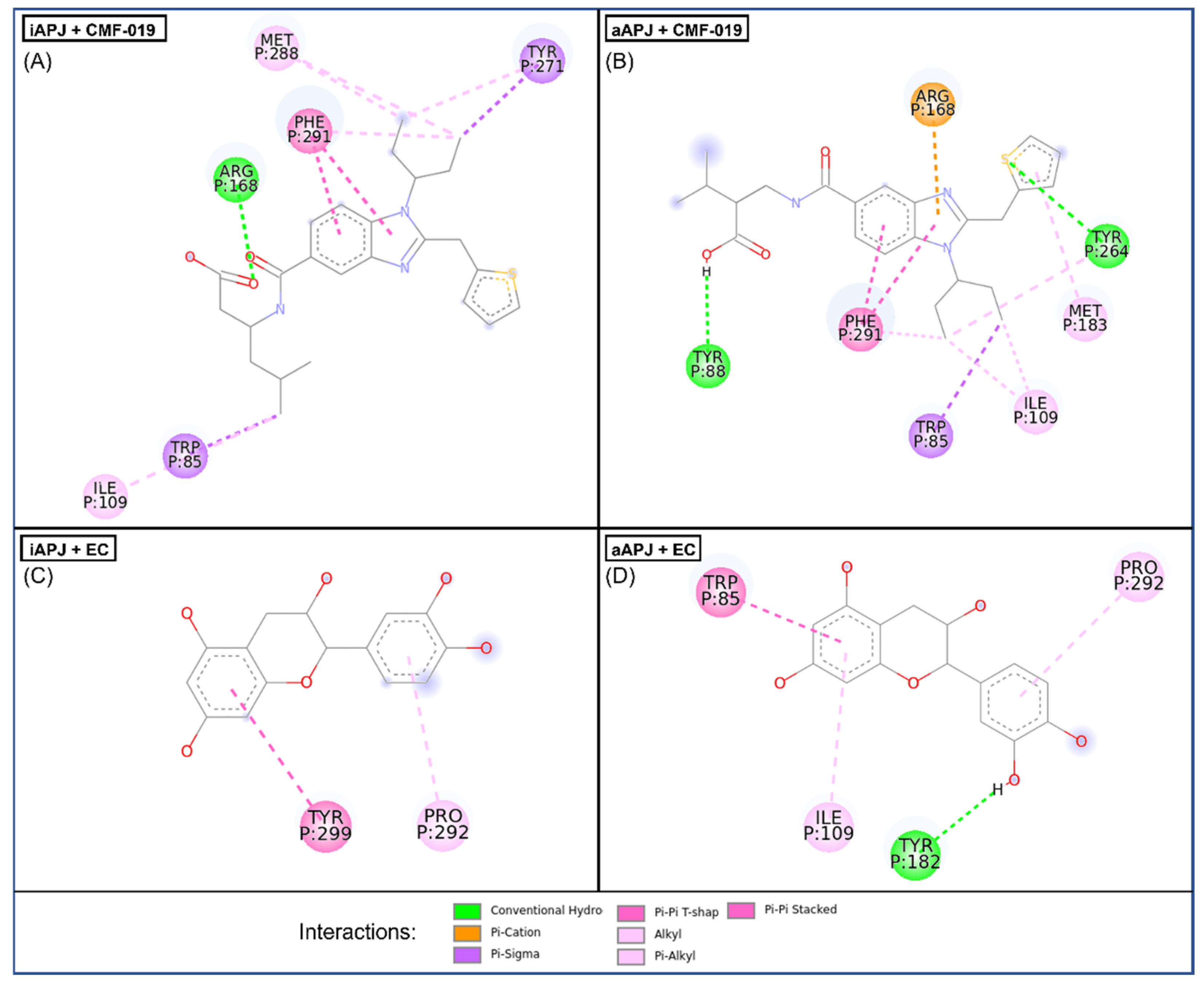
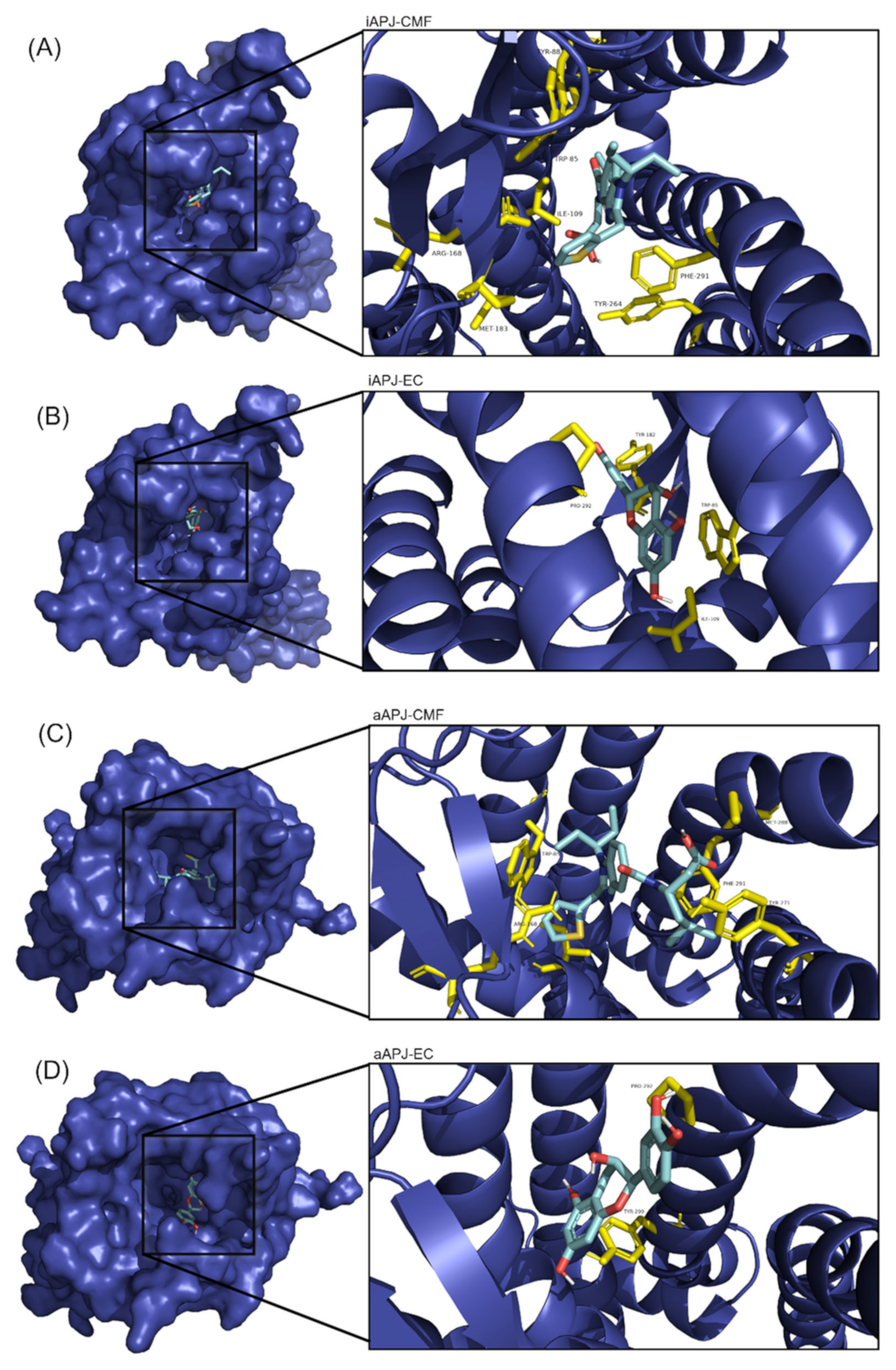
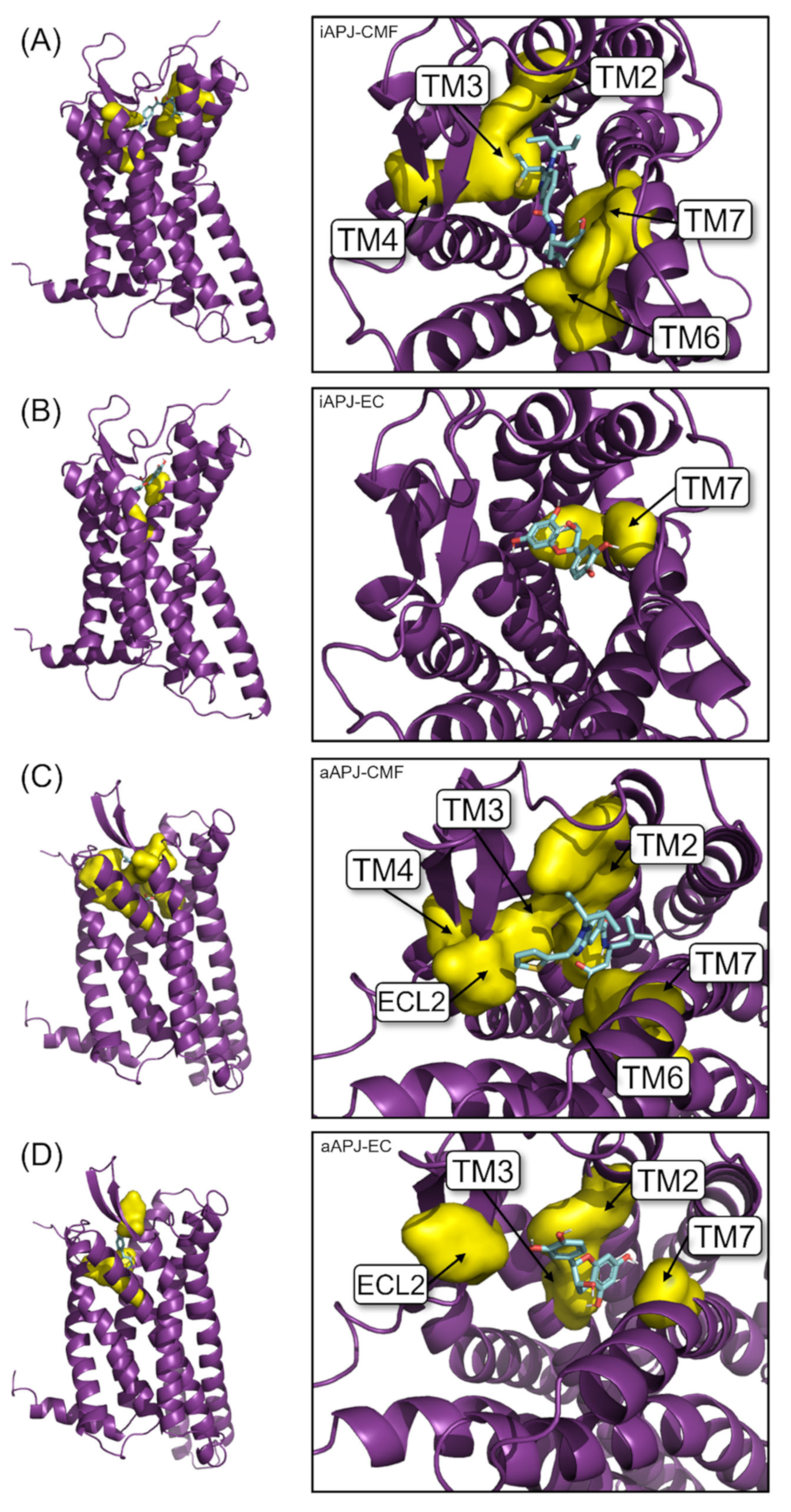
2.5. Molecular Docking
2.6. Molecular Dynamics (MD)
2.6.1. RMSD Analysis
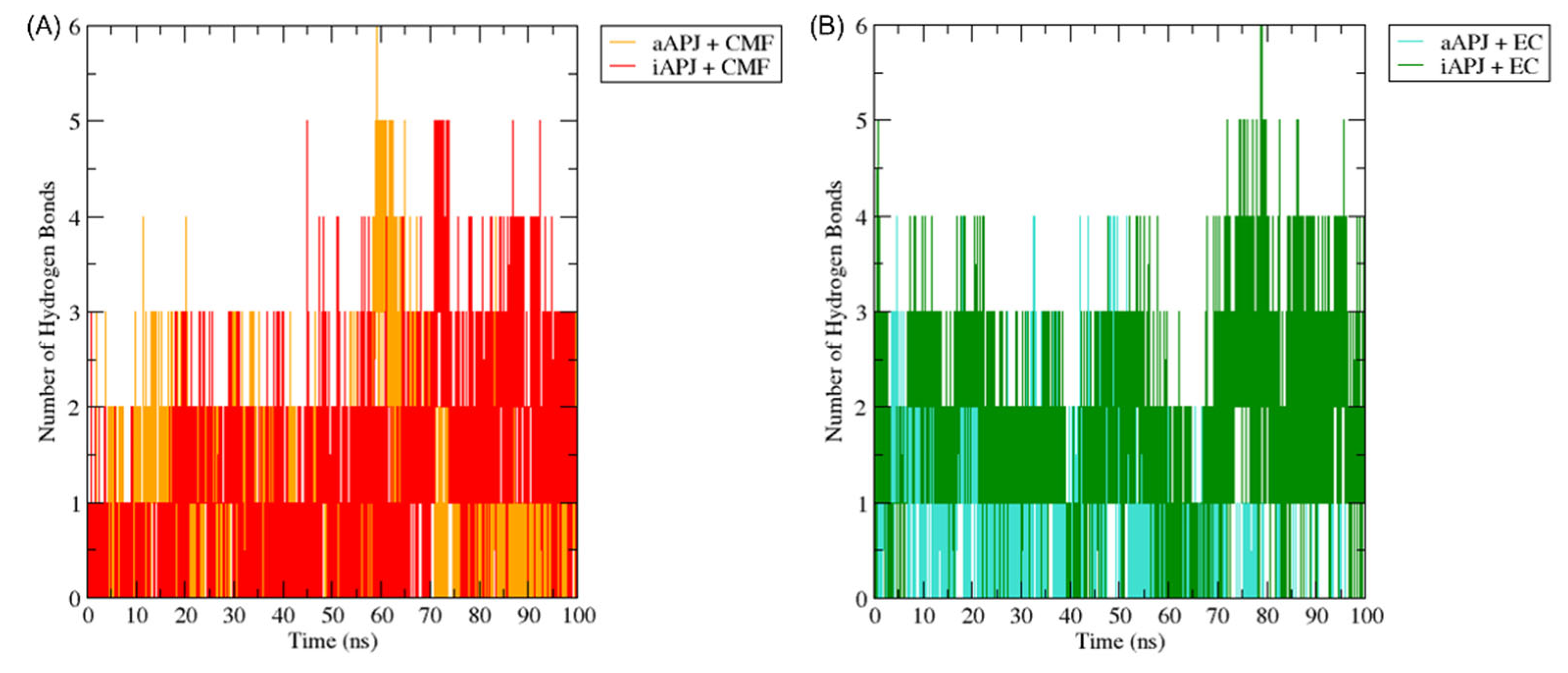
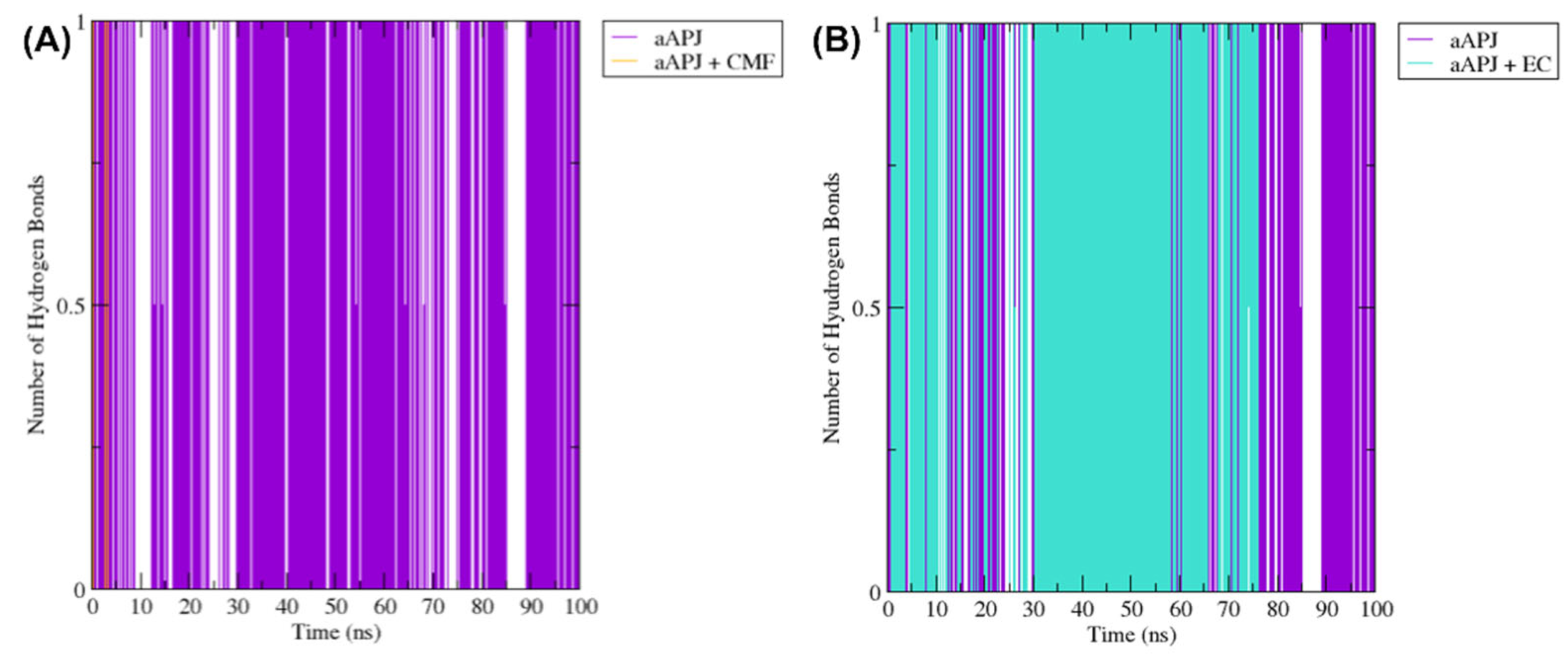
2.6.2. Hydrogen Bonds Analysis
2.7. Molecular Features Pointing to Biased Agonism (β-Arrestin Pathway)
2.7.1. Hydrogen Bond between Tyr221 and Tyr309
2.7.2. Cluster Analysis and Clash with G Protein
2.8. Binding Free Energy Calculations (MM/GBSA)
2.9. Effect of β-Arrestin Pathway Inhibition of Akt Phosphorylation
3. Discussion
4. Materials and Methods
4.1. Protein Isolation from Mouse Skeletal Muscle
4.2. Akt Phosphorylation in C2C12 Assay
4.3. β-Arrestin Recruitment Assay
4.4. G Protein Pathways in EPI-Induced Activation of APLNR
4.5. Molecular Docking
4.6. Molecular Dynamics
4.7. Binding Free Energy Calculations
5. Conclusions
6. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taub, P.R.; Ramirez-Sanchez, I.; Ciaraldi, T.P.; Gonzalez-Basurto, S.; Coral-Vazquez, R.; Perkins, G.; Hogan, M.; Maisel, A.S.; Henry, R.R.; Ceballos, G.; et al. Perturbations in skeletal muscle sarcomere structure in patients with heart failure and Type 2 diabetes: Restorative effects of (-)-epicatechinrich cocoa. Clin. Sci. 2013, 125, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Sanchez, I.; Taub, P.R.; Ciaraldi, T.P.; Nogueira, L.; Coe, T.; Perkins, G.; Hogan, M.; Maisel, A.S.; Henry, R.R.; Ceballos, G.; et al. (-)-Epicatechin rich cocoa mediated modulation of oxidative stress regulators in skeletal muscle of heart failure and type 2 diabetes patients. Int. J. Cardiol. 2013, 168, 3982–3990. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, L.; Ramirez-Sanchez, I.; Perkins, G.A.; Murphy, A.; Taub, P.R.; Ceballos, G.; Villarreal, F.J.; Hogan, M.C.; Malek, M.H. (-)-Epicatechin enhances fatigue resistance and oxidative capacity in mouse muscle. J. Physiol. 2011, 589, 4615–4631. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Salmeán, G.; Ortiz-Vilchis, P.; Vacaseydel, C.M.; Garduño-Siciliano, L.; Chamorro-Cevallos, G.; Meaney, E.; Villafaña, S.; Villarreal, F.; Ceballos, G.; Ramírez-Sánchez, I. Effects of (-)-epicatechin on a diet-induced rat model of cardiometabolic risk factors. Eur. J. Pharmacol. 2014, 728, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Ulloa, A.; Miranda-Cervantes, A.; Licea-Navarro, A.; Mansour, C.; Beltrán-Partida, E.; Donis-Maturano, L.; de la Herrán, H.C.D.; Villarreal, F.; Álvarez-Delgado, C. (-)-Epicatechin Stimulates Mitochondrial Biogenesis and Cell Growth in C2C12 Myotubes via the G-Protein Coupled Estrogen Receptor. Eur. J. Pharmacol. 2018, 822, 95. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, A.M.; Mendez-Luna, D.; Beltran-Partida, E.; Castillo, C.; Guevara, G.; Ramirez-Sanchez, I.; Correa-Basurto, J.; Ceballos, G.; Villarreal, F. The effects of (-)-epicatechin on endothelial cells involve the G protein-coupled estrogen receptor (GPER). Pharmacol. Res. 2015, 100, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Flores, M.; Portilla-Martínez, A.; Cabrera-Pérez, F.; Nájera, N.; Meaney, E.; Villarreal, F.; Pérez-Durán, J.; Ceballos, G. PXR is a target of (-)-epicatechin in skeletal muscle. Heliyon 2020, 6, e05357. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O.; Physiology, C.; Engineering, M. Molecular Dynamics Simulation for All. Neuron 2019, 99, 1129. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef]
- Dror, R.O.; Pan, A.C.; Arlow, D.H.; Borhani, D.W.; Maragakis, P.; Shan, Y.; Xu, H.; Shaw, D.E. Pathway and Mech-anism of Drug Binding to G-Protein-Coupled Receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 13118. [Google Scholar] [CrossRef]
- Gupta, G.S.; Gupta, A.; Gupta, R.K. P-Type Lectins: Cation-Dependent Mannose-6-Phosphate Receptor. Anim. Lectins Form Funct. Clin. Appl. 2012, 1, 9783709110. [Google Scholar]
- Teixeira, A.L.; Fernandes, I.A.; Vianna, L.C. GABA A Receptors Modulate Sympathetic Vasomotor Outflow and the Pressor Response to Skeletal Muscle Metaboreflex Activation in Humans. J. Physiol. 2019, 597, 4139. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Park, J.-Y.; Kim, K. NMDA receptor-mediated calcium influx plays an essential role in myoblast fusion. FEBS Lett. 2004, 578, 47–52. [Google Scholar] [CrossRef]
- Biondi, O.; Branchu, J.; Sanchez, G.; Lancelin, G.; Deforges, S.; Lopes, P.; Pariset, C.; Lécolle, S.; Côté, L.; Chanoine, C.; et al. In Vivo NMDA Receptor Activation Accelerates Motor Unit Maturation, Protects Spinal Motor Neurons, and Enhances SMN2 Gene Expression in Severe Spinal Muscular Atrophy Mice. J. Neurosci. 2010, 30, 11288. [Google Scholar] [CrossRef] [PubMed]
- Biondi, O.; Grondard, C.; Lecolle, S.; Deforges, S.; Pariset, C.; Lopes, P.; Cifuentes-Diaz, C.; Li, H.; Della Gaspera, B.; Chanoine, C.; et al. Exercise-Induced Activation of NMDA Receptor Promotes Motor Unit Development and Survival in a Type 2 Spinal Muscular Atrophy Model Mouse. J. Neurosci. 2008, 28, 953–962. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, A.M.; Lolait, S.J.; Harris, L.E.; Pope, G.R. The Apelin Receptor APJ: Journey from an Orphan to a Multi-faceted Regulator of Homeostasis. J. Endocrinol. 2013, 219, R13–R35. [Google Scholar] [CrossRef] [PubMed]
- Vinel, C.; Lukjanenko, L.; Batut, A.; Deleruyelle, S.; Pradère, J.-P.; Le Gonidec, S.; Dortignac, A.; Geoffre, N.; Pereira, O.; Karaz, S.; et al. The exerkine apelin reverses age-associated sarcopenia. Nat. Med. 2018, 24, 1360–1371. [Google Scholar] [CrossRef]
- Son, J.S.; Chae, S.A.; Wang, H.; Chen, Y.; Iniguez, A.B.; de Avila, J.M.; Jiang, Z.; Zhu, M.J.; Du, M. Maternal Inac-tivity Programs Skeletal Muscle Dysfunction in Offspring Mice by Attenuating Apelin Signaling and Mitochondrial Bio-genesis. Cell Rep. 2020, 33, 108461. [Google Scholar] [CrossRef]
- Frier, B.C.; Williams, D.B.; Wright, D.C. The effects of apelin treatment on skeletal muscle mitochondrial content. Am. J. Physiol. Integr. Comp. Physiol. 2009, 297, R1761–R1768. [Google Scholar] [CrossRef]
- Son, J.S.; Chae, S.A.; Zhao, L.; Wang, H.; de Avila, J.M.; Zhu, M.J.; Jiang, Z.; Du, M. Maternal Exercise Intergenera-tionally Drives Muscle-Based Thermogenesis via Activation of Apelin-AMPK Signaling. EBioMedicine 2022, 76, 103842. [Google Scholar] [CrossRef] [PubMed]
- Potts, J.T.; Paton, J.F.R.; Mitchell, J.H.; Garry, M.G.; Kline, G.; Anguelov, P.T.; Lee, S.M. Contraction-Sensitive Skeletal Muscle Afferents Inhibit Arterial Baroreceptor Signalling in the Nucleus of the Solitary Tract: Role of Intrinsic GABA In-terneurons. Neuroscience 2003, 119, 201. [Google Scholar] [CrossRef]
- Banks, G.B.; Kanjhan, R.; Wiese, S.; Kneussel, M.; Wong, L.M.; O’Sullivan, G.; Sendtner, M.; Bellingham, M.C.; Betz, H.; Noakes, P.G. Glycinergic and GABAergic Synaptic Activity Differentially Regulate Motoneuron Survival and Skeletal Muscle Innervation. J. Neurosci. 2005, 25, 1249–1259. [Google Scholar] [CrossRef]
- Stamler, J.S.; Meissner, G. Physiology of Nitric Oxide in Skeletal Muscle. Physiol. Rev. 2001, 81, 209–237. [Google Scholar] [CrossRef]
- Castan-Laurell, I.; Dray, C.; Knauf, C.; Kunduzova, O.; Valet, P. Apelin, a promising target for type 2 diabetes treatment? Trends Endocrinol. Metab. 2012, 23, 234–241. [Google Scholar] [CrossRef]
- Hüttemann, M.; Lee, I.; Perkins, G.A.; Britton, S.L.; Koch, L.G.; Malek, M.H. (-)-Epicatechin is associated with increased angiogenic and mitochondrial signalling in the hindlimb of rats selectively bred for innate low running capacity. Clin. Sci. 2013, 124, 663–674. [Google Scholar] [CrossRef]
- Lee, I.; Hüttemann, M.; Kruger, A.; Bollig-Fischer, A.; Malek, M.H. (-)-Epicatechin combined with 8 weeks of treadmill exercise is associated with increased angiogenic and mitochondrial signaling in mice. Front. Pharmacol. 2015, 6, 43. [Google Scholar] [CrossRef]
- Attané, C.; Foussal, C.; Le Gonidec, S.; Benani, A.; Daviaud, D.; Wanecq, E.; Guzmán-Ruiz, R.; Dray, C.; Bezaire, V.; Rancoule, C.; et al. Apelin Treatment Increases Complete Fatty Acid Oxidation, Mitochondrial Oxidative Capacity, and Biogenesis in Muscle of Insulin-Resistant Mice. Diabetes 2012, 61, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, A.M.; Nogueira, L.; Rodriguez, A.; Barboza, J.; Hogan, M.C.; Ceballos, G.; Villarreal, F.; Ramirez-Sanchez, I. Recovery of Indicators of Mitochondrial Biogenesis, Oxidative Stress, and Aging with (-)-Epicatechin in Senile Mice. J. Gerontol. Ser. A 2015, 70, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, W.; Zhang, P.; Chen, K.; Zhao, L.; Li, J.; Wei, M.; Liu, M. Apelin-13 stimulates angiogenesis by promoting cross-talk between AMP-activated protein kinase and Akt signaling in myocardial microvascular endothelial cells. Mol. Med. Rep. 2014, 9, 1590–1596. [Google Scholar] [CrossRef]
- Ramirez-Sanchez, I.; Maya, L.; Ceballos, G.; Villarreal, F. (-)-Epicatechin Activation of Endothelial Cell Endothelial Nitric Oxide Synthase, Nitric Oxide, and Related Signaling Pathways. Hypertension 2010, 55, 1398. [Google Scholar] [CrossRef]
- Ramirez-Sanchez, I.; Ceballos, G.; Moreno-Ulloa, A.; Ciaraldi, T.P.; Henry, R.R.; Villarreal, F. Evaluation and Com-parison of Epicatechin Epimer Effects on Skeletal Muscle Structure, Function, and Regulators of Metabolism. Diabetes 2014, 63, A478. [Google Scholar]
- Yue, P.; Jin, H.; Aillaud, M.; Deng, A.C.; Azuma, J.; Asagami, T.; Kundu, R.K.; Reaven, G.M.; Quertermous, T.; Tsao, P.S. Apelin is necessary for the maintenance of insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E59–E67. [Google Scholar] [CrossRef]
- Lee, S.J.; Leem, Y.-E.; Go, G.-Y.; Choi, Y.; Song, Y.J.; Kim, I.; Kim, D.Y.; Kim, Y.K.; Seo, D.-W.; Kang, J.-S.; et al. Epicatechin Elicits MyoD-Dependent Myoblast Differentiation and Myogenic Conversion of Fibroblasts. PLoS ONE 2017, 12, e0175271. [Google Scholar] [CrossRef] [PubMed]
- Taub, P.R.; Ramirez-Sanchez, I.; Ciaraldi, T.P.; Perkins, G.; Murphy, A.N.; Naviaux, R.; Hogan, M.; Maisel, A.S.; Henry, R.R.; Ceballos, G.; et al. Alterations in Skeletal Muscle Indicators of Mitochondrial Structure and Biogenesis in Patients with Type 2 Diabetes and Heart Failure: Effects of Epicatechin Rich Cocoa. Clin. Transl. Sci. 2012, 5, 43–47. [Google Scholar] [CrossRef] [PubMed]
- los Santos, S.D.; García-Pérez, V.; Hernández-Reséndiz, S.; Palma-Flores, C.; González-Gutiérrez, C.J.; Zazueta, C.; Canto, P.; Coral-Vázquez, R.M. (-)-Epicatechin Induces Physiological Cardiac Growth by Activation Of The Pi3k/Akt Pathway In Mice. Mol. Nutr. Food Res. 2016, 61, 1600343. [Google Scholar] [CrossRef]
- Matsumura, Y.; Nakagawa, Y.; Mikome, K.; Yamamoto, H.; Osakabe, N. Enhancement of Energy Expenditure following a Single Oral Dose of Flavan-3-Ols Associated with an Increase in Catecholamine Secretion. PLoS ONE 2014, 9, e112180. [Google Scholar] [CrossRef]
- Black, J.W.; Leff, P. Operational Models of Pharmacological Agonism. Proc. R. Soc. London Ser. B. Biol. Sci. 1983, 220, 141. [Google Scholar]
- Maloney, P.R.; Khan, P.; Hedrick, M.; Gosalia, P.; Milewski, M.; Li, L.; Roth, G.P.; Sergienko, E.; Suyama, E.; Sugarman, E.; et al. Discovery of 4-Oxo-6-((Pyrimidin-2-Ylthio)Methyl)-4H-Pyran-3-Yl 4-Nitrobenzoate (ML221) as a Functional Antagonist of the Apelin (APJ) Receptor. Bioorg. Med. Chem. Lett. 2012, 22, 21. [Google Scholar] [CrossRef]
- BIOVIA DS. Discovery Studio; 2017R2; BIOVIA DS: San Diego, CA, USA, 2022. [Google Scholar]
- Derewenda, Z.S.; Lee, L.; Derewenda, U. The Occurrence of C-H ⋯ O Hydrogen Bonds in Proteins. J. Mol. Biol. 1995, 252, 248. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; He, X.; Yang, Z.; Chai, Z.; Zhou, S.; Wang, J.; Rehman, A.U.; Ni, D.; Pu, J.; Sun, J.; et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat. Commun. 2021, 12, 4721. [Google Scholar] [CrossRef] [PubMed]
- Sticke, D.F.; Presta, L.G.; Dill, K.A.; Rose, G.D. Hydrogen bonding in globular proteins. J. Mol. Biol. 1992, 226, 1143–1159. [Google Scholar] [CrossRef]
- Humphrey, K.; Dalke, W.; Schulten, A. VMD—Visual Molecular Dynamics. J. Molec. Graph. 1996, 1, 33. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Salahudeen, M.S.; Nishtala, P.S. An overview of pharmacodynamic modelling, ligand-binding approach and its application in clinical practice. Saudi Pharm. J. 2017, 25, 165–175. [Google Scholar] [CrossRef]
- Gesztelyi, R.; Zsuga, J.; Kemeny-Beke, A.; Varga, B.; Juhasz, B.; Tosaki, A. The Hill Equation and the Origin of Quanti-tative Pharmacology. Arch. Hist. Exact Sci. 2012, 66, 427. [Google Scholar] [CrossRef]
- Read, C.; Fitzpatrick, C.M.; Yang, P.; Kuc, R.E.; Maguire, J.J.; Glen, R.C.; Foster, R.E.; Davenport, A.P. Cardiac Action of the First G Protein Biased Small Molecule Apelin Agonist. Biochem. Pharmacol. 2016, 116, 63. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yue, Y.; Ma, Y.; Zhang, Q.; Zhou, Q.; Song, Y.; Shen, Y.; Li, X.; Ma, X.; Li, C. Structural Basis for Apelin Control of the Human Apelin Receptor. Structure 2017, 25, 858. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Dong, C.; Wu, G.; Lambert, N.A. Inactive-State Preassembly of Gq-Coupled Receptors and G q Heterotrimers. Nat. Chem. Biol. 2011, 7, 740. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.; Adams, C.E.; Liu, H.; Zhang, C.; Levy, F.O.; Andressen, K.W.; Lambert, N.A. An inactive receptor-G protein complex maintains the dynamic range of agonist-induced signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 30755–30762. [Google Scholar] [CrossRef] [PubMed]
- Ashokan, A.; Harisankar, H.S.; Kameswaran, M.; Aradhyam, G.K. Critical APJ receptor residues in extracellular domains that influence effector selectivity. FEBS J. 2021, 288, 6543–6562. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef] [PubMed]
- Read, C.; Nyimanu, D.; Yang, P.; Kuc, R.E.; Williams, T.L.; Fitzpatrick, C.M.; Foster, R.; Glen, R.C.; Maguire, J.J.; Davenport, A.P. The G Protein Biased Small Molecule Apelin Agonist CMF-019 Is Disease Modifying in Endothelial Cell Apoptosis In Vitro and Induces Vasodilatation Without Desensitisation In Vivo. Front. Pharmacol. 2021, 11, 588669. [Google Scholar] [CrossRef] [PubMed]
- Suomivuori, C.-M.; Latorraca, N.R.; Wingler, L.M.; Eismann, S.; King, M.C.; Kleinhenz, A.L.W.; Skiba, M.A.; Staus, D.P.; Kruse, A.C.; Lefkowitz, R.J.; et al. Molecular mechanism of biased signaling in a prototypical G protein–coupled receptor. Science 2020, 367, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Yamano, Y.; Sano, T.; Nakagomi, Y.; Wada, M.; Inagami, T. Role of the Conserved DRY Motif on G Protein Activation of Rat Angiotensin II Receptor Type 1A. Biochem. Biophys. Res. Commun. 2002, 292, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and Auto-DockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Flores, M.; Portilla-Martínez, A.; González-Ruiz, C.; Villarreal, F.; Meaney, E.; Nájera, N.; Ceballos, G. Catechins Modulate the Activity of Mu Opioid Receptor (ΜOR): An In Silico Approach. Inform. Med. Unlocked 2020, 20, 100431. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Van Der Spoel, D.; Van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314. [Google Scholar] [CrossRef] [PubMed]


| Protein | Function in Skeletal Muscle |
|---|---|
| Apelin receptor | Apelin signaling through APJ enhances muscle function by triggering mitochondriogenesis, autophagy, and anti-inflammatory pathways in myofibers as well as enhancing the regenerative capacity by targeting muscle stem cells. It mirrors the effects of maternal exercise on mitochondrial biogenesis and fetal muscle development. It also promotes non-shivering thermogenesis gene expression in fetal and offspring muscle impaired due to maternal obesity, which intergenerationally protects offspring from diet-induced obesity and metabolic disorders [17,18,19,20] |
| Cation-dependent mannose-6-phosphate receptor | Not reported |
| Gamma-aminobutyric acid receptor subunit beta-2 | GABAergic receptors contribute to the cardiovascular responses during the activation of group III/IV skeletal muscle afferent fibers [21]. Modulates sympathetic vasomotor outflow and the pressor responses to activation of metabolically sensitive skeletal muscle afferent fibers, and it also assists the development of motor circuit activity, motoneuron survival, and muscle innervation [12,22]. |
| Glutamate receptor ionotropic, NMDA 2A, and glutamate receptor ionotropic, NMDA 2D | NMDA receptor is found in muscle differentiation, promotes Ca2+ influx in myoblasts, thereby triggering myoblast fusion. It mediates the “nerve–muscle” system, due to the enhancement of NO synthesis under the influence of a glutamate. It promotes acceleration of motor neuron development and is directly involved in skeletal muscle maturation. Exercise triggers NMDA signaling in motor neurons. May accelerate the development of the motor units [13,14,15,23]. |
| Apelin | (-)-Epicatechin | ||
|---|---|---|---|
| Experiment Models | Pathway or Molecule | Experiment Models | Pathway or Molecule |
| Developing cardiovascular in mouse, xenopus, and zebrafish embryos | ↑MEF2 [24] | Hindlimb muscles from exercised C57BL/6 mice. | ↑MEF2 [25] |
| Chow-fed rat triceps | ↑PGC-1β, ↑activity of cytochrome C oxidase [24] | Old and trained C57BL/6N mice | ↑PGC-1β, ↑activity of cytochrome C oxidase [26] |
| In HFD mice | ↑PGC-1α, TFAM [27] | C57BL/6 mice quadriceps | ↑PGC-1α, TFAM [28] |
| Myocardial microvascular endothelial cells (MMVEC) | ↑eNOS [29] | Human coronary artery endothelial cells (HCAEC) | ↑eNOS [30] |
| In HFD mice | ↑PGC1α, AMPK [27] | Human quadriceps | ↑PGC1α, AMPK [31]. |
| C2C12 skeletal myotubes | ↑Akt phosphorylation [32] | C2C12 skeletal myotubes | ↑Akt phosphorylation [33] |
| Obese and insulin-resistant mice | ↑Mitochondrial biogenesis, PGC1α, NRF-1, TFAM [24] | Human muscle biopsies from quadriceps femoris | ↑Mitochondrial biogenesis, PGC1α, NRF-1, TFAM [34] |
| Muscle cells from young and aged human donors | ↑mTOR, and P70S6K, Akt phosphorylation [17] | Male CD-1 mice. | ↑mTOR, and P70S6K, Akt phosphorylation [35] |
| Male Wistar rats | ↑Activity of Citrate Synthase [19] | C2C12 skeletal myotubes | ↑Activity of Citrate Synthase [5] |
| Different rodent models | ↑UCP3 [24] | Male ICR mice | ↑UCP3 [36] |
| Ligand | IC50 | Tau (τ) | KA | Bmax | Kd | h |
|---|---|---|---|---|---|---|
| Apelin | - | - | - | 801,286 | 2.508 × 10−11 M | 0.2202 |
| EC | - | 21.8 | 20.8 | 810,100 | 1.755 × 10−12 M | 0.3432 |
| EC + ML221 | 6.93 × 10−11 M | 1.53 | 0.53 | 733,981 | 1.012 × 10−9 M | 0.2134 |
| Protein | Ligand | ∆G | Residue Interaction |
|---|---|---|---|
| Active APLNR | EC | −8.2 ± 0.005 | Trp85, Ile109, Tyr182, Pro292 |
| CMF-019 | −9 ± 0.276 | Trp85, Tyr88, Ile109, Arg168, Met183, Tyr264, Phe291 | |
| Inactive APLNR | EC | −7.3 ± 0.028 | Pro292, Tyr299 |
| CMF-019 | −8.1 ± 0.315 | Trp85, Ile109, Arg168, Tyr271, Met288, Phe291 |
| EC | CMF | |||
|---|---|---|---|---|
| Trajectory | Tyr309O-HTyr221 | Tyr221O-HTyr309 | Tyr309O-HTyr221 | Tyr221O-HTyr309 |
| 10 ns | 3.04 | 2.22 | 4.15 | 3.96 |
| 20 ns | 3.41 | 2.31 | 3.85 | 3.57 |
| 30 ns | 3.08 | 2.11 | 3.42 | 3.38 |
| 40 ns | 4.25 | 3.41 | 4.66 | 3.99 |
| 50 ns | 2.7 | 2.94 | 4.85 | 4.06 |
| 60 ns | 2.18 | 2.8 | 5.52 | 4.68 |
| 70 ns | 2.15 | 3.07 | 10.31 | 11.13 |
| 80 ns | 3.49 | 4.34 | 6.42 | 5.72 |
| 90 ns | 3.3 | 3.29 | 9.31 | 9.95 |
| 100 ns | 2.12 | 2.92 | 8.67 | 9.82 |
| Protein | Ligand | Calculated Free Energy of Decomposition (kcal/mol) | ||||
|---|---|---|---|---|---|---|
| ∆G Binding | ∆E vdW | ∆E Elect | ∆G Gas | ∆G Solv | ||
| Active APLNR | EC | −11.61 ± 0.71 | −19.41 ± 0.75 | −9.35 ± 1.59 | −28.76 ± 1.92 | 17.15 ± 1.50 |
| CMF | −32.25 ± 0.83 | −46.50 ± 0.85 | −27.89 ± 1.84 | −74.38 ± 2.24 | 42.14 ± 1.81 | |
| Inactive APLNR | EC | −22.59 ± 0.76 | −31.47 ± 0.57 | −39.02 ± 2.55 | −70.49 ± 2.55 | 47.15 ± 2.06 |
| CMF | −31.50 ± 1.11 | −43.08 ± 0.64 | −13.69 ± 2.28 | −56.77 ± 244 | 25.27 ± 1.56 | |
| Receptor | Ligand | Residue | ∆G Binding | Std. Err. |
|---|---|---|---|---|
| Active APLNR | EC | ILE 8 | −0.0746 | 0.0199 |
| TYR 12 | −0.0143 | 0.0091 | ||
| PHE 55 | −0.0029 | 0.0043 | ||
| LEU 59 | −0.0143 | 0.0050 | ||
| TRP 62 | −0.2029 | 0.0805 | ||
| TYR 65 | −0.3447 | 0.1151 | ||
| THE 66 | −0.2534 | 0.0692 | ||
| ASP 69 | −0.0297 | 0.0514 | ||
| TYR 70 | −0.3177 | 0.0821 | ||
| SER 83 | −0.0045 | 0.0053 | ||
| ILE 86 | −0.0318 | 0.0213 | ||
| PHE 87 | −0.0024 | 0.0034 | ||
| ARG 145 | −0.0685 | 0.0400 | ||
| CYS 158 | −0.3188 | 0.0918 | ||
| TYR 159 | −0.2368 | 0.0779 | ||
| MET 262 | −0.6250 | 0.1350 | ||
| PHE 265 | −0.2304 | 0.0678 | ||
| PRO 266 | −0.1681 | 0.0603 | ||
| THR 269 | −0.0327 | 0.0183 | ||
| TYR 273 | 0.0056 | 0.0020 | ||
| EC | −4.6858 | 0.3731 | ||
| CMF | ILE 8 | −0.0144 | 0.0037 | |
| TYR 12 | 0.0815 | 0.0190 | ||
| LEU 59 | −0.0177 | 0.0148 | ||
| TRP 62 | −0.6329 | 0.1180 | ||
| TYR 65 | −0.2503 | 0.0843 | ||
| THR 66 | −0.0272 | 0.0273 | ||
| ASP 69 | 0.0094 | 0.0062 | ||
| TYR 70 | −0.2867 | 0.0614 | ||
| ILE 86 | −0.4727 | 0.0809 | ||
| PHE 87 | −0.2232 | 0.0742 | ||
| ARG 145 | −0.7602 | 0.1734 | ||
| CYS 158 | −0.3870 | 0.0974 | ||
| TYR 159 | −0.6344 | 0.1119 | ||
| MET 160 | −0.1602 | 0.0540 | ||
| TYR 240 | −0.3717 | 0.0971 | ||
| LYS 243 | −1.4040 | 0.1433 | ||
| TYR 246 | −1.7127 | 0.1099 | ||
| LEU 261 | −0.8716 | 0.0898 | ||
| MET 262 | −1.5429 | 0.0996 | ||
| ASN 263 | 0.0067 | 0.0056 | ||
| PHE 265 | −2.0900 | 0.1014 | ||
| PRO 266 | 0.0297 | 0.0364 | ||
| THR 269 | −0.1933 | 0.0418 | ||
| CMF | −17.9118 | 0.4758 | ||
| Inactive APLNR | EC | ILE 14 | −0.0112 | 0.0054 |
| TYR 18 | 0.0087 | 0.0078 | ||
| TRP 68 | −0.0609 | 0.0573 | ||
| TYR 71 | −0.0291 | 0.0292 | ||
| THR 72 | −0.0155 | 0.0151 | ||
| TYR 76 | −0.1145 | 0.0398 | ||
| SER 89 | −0.0038 | 0.0017 | ||
| ILE 92 | −0.0270 | 0.0132 | ||
| PHE 93 | −0.0593 | 0.0196 | ||
| ARG 151 | 0.0188 | 0.0218 | ||
| LEU 270 | −0.2012 | 0.0304 | ||
| MET 271 | −2.6193 | 0.1652 | ||
| ASN 272 | −0.0825 | 0.0170 | ||
| PHE 274 | −1.3984 | 0.0695 | ||
| PRO 275 | −0.3658 | 0.0470 | ||
| THR 278 | −0.0826 | 0.0428 | ||
| TYR 282 | −0.0362 | 0.0480 | ||
| EC 316 | −10.7886 | 0.4734 | ||
| CMF | ILE 14 | −0.0770 | 0.0092 | |
| TYR 18 | 0.1837 | 0.1171 | ||
| TRP 68 | −2.1810 | 0.2573 | ||
| TYR 71 | −1.4010 | 0.3388 | ||
| THR 72 | −0.1017 | 0.0404 | ||
| TYR 76 | −1.1871 | 0.1812 | ||
| SER 88 | −0.1652 | 0.0353 | ||
| SER 89 | −0.4662 | 0.1090 | ||
| TYR 90 | −0.0075 | 0.0091 | ||
| ILE 92 | −0.9967 | 0.1781 | ||
| PHE 93 | −0.2054 | 0.0779 | ||
| VAL 147 | −0.0611 | 0.0122 | ||
| ARG 151 | −0.8365 | 0.4741 | ||
| CYS 164 | −0.6322 | 0.1208 | ||
| TYR 165 | −0.2165 | 0.0673 | ||
| MET 166 | −0.0406 | 0.0267 | ||
| TYR 168 | 0.0008 | 0.0158 | ||
| GLU 181 | 0.4742 | 0.0621 | ||
| GLY 185 | −0.0146 | 0.0146 | ||
| TYR 247 | −0.0855 | 0.0354 | ||
| VAL 250 | −0.0690 | 0.0356 | ||
| LYS 251 | 0.0795 | 0.1024 | ||
| TYR 254 | −0.3275 | 0.1917 | ||
| MET 255 | −0.0609 | 0.0648 | ||
| LEU 270 | −0.2245 | 0.1747 | ||
| MET 271 | −0.6861 | 0.1217 | ||
| ILE 273 | −0.0747 | 0.0065 | ||
| PHE 274 | −1.3556 | 0.2144 | ||
| PRO 275 | −1.2212 | 0.0936 | ||
| THR 278 | −0.9633 | 0.1438 | ||
| TYR 282 | −0.0507 | 0.1013 | ||
| CMF | −16.2988 | 0.8901 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Portilla-Martínez, A.; Ortiz-Flores, M.Á.; Meaney, E.; Villarreal, F.; Nájera, N.; Ceballos, G. (-)-Epicatechin Is a Biased Ligand of Apelin Receptor. Int. J. Mol. Sci. 2022, 23, 8962. https://doi.org/10.3390/ijms23168962
Portilla-Martínez A, Ortiz-Flores MÁ, Meaney E, Villarreal F, Nájera N, Ceballos G. (-)-Epicatechin Is a Biased Ligand of Apelin Receptor. International Journal of Molecular Sciences. 2022; 23(16):8962. https://doi.org/10.3390/ijms23168962
Chicago/Turabian StylePortilla-Martínez, Andrés, Miguel Ángel Ortiz-Flores, Eduardo Meaney, Francisco Villarreal, Nayelli Nájera, and Guillermo Ceballos. 2022. "(-)-Epicatechin Is a Biased Ligand of Apelin Receptor" International Journal of Molecular Sciences 23, no. 16: 8962. https://doi.org/10.3390/ijms23168962
APA StylePortilla-Martínez, A., Ortiz-Flores, M. Á., Meaney, E., Villarreal, F., Nájera, N., & Ceballos, G. (2022). (-)-Epicatechin Is a Biased Ligand of Apelin Receptor. International Journal of Molecular Sciences, 23(16), 8962. https://doi.org/10.3390/ijms23168962






