Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase
Abstract
:1. Introduction
2. Results
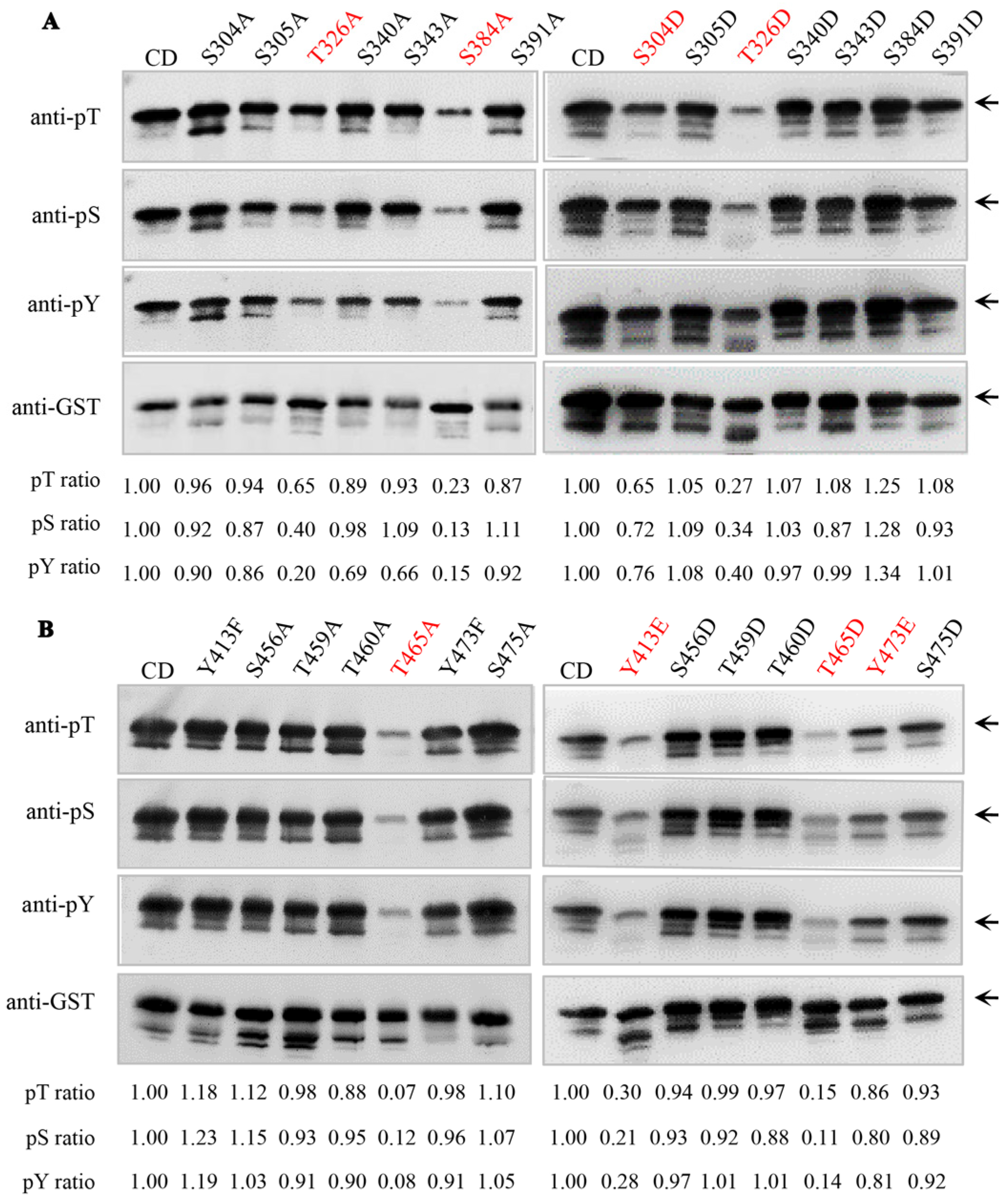
2.1. Identification of Key Autophosphorylation Residues of Senescence-Associated Receptor-like Kinase AtSARK
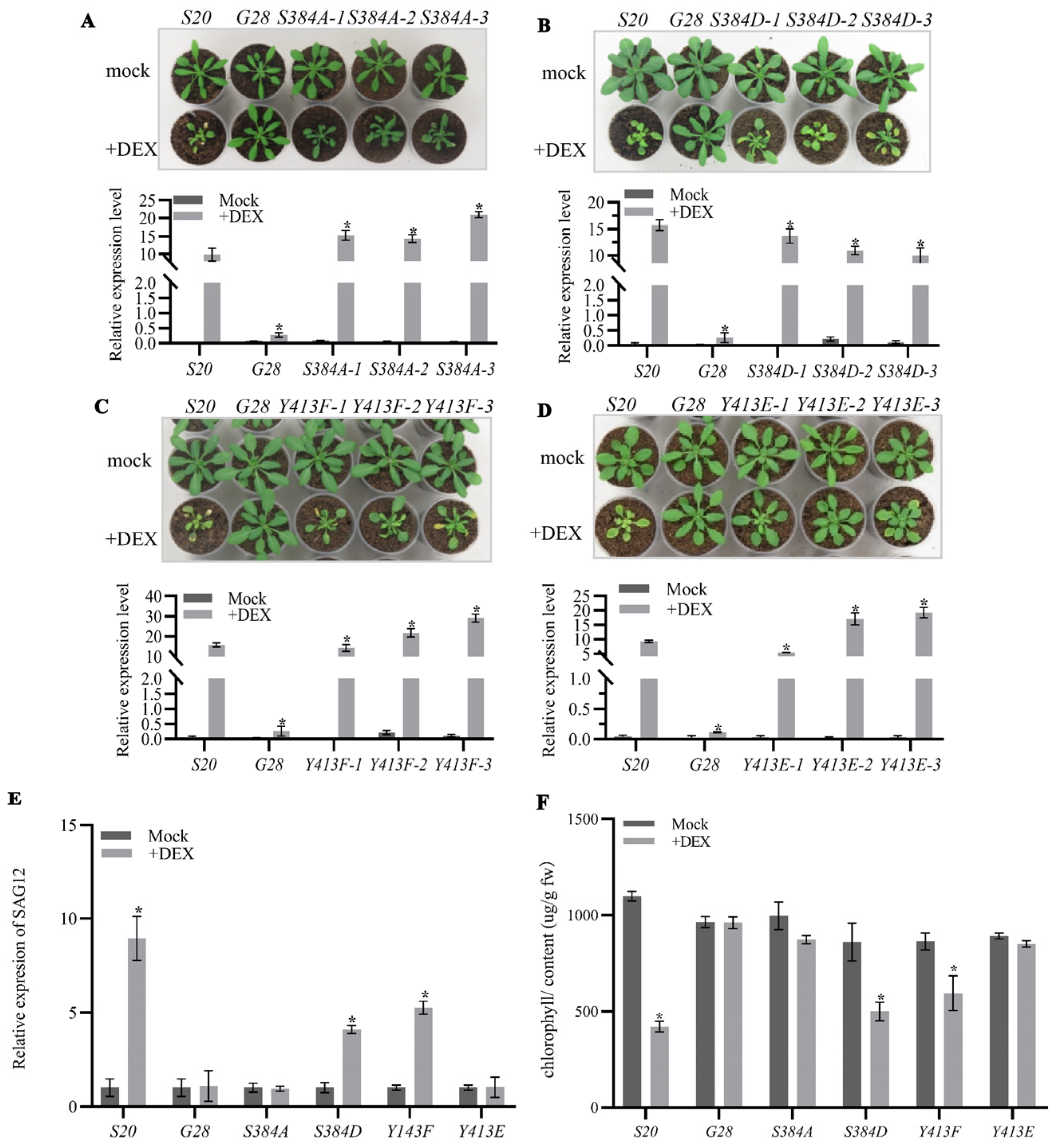
2.2. The Autophosphorylation of AtSARK Is Essential for Its Function in Promoting Leaf Senescence
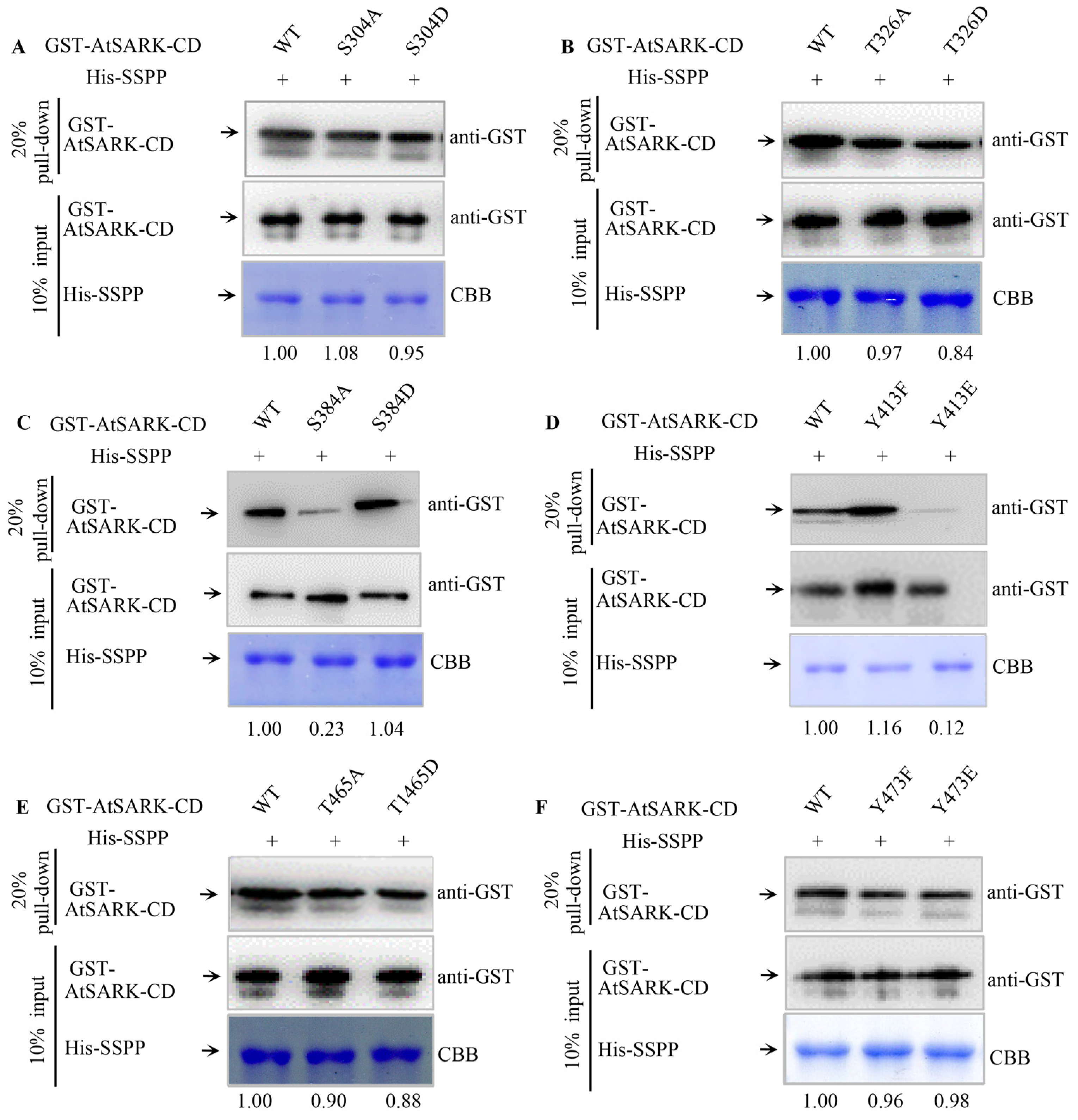
2.3. S384 and Y413 Play Important Roles in the Interaction between AtSARK and Senescence-Suppressed Protein Phosphatase SSPP
3. Discussion
3.1. Autophosphorylation of AtSARK Is Required for Its Functions in Promoting Leaf Senescence
3.2. The Phosphorylated S384 Residue of AtSARK Is the Main Dephosphorylation Site for SSPP Whereas the Phosphorylation of Y413 Residue Fine-Tunes the Interaction between AtSARK and SSPP
4. Materials and Methods
4.1. Liquid Chromatography-tandem Mass Spectrometry (LC–MS/MS) Analysis and Phosphosite Identification
4.2. Site-Directed Mutagenesis of AtSARK and Recombinant Protein Purification
4.3. In Vitro Autophosphorylation Assay
4.4. Pull-Down Assay
4.5. Plant Growth and Generation of Transgenic Lines
4.6. RNA Extraction and RT-PCR Analysis of Gene Expression
4.7. Determination of Chlorophyll Content
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death-regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, D.H. Functional roles of protein phosphatase 4 in multiple aspects of cellular physiology: A friend and a foe. BMB Rep. 2020, 53, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, Y.; Xu, F.; Zhang, Y. Differential requirement of BAK1 C-terminal tail in development and immunity. J. Integr. Plant Biol. 2017, 60, 270–275. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Suzuki, M.; Shibuya, M.; Shimada, H.; Motoyama, N.; Nakashima, M.; Takahashi, S.; Suto, K.; Yoshida, I.; Matsui, S.; Tsujimoto, N.; et al. Autophosphorylation of specific threonine and tyrosine residues in Arabidopsis CERK1 is essential for the activation of chitin-induced immune signaling. Plant Cell Physiol. 2016, 57, 2312–2322. [Google Scholar] [CrossRef]
- Cai, W.; Zhang, D. The role of receptor-like kinases in regulating plant male reproduction. Plant Reprod. 2018, 31, 77–87. [Google Scholar] [CrossRef]
- Van Bentem, S.I.F.; Hirt, H. Protein tyrosine phosphorylation in plants: More abundant than expected? Trends Plant Sci. 2009, 14, 71–76. [Google Scholar] [CrossRef]
- Macho, A.P.; Lozano-Duran, R.; Zipfel, C. Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 2015, 20, 269–272. [Google Scholar] [CrossRef]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-dependent action of an ABA-inducible receptor kinase, PPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef]
- Chen, L.J.; Wuriyanghan, H.; Zhang, Y.Q.; Duan, K.X.; Chen, H.W.; Li, Q.T.; Lu, X.; He, S.J.; Ma, B.; Zhang, W.K.; et al. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef]
- Li, X.; Ahmad, S.; Ali, A.; Guo, C.; Li, H.; Yu, J.; Zhang, Y.; Gao, X.; Guo, Y. Characterization of somatic embryogenesis receptor-like kinase 4 as a negative regulator of leaf senescence in Arabidopsis. Cells 2019, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, K.; Ali, A.; Guo, Y. AtWAKL10, a cell wall associated receptor-like kinase, negatively regulates leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 4885. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, H.; Liu, G.; Ma, W.; Li, C.; Huo, H. PpSARK regulates moss senescence and salt tolerance through ABA related pathway. Int. J. Mol. Sci. 2018, 19, 2609. [Google Scholar] [CrossRef] [PubMed]
- Hajouj, T.; Michelis, R.; Gepstein, S. Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 2000, 124, 1305–1314. [Google Scholar] [CrossRef]
- Xu, F.; Meng, T.; Li, P.L.; Yu, Y.; Cui, Y.; Wang, Y.; Gong, Q.; Wang, N.N. A soybean dual-specificity kinase, GmSARK, and its Arabidopsis homolog, AtSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 2011, 157, 2131–2153. [Google Scholar] [CrossRef]
- Li, X.P.; Gan, R.; Li, P.L.; Ma, Y.Y.; Zhang, L.W.; Zhang, R.; Wang, Y.; Wang, N.N. Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol. Biol. 2006, 61, 829–844. [Google Scholar] [CrossRef]
- Courtney, T.M.; Deiters, A. Optical control of protein phosphatase function. Nat. Commun. 2019, 10, 4384. [Google Scholar] [CrossRef]
- Luo, X.; Wu, W.; Liang, Y.; Xu, N.; Wang, Z.; Zou, H.; Liu, J. Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 2020, 39, e102856. [Google Scholar] [CrossRef]
- Gepstein, S.; Sabehi, G.; Carp, M.J.; Hajouj, T.; Nesher, M.F.O.; Yariv, I.; Dor, C.; Bassani, M. Large-scale identification of leaf senescence-associated genes. Plant J. 2003, 36, 629–642. [Google Scholar] [CrossRef]
- Robinson, W.D.; Carson, I.; Ying, S.; Ellis, K.; Plaxton, W.C. Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization. New Phytol. 2012, 196, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, S.S. An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 2012, 158, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Durian, G.; Jeschke, V.; Rahikainen, M.; Vuorinen, K.; Gollan, P.J.; Brosche, M.; Salojarvi, J.; Glawischnig, E.; Winter, Z.; Li, S.; et al. Protein phosphatase 2A-B’γ controls Botrytis cinerea resistance and developmental leaf senescence. Plant Physiol. 2020, 182, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Cui, Y.; Xu, F.; Xu, X.; Gao, G.; Wang, Y.; Guo, Z.; Wang, D.; Wang, N.N. Senescence-suppressed protein phosphatase directly interacts with the cytoplasmic domain of senescence-associated receptor-like kinase and negatively regulates leaf senescence in Arabidopsis. Plant Physiol. 2015, 169, 1275–1291. [Google Scholar] [CrossRef]
- Aoyama, T.; Chua, N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997, 11, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.R.; Shah, S.; Rao, A.G. Investigation of autophosphorylation sites of plant receptor kinases and phosphorylation of interacting partners. In Plant Receptor Kinases: Methods and Protocols, Methods in Molecular Biology; Aalen, R., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1621, pp. 121–130. [Google Scholar] [CrossRef]
- Cao, M.; Meng, D.; Wang, L.; Bei, S.; Snell, W.J.; Pan, J. Activation loop phosphorylation of a protein kinase is a molecular marker of organelle size that dynamically reports flagellar length. Proc. Natl. Acad. Sci. USA 2013, 110, 12337–12342. [Google Scholar] [CrossRef]
- Sugano, S.; Maeda, S.; Hayashi, N.; Kajiawara, H.; Inoue, H.; Jiang, C.J.; Takatsuji, H.; Mori, M. Tyrosine phosphorylation of a receptor-like cytoplasmic kinase, BSR1, plays a crucial role in resistance to multiple pathogens in rice. Plant J. 2018, 96, 1137–1147. [Google Scholar] [CrossRef]
- Noh, Y.; Amasino, R.M. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 1999, 41, 181–194. [Google Scholar] [CrossRef]
- Wen, Z.; Mei, Y.; Zhou, J.; Cui, Y.; Wang, D.; Wang, N.N. SAUR49 can positively regulate leaf senescence by suppressing SSPP in Arabidopsis. Plant Cell Physiol. 2020, 61, 644–658. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.; Mei, Y.; Wang, D.; Xiao, D.; Tang, X.; Gong, Y.; Xu, X.; Wang, N.N. Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase. Int. J. Mol. Sci. 2022, 23, 8873. https://doi.org/10.3390/ijms23168873
Guo Z, Mei Y, Wang D, Xiao D, Tang X, Gong Y, Xu X, Wang NN. Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase. International Journal of Molecular Sciences. 2022; 23(16):8873. https://doi.org/10.3390/ijms23168873
Chicago/Turabian StyleGuo, Zhaoxia, Yuanyuan Mei, Dan Wang, Dong Xiao, Xianglin Tang, Yaru Gong, Xinxin Xu, and Ning Ning Wang. 2022. "Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase" International Journal of Molecular Sciences 23, no. 16: 8873. https://doi.org/10.3390/ijms23168873
APA StyleGuo, Z., Mei, Y., Wang, D., Xiao, D., Tang, X., Gong, Y., Xu, X., & Wang, N. N. (2022). Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase. International Journal of Molecular Sciences, 23(16), 8873. https://doi.org/10.3390/ijms23168873





