An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study
Abstract
:1. Introduction
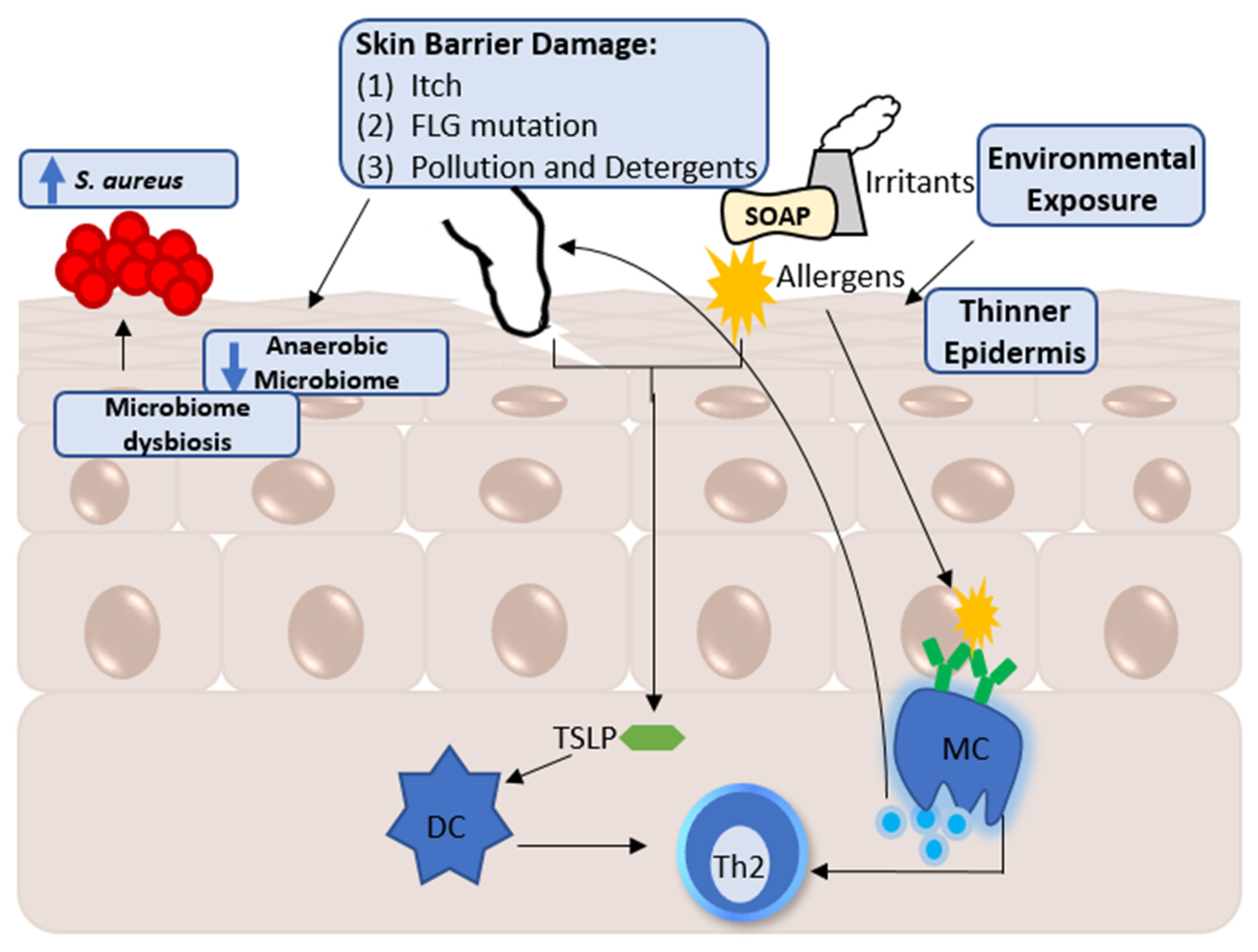
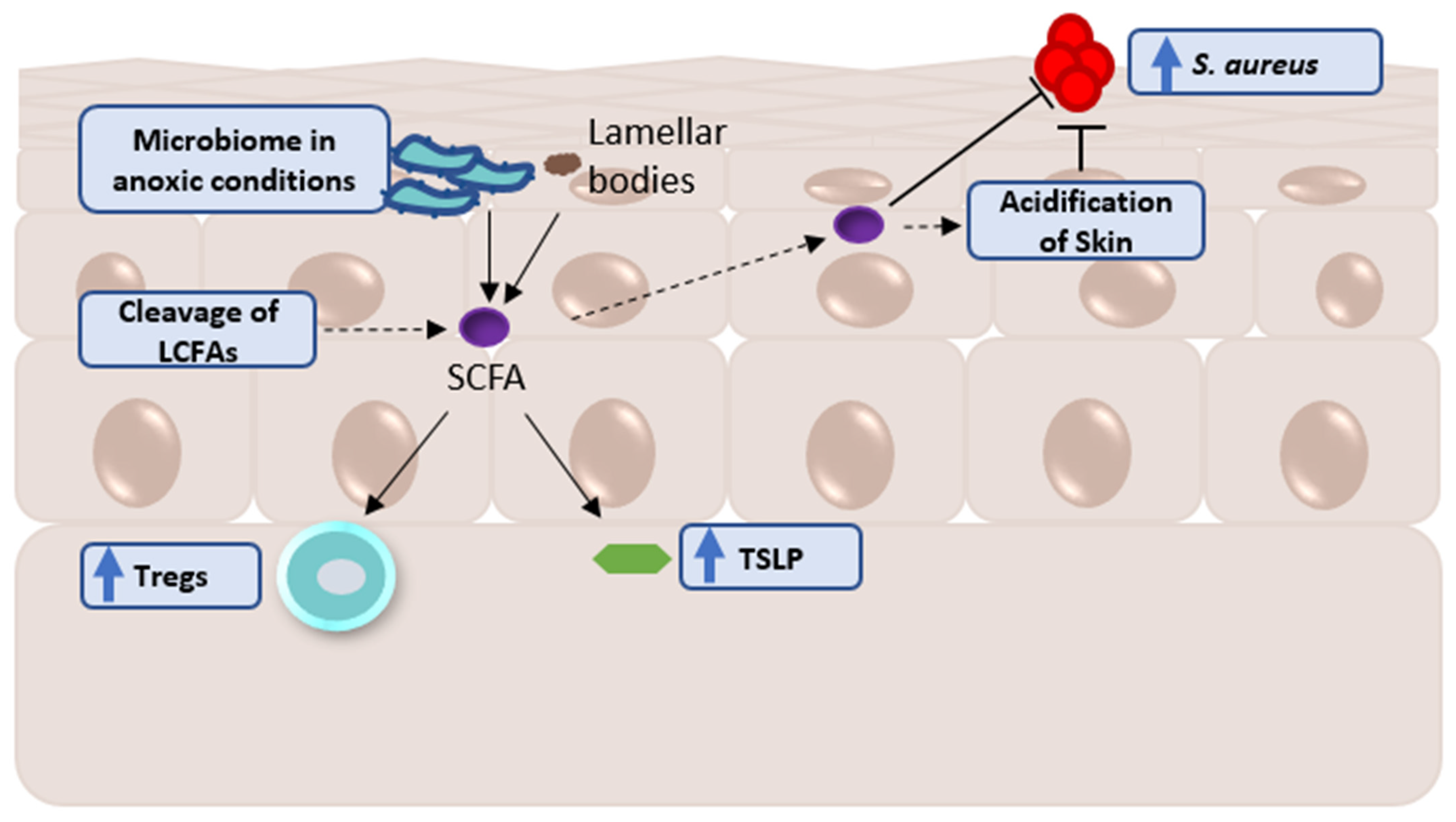
2. Factors Influencing the Skin Metabolome
A Note Regarding Skin Metabolomic Collection and Processing
3. Current Understanding in the Metabolomics of Atopic Eczema
3.1. Findings within the Skin
3.2. Findings within the Blood
3.3. Infantile AE Findings as a Precursor for the AE Metabolome
| References | Matrices | Target | Sample Size | Findings |
|---|---|---|---|---|
| [109] | Serum | Eicosanoids and UT | 41 AE 22 HE 42 AE 23 HE | Metabolomics profiles separated according to IgE levels ↓ Glycine in AE vs. HE ↓ Taurine in AE vs. HE ↑ Unsaturated fatty acid in AE vs. HE ↑ Carnitines, FFA, sphingomyelins, and lactic acid in high IgE AE vs. low IgE AE and HE |
| [110] | Urine | UT | 20 AE 12 HE | By supervised statistics, there is a prominent distinction between AE and HE ↑ Creatinine, creatine, citrate, formate, 2-hydroxybutyrate, dimethylglycine, and lactate in AE vs. HE ↓ Betaine, glycine, and alanine in AE vs. HE |
| [111] | Tape strips | Lipids | 28 AE 32 HE | ↓ Glyceroglycolipids in AE LS vs. HE ↓ Sphingomyelin in AE LS vs. HE ↑ Glycerophospholipids in AE LS vs. HE |
| [104] | Fecal matter | SCFA | 24 AE 33 HE | ↑ Butyrate in HE and persistent AE vs. transient AE ↑ Valerate in HE and persistent AE vs. transient AE No difference in acetate and propionate between the groups |
| [108] | Urine | UT | 455 Children | Propyl-parabens presence is associated with aeroallergen sensitization but not with AE Propyl-paraben is associated with AE severity ↑ Amino acids in general within the high propyl-paraben group ↑ Picolinic acid in high propyl-paraben group ↓ 2-palmitoylglycerol in high propyl-paraben group |
| [107] | Serum | Metabolites in Biocrates Absolute IDQ® P180 kit | 495 Newborns 449 1-year-olds | ↑ Hexose levels in newborns and 1-year old AE Amino acids are negatively correlated with inflammasome expression Lysophosphatidylcholines negatively correlate with inflammasome expression |
| [106] | Breast milk of AE mothers | UT | 75 AE 75 HE | ↑ LCSFA in AE mothers’ vs. HE mothers’ milk |
| [105] | Fecal matter | SCFA and UT | 33 AE 30 HE | Allergy sensitization is an endotype of AE ↓ Butyrate and propionate in infants that later developed eczema |
| [112] | Plasma | Vitamin D | 4327 2-year-olds | Vitamin D does not predict the development of AE |
4. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bieber, T.; Akdis, C.; Lauener, R.; Traidl-Hoffmann, C.; Schmid-Grendelmeier, P.; Schäppi, G.; Allam, J.-P.; Apfelbacher, C.; Augustin, M.; Beck, L.; et al. Global Allergy Forum and 3rd Davos Declaration 2015: Atopic Dermatitis/Eczema: Challenges and Opportunities toward Precision Medicine. Allergy 2016, 71, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Asher, M.I.; Montefort, S.; Björkstén, B.; Lai, C.K.W.; Strachan, D.P.; Weiland, S.K.; Williams, H.; ISAAC Phase Three Study Group. Worldwide Time Trends in the Prevalence of Symptoms of Asthma, Allergic Rhinoconjunctivitis, and Eczema in Childhood: ISAAC Phases One and Three Repeat Multicountry Cross-Sectional Surveys. Lancet Lond. Engl. 2006, 368, 733–743. [Google Scholar] [CrossRef]
- Williams, H.; Stewart, A.; von Mutius, E.; Cookson, W.; Anderson, H.R. Is Eczema Really on the Increase Worldwide? J. Allergy Clin. Immunol. 2008, 121, 947–954.e15. [Google Scholar] [CrossRef] [PubMed]
- Hadi, H.A.; Tarmizi, A.I.; Khalid, K.A.; Gajdács, M.; Aslam, A.; Jamshed, S. The Epidemiology and Global Burden of Atopic Dermatitis: A Narrative Review. Life 2021, 11, 936. [Google Scholar] [CrossRef]
- Nutten, S. Atopic Dermatitis: Global Epidemiology and Risk Factors. Ann. Nutr. Metab. 2015, 66, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Ujiie, H.; Rosmarin, D.; Schön, M.P.; Ständer, S.; Boch, K.; Metz, M.; Maurer, M.; Thaci, D.; Schmidt, E.; Cole, C.; et al. Unmet Medical Needs in Chronic, Non-Communicable Inflammatory Skin Diseases. Front. Med. 2022, 9, 875492. [Google Scholar] [CrossRef] [PubMed]
- Barbarot, S.; Auziere, S.; Gadkari, A.; Girolomoni, G.; Puig, L.; Simpson, E.L.; Margolis, D.J.; de Bruin-Weller, M.; Eckert, L. Epidemiology of Atopic Dermatitis in Adults: Results from an International Survey. Allergy 2018, 73, 1284–1293. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.H.; Patel, K.R.; Singam, V.; Rastogi, S.; Silverberg, J.I. A Systematic Review and Meta-Analysis of the Prevalence and Phenotype of Adult-Onset Atopic Dermatitis. J. Am. Acad. Dermatol. 2019, 80, 1526–1532.e7. [Google Scholar] [CrossRef]
- Bantz, S.K.; Zhu, Z.; Zheng, T. The Atopic March: Progression from Atopic Dermatitis to Allergic Rhinitis and Asthma. J. Clin. Cell. Immunol. 2014, 5, 202. [Google Scholar] [CrossRef] [Green Version]
- Silverwood, R.J.; Forbes, H.J.; Abuabara, K.; Ascott, A.; Schmidt, M.; Schmidt, S.A.J.; Smeeth, L.; Langan, S.M. Severe and Predominantly Active Atopic Eczema in Adulthood and Long Term Risk of Cardiovascular Disease: Population Based Cohort Study. BMJ 2018, 361, k1786. [Google Scholar] [CrossRef] [Green Version]
- Ferrucci, S.M.; Tavecchio, S.; Angileri, L.; Surace, T.; Berti, E.; Buoli, M. Factors Associated with Affective Symptoms and Quality of Life in Patients with Atopic Dermatitis. Acta Derm. Venereol. 2021, 101, adv00590. [Google Scholar] [CrossRef] [PubMed]
- Bieber, T. Atopic Dermatitis. N. Engl. J. Med. 2008, 358, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, Z.; Zhang, H.; Guo, Y.; Yao, Z. Update on the Pathogenesis and Therapy of Atopic Dermatitis. Clinic Rev Allerg Immunol 2021, 61, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Luschkova, D.; Zeiser, K.; Ludwig, A.; Traidl-Hoffmann, C. Atopic Eczema Is an Environmental Disease. Allergol. Sel. 2021, 5, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, H.; Ebise, H.; Tazawa, T.; Tanaka, K.; Sugiura, Y.; Uehara, M.; Kikuchi, K.; Kimura, T. Large-Scale DNA Microarray Analysis of Atopic Skin Lesions Shows Overexpression of an Epidermal Differentiation Gene Cluster in the Alternative Pathway and Lack of Protective Gene Expression in the Cornified Envelope. Br. J. Dermatol. 2005, 152, 146–149. [Google Scholar] [CrossRef]
- Palmer, C.N.A.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.D.; et al. Common Loss-of-Function Variants of the Epidermal Barrier Protein Filaggrin Are a Major Predisposing Factor for Atopic Dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef]
- Barnes, K.C. An Update on the Genetics of Atopic Dermatitis: Scratching the Surface in 2009. J. Allergy Clin. Immunol. 2010, 125, 16–29.e11. [Google Scholar] [CrossRef]
- Agrawal, R.; Woodfolk, J.A. Skin Barrier Defects in Atopic Dermatitis. Curr. Allergy Asthma Rep. 2014, 14, 433. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Imanishi, I. Epithelial–Immune Crosstalk with the Skin Microbiota in Homeostasis and Atopic Dermatitis—A Mini Review. Vet. Dermatol. 2021, 32, 533-e147. [Google Scholar] [CrossRef]
- Tabata, N.; Tagami, H.; Kligman, A.M. A Twenty-Four-Hour Occlusive Exposure to 1% Sodium Lauryl Sulfate Induces a Unique Histopathologic Inflammatory Response in the Xerotic Skin of Atopic Dermatitis Patients. Acta Derm. Venereol. 1998, 78, 244–247. [Google Scholar] [CrossRef] [Green Version]
- Flohr, C.; Pascoe, D.; Williams, H.C. Atopic Dermatitis and the “Hygiene Hypothesis”: Too Clean to Be True? Br. J. Dermatol. 2005, 152, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K. The Role of Air Pollutants in Atopic Dermatitis. J. Allergy Clin. Immunol. 2014, 134, 993–999. [Google Scholar] [CrossRef]
- Wong, T.-Y. Smog Induces Oxidative Stress and Microbiota Disruption. J. Food Drug Anal. 2017, 25, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Celebi Sozener, Z.; Ozdel Ozturk, B.; Cerci, P.; Turk, M.; Gorgulu Akin, B.; Akdis, M.; Altiner, S.; Ozbey, U.; Ogulur, I.; Mitamura, Y.; et al. Epithelial Barrier Hypothesis: Effect of the External Exposome on the Microbiome and Epithelial Barriers in Allergic Disease. Allergy 2022, 77, 1418–1449. [Google Scholar] [CrossRef] [PubMed]
- Ong, P.Y.; Ohtake, T.; Brandt, C.; Strickland, I.; Boguniewicz, M.; Ganz, T.; Gallo, R.L.; Leung, D.Y.M. Endogenous Antimicrobial Peptides and Skin Infections in Atopic Dermatitis. N. Engl. J. Med. 2002, 347, 1151–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-Host Interplay in Atopic Dermatitis and Psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona, I.D.; Cho, S.H.; Leung, D.Y.M. Role of Bacterial Superantigens in Atopic Dermatitis: Implications for Future Therapeutic Strategies. Am. J. Clin. Dermatol. 2006, 7, 273–279. [Google Scholar] [CrossRef]
- Hülpüsch, C.; Tremmel, K.; Hammel, G.; Bhattacharyya, M.; de Tomassi, A.; Nussbaumer, T.; Neumann, A.U.; Reiger, M.; Traidl-Hoffmann, C. Skin PH-Dependent Staphylococcus Aureus Abundance as Predictor for Increasing Atopic Dermatitis Severity. Allergy 2020, 75, 2888–2898. [Google Scholar] [CrossRef]
- Al-Jaberi, H.; Marks, R. Studies of the Clinically Uninvolved Skin in Patients with Dermatitis. Br. J. Dermatol. 1984, 111, 437–443. [Google Scholar] [CrossRef]
- Gavrilova, T. Immune Dysregulation in the Pathogenesis of Atopic Dermatitis. Dermat. Contact Atopic Occup. Drug 2018, 29, 57–62. [Google Scholar] [CrossRef]
- Yang, L.; Fu, J.; Zhou, Y. Research Progress in Atopic March. Front. Immunol. 2020, 11, 1907. [Google Scholar] [CrossRef] [PubMed]
- Werfel, T.; Allam, J.-P.; Biedermann, T.; Eyerich, K.; Gilles, S.; Guttman-Yassky, E.; Hoetzenecker, W.; Knol, E.; Simon, H.-U.; Wollenberg, A.; et al. Cellular and Molecular Immunologic Mechanisms in Patients with Atopic Dermatitis. J. Allergy Clin. Immunol. 2016, 138, 336–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman-Yassky, E.; Krueger, J.G.; Lebwohl, M.G. Systemic Immune Mechanisms in Atopic Dermatitis and Psoriasis with Implications for Treatment. Exp. Dermatol. 2018, 27, 409–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnowicki, T.; He, H.; Krueger, J.G.; Guttman-Yassky, E. Atopic Dermatitis Endotypes and Implications for Targeted Therapeutics. J. Allergy Clin. Immunol. 2019, 143, 1–11. [Google Scholar] [CrossRef]
- Voss, M.; Kotrba, J.; Gaffal, E.; Katsoulis-Dimitriou, K.; Dudeck, A. Mast Cells in the Skin: Defenders of Integrity or Offenders in Inflammation? Int. J. Mol. Sci. 2021, 22, 4589. [Google Scholar] [CrossRef]
- Müller, C.; Dietz, I.; Tziotis, D.; Moritz, F.; Rupp, J.; Schmitt-Kopplin, P. Molecular Cartography in Acute Chlamydia Pneumoniae Infections--a Non-Targeted Metabolomics Approach. Anal. Bioanal. Chem. 2013, 405, 5119–5131. [Google Scholar] [CrossRef]
- Elpa, D.P.; Chiu, H.-Y.; Wu, S.-P.; Urban, P.L. Skin Metabolomics. Trends Endocrinol. Metab. 2021, 32, 66–75. [Google Scholar] [CrossRef]
- Afghani, J.; Huelpuesch, C.; Reiger, M.; Schmitt-Kopplin, P.; Traidl-Hoffmann, C.; Mueller, C. Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive Wet-Prep Sampling. Metabolites 2021, 11, 415. [Google Scholar] [CrossRef]
- Dalgard, F.J.; Gieler, U.; Tomas-Aragones, L.; Lien, L.; Poot, F.; Jemec, G.B.E.; Misery, L.; Szabo, C.; Linder, D.; Sampogna, F.; et al. The Psychological Burden of Skin Diseases: A Cross-Sectional Multicenter Study among Dermatological out-Patients in 13 European Countries. J. Investig. Dermatol. 2015, 135, 984–991. [Google Scholar] [CrossRef] [Green Version]
- Nestle, F.O.; Di Meglio, P.; Qin, J.-Z.; Nickoloff, B.J. Skin Immune Sentinels in Health and Disease. Nat. Rev. Immunol. 2009, 9, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.V.; Soulika, A.M. The Dynamics of the Skin’s Immune System. Int. J. Mol. Sci. 2019, 20, 1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quaresma, J.A.S. Organization of the Skin Immune System and Compartmentalized Immune Responses in Infectious Diseases. Clin. Microbiol. Rev. 2019, 32, e00034-18. [Google Scholar] [CrossRef]
- Swaney, M.H.; Kalan, L.R. Living in Your Skin: Microbes, Molecules, and Mechanisms. Infect. Immun. 2021, 89, e00695-20. [Google Scholar] [CrossRef]
- Bouslimani, A.; da Silva, R.; Kosciolek, T.; Janssen, S.; Callewaert, C.; Amir, A.; Dorrestein, K.; Melnik, A.V.; Zaramela, L.S.; Kim, J.-N.; et al. The Impact of Skin Care Products on Skin Chemistry and Microbiome Dynamics. BMC Biol. 2019, 17, 47. [Google Scholar] [CrossRef]
- Misra, N.; Clavaud, C.; Guinot, F.; Bourokba, N.; Nouveau, S.; Mezzache, S.; Palazzi, P.; Appenzeller, B.M.R.; Tenenhaus, A.; Leung, M.H.Y.; et al. Multi-Omics Analysis to Decipher the Molecular Link between Chronic Exposure to Pollution and Human Skin Dysfunction. Sci. Rep. 2021, 11, 18302. [Google Scholar] [CrossRef]
- Randhawa, M.; Southall, M.; Samaras, S.T. Metabolomic Analysis of Sun Exposed Skin. Mol. Biosyst. 2013, 9, 2045–2050. [Google Scholar] [CrossRef]
- Jung, E.S.; Park, J.I.; Park, H.; Holzapfel, W.; Hwang, J.S.; Lee, C.H. Seven-Day Green Tea Supplementation Revamps Gut Microbiome and Caecum/Skin Metabolome in Mice from Stress. Sci. Rep. 2019, 9, 18418. [Google Scholar] [CrossRef] [Green Version]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An Integrative Metabolomics and Transcriptomics Study to Identify Metabolic Alterations in Aged Skin of Humans in vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef] [Green Version]
- Hooton, K.; Han, W.; Li, L. Comprehensive and Quantitative Profiling of the Human Sweat Submetabolome Using High-Performance Chemical Isotope Labeling LC–MS. Available online: https://pubs.acs.org/doi/pdf/10.1021/acs.analchem.6b01930 (accessed on 8 July 2020).
- Sitter, B.; Johnsson, M.K.; Halgunset, J.; Bathen, T.F. Metabolic Changes in Psoriatic Skin under Topical Corticosteroid Treatment. BMC Dermatol. 2013, 13, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, E.; Murota, H.; Mori, Y.; Yoshioka, Y.; Nomura, Y.; Munetsugu, T.; Yokozeki, H.; Katayama, I. Sweat Glucose and GLUT2 Expression in Atopic Dermatitis: Implication for Clinical Manifestation and Treatment. PLoS ONE 2018, 13, e0195960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, D.K.; Sinclair, E.; Xu, Y.; Sarkar, D.; Walton-Doyle, C.; Liscio, C.; Banks, P.; Milne, J.; Silverdale, M.; Kunath, T.; et al. Discovery of Volatile Biomarkers of Parkinson’s Disease from Sebum. ACS Cent. Sci. 2019, 5, 599–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, E.; Trivedi, D.K.; Sarkar, D.; Walton-Doyle, C.; Milne, J.; Kunath, T.; Rijs, A.M.; de Bie, R.M.A.; Goodacre, R.; Silverdale, M.; et al. Metabolomics of Sebum Reveals Lipid Dysregulation in Parkinson’s Disease. Nat. Commun. 2021, 12, 1592. [Google Scholar] [CrossRef]
- Calderón-Santiago, M.; Priego-Capote, F.; Turck, N.; Robin, X.; Jurado-Gámez, B.; Sanchez, J.C.; Luque de Castro, M.D. Human Sweat Metabolomics for Lung Cancer Screening. Anal. Bioanal. Chem. 2015, 407, 5381–5392. [Google Scholar] [CrossRef]
- Jarmusch, A.K.; Elijah, E.O.; Vargas, F.; Bouslimani, A.; da Silva, R.R.; Ernst, M.; Wang, M.; Del Rosario, K.K.; Dorrestein, P.C.; Tsunoda, S.M. Initial Development toward Non-Invasive Drug Monitoring via Untargeted Mass Spectrometric Analysis of Human Skin. Anal. Chem. 2019, 91, 8062–8069. [Google Scholar] [CrossRef]
- Bittremieux, W.; Advani, R.S.; Jarmusch, A.K.; Aguirre, S.; Lu, A.; Dorrestein, P.C.; Tsunoda, S.M. Physicochemical Properties Determining Drug Detection in Skin. Clin. Transl. Sci. 2021, 15, 761–770. [Google Scholar] [CrossRef]
- Perrier, E.; Demazières, A.; Girard, N.; Pross, N.; Osbild, D.; Metzger, D.; Guelinckx, I.; Klein, A. Circadian Variation and Responsiveness of Hydration Biomarkers to Changes in Daily Water Intake. Eur. J. Appl. Physiol. 2013, 113, 2143–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.L.; de la Mata, A.P.; Dias, R.P.; Harynuk, J.J. Towards Standardization of Data Normalization Strategies to Improve Urinary Metabolomics Studies by GC×GC-TOFMS. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Hahnefeld, L.; Gurke, R.; Thomas, D.; Schreiber, Y.; Schäfer, S.M.G.; Trautmann, S.; Snodgrass, I.F.; Kratz, D.; Geisslinger, G.; Ferreirós, N. Implementation of Lipidomics in Clinical Routine: Can Fluoride/Citrate Blood Sampling Tubes Improve Preanalytical Stability? Talanta 2020, 209, 120593. [Google Scholar] [CrossRef]
- Ang, J.E.; Revell, V.; Mann, A.; Mäntele, S.; Otway, D.T.; Johnston, J.D.; Thumser, A.E.; Skene, D.J.; Raynaud, F. Identification of Human Plasma Metabolites Exhibiting Time-of-Day Variation Using an Untargeted Liquid Chromatography–Mass Spectrometry Metabolomic Approach. Chronobiol. Int. 2012, 29, 868–881. [Google Scholar] [CrossRef] [Green Version]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Christensen, G.J.M.; Nakatsuji, T.; et al. Molecular Cartography of the Human Skin Surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiemsma, L.T.; Reynolds, L.A.; Turvey, S.E.; Finlay, B.B. The Hygiene Hypothesis: Current Perspectives and Future Therapies. Immunotargets Ther. 2015, 4, 143–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strachan, D.P. Hay Fever, Hygiene, and Household Size. BMJ 1989, 299, 1259–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloomfield, S.; Stanwell-Smith, R.; Crevel, R.; Pickup, J. Too Clean, or Not Too Clean: The Hygiene Hypothesis and Home Hygiene. Clin. Exp. Allergy 2006, 36, 402–425. [Google Scholar] [CrossRef]
- Ege, M.J.; Mayer, M.; Normand, A.-C.; Genuneit, J.; Cookson, W.O.C.M.; Braun-Fahrländer, C.; Heederik, D.; Piarroux, R.; von Mutius, E.; GABRIELA Transregio 22 Study Group. Exposure to Environmental Microorganisms and Childhood Asthma. N. Engl. J. Med. 2011, 364, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, N.; Sato, W.J.; Kelly, A.; Ganguli-Indra, G.; Indra, A.K. Epidermal Lipids: Key Mediators of Atopic Dermatitis Pathogenesis. Trends Mol. Med. 2019, 25, 551–562. [Google Scholar] [CrossRef] [PubMed]
- van Smeden, J.; Bouwstra, J.A. Stratum Corneum Lipids: Their Role for the Skin Barrier Function in Healthy Subjects and Atopic Dermatitis Patients. Curr. Probl. Dermatol. 2016, 49, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Ilves, L.; Ottas, A.; Kaldvee, B.; Abram, K.; Soomets, U.; Zilmer, M.; Jaks, V.; Kingo, K. Metabolomic Analysis of Skin Biopsies from Patients with Atopic Dermatitis Reveals Hallmarks of Inflammation, Disrupted Barrier Function and Oxidative Stress. Acta Derm. Venereol. 2021, 101, adv00407. [Google Scholar] [CrossRef] [PubMed]
- Emmert, H.; Baurecht, H.; Thielking, F.; Stölzl, D.; Rodriguez, E.; Harder, I.; Proksch, E.; Weidinger, S. Stratum Corneum Lipidomics Analysis Reveals Altered Ceramide Profile in Atopic Dermatitis Patients across Body Sites with Correlated Changes in Skin Microbiome. Exp. Dermatol. 2021, 30, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Töröcsik, D.; Weise, C.; Gericke, J.; Szegedi, A.; Lucas, R.; Mihaly, J.; Worm, M.; Rühl, R. Transcriptomic and Lipidomic Profiling of Eicosanoid/Docosanoid Signalling in Affected and Non-Affected Skin of Human Atopic Dermatitis Patients. Exp. Dermatol. 2019, 28, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, K.; Hassoun, L.A.; Foolad, N.; Pedersen, T.L.; Sivamani, R.K.; Newman, J.W. Sweat Lipid Mediator Profiling: A Noninvasive Approach for Cutaneous Research. J. Lipid Res. 2017, 58, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Blunder, S.; Rühl, R.; Moosbrugger-Martinz, V.; Krimmel, C.; Geisler, A.; Zhu, H.; Crumrine, D.; Elias, P.M.; Gruber, R.; Schmuth, M.; et al. Alterations in Epidermal Eicosanoid Metabolism Contribute to Inflammation and Impaired Late Differentiation in FLG-Mutated Atopic Dermatitis. J. Investig. Dermatol. 2017, 137, 706–715. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Villarreal, M.; Stewart, S.; Choi, J.; Ganguli-Indra, G.; Babineau, D.C.; Philpot, C.; David, G.; Yoshida, T.; Boguniewicz, M.; et al. Altered Composition of Epidermal Lipids Correlates with Staphylococcus Aureus Colonization Status in Atopic Dermatitis. Br. J. Dermatol. 2017, 177, e125–e127. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, L.; Kragballe, K. Abnormalities in Epidermal Lipid Metabolism in Patients with Atopic Dermatitis. J. Investig. Dermatol. 1991, 96, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzicka, T.; Simmet, T.; Peskar, B.A.; Ring, J. Skin Levels of Arachidonic Acid-Derived Inflammatory Mediators and Histamine in Atopic Dermatitis and Psoriasis. J. Investig. Dermatol. 1986, 86, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-J.; Kim, S.-E.; Shin, K.-O.; Park, K.; Lee, S.E. Dupilumab Therapy Improves Stratum Corneum Hydration and Skin Dysbiosis in Patients with Atopic Dermatitis. Allergy Asthma Immunol. Res. 2021, 13, 762. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-J.; Chang, T.-W.; Jiang, Y.; Kao, H.-J.; Chiou, B.-H.; Kao, M.-S.; Huang, C.-M. Commensal Staphylococcus Aureus Provokes Immunity to Protect against Skin Infection of Methicillin-Resistant Staphylococcus Aureus. Int. J. Mol. Sci. 2018, 19, 1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callewaert, C.; Nakatsuji, T.; Knight, R.; Kosciolek, T.; Vrbanac, A.; Kotol, P.; Ardeleanu, M.; Hultsch, T.; Guttman-Yassky, E.; Bissonnette, R.; et al. IL-4Rα Blockade by Dupilumab Decreases Staphylococcus Aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J. Investig. Dermatol. 2020, 140, 191–202.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feingold, K.R.; Elias, P.M. Role of Lipids in the Formation and Maintenance of the Cutaneous Permeability Barrier. Biochim. Biophys. Acta 2014, 1841, 280–294. [Google Scholar] [CrossRef]
- Elias, P.M. Stratum Corneum Architecture, Metabolic Activity and Interactivity with Subjacent Cell Layers. Exp. Dermatol. 1996, 5, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Holland, K.T.; Greenman, J.; Cunliffe, W.J. Growth of Cutaneous Propionibacteria on Synthetic Medium; Growth Yields and Exoenzyme Production. J. Appl. Bacteriol. 1979, 47, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, H.; Henne, A.; Hoster, F.; Liesegang, H.; Wiezer, A.; Strittmatter, A.; Hujer, S.; Dürre, P.; Gottschalk, G. The Complete Genome Sequence of Propionibacterium Acnes, a Commensal of Human Skin. Science 2004, 305, 671–673. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chiang, H.-I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The Microbiome Extends to Subepidermal Compartments of Normal Skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, H.; Matsuda, A.; Jung, K.; Karasawa, K.; Matsuda, K.; Oida, K.; Ishizaka, S.; Ahn, G.; Amagai, Y.; Moon, C.; et al. Skin PH Is the Master Switch of Kallikrein 5-Mediated Skin Barrier Destruction in a Murine Atopic Dermatitis Model. J. Investig. Dermatol. 2016, 136, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Stücker, M.; Struk, A.; Altmeyer, P.; Herde, M.; Baumgärtl, H.; Lübbers, D.W. The Cutaneous Uptake of Atmospheric Oxygen Contributes Significantly to the Oxygen Supply of Human Dermis and Epidermis. J. Physiol. 2002, 538, 985–994. [Google Scholar] [CrossRef]
- Gong, J.Q.; Lin, L.; Lin, T.; Hao, F.; Zeng, F.Q.; Bi, Z.G.; Yi, D.; Zhao, B. Skin Colonization by Staphylococcus Aureus in Patients with Eczema and Atopic Dermatitis and Relevant Combined Topical Therapy: A Double-Blind Multicentre Randomized Controlled Trial. Br. J. Dermatol. 2006, 155, 680–687. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.-M. Staphylococcus Epidermidis in the Human Skin Microbiome Mediates Fermentation to Inhibit the Growth of Propionibacterium Acnes: Implications of Probiotics in Acne Vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef] [Green Version]
- Lambers, H.; Piessens, S.; Bloem, A.; Pronk, H.; Finkel, P. Natural Skin Surface PH Is on Average below 5, Which Is Beneficial for Its Resident Flora. Int. J. Cosmet. Sci. 2006, 28, 359–370. [Google Scholar] [CrossRef]
- Schwarz, A.; Bruhs, A.; Schwarz, T. The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System. J. Investig. Dermatol. 2017, 137, 855–864. [Google Scholar] [CrossRef] [Green Version]
- Krejner, A.; Bruhs, A.; Mrowietz, U.; Wehkamp, U.; Schwarz, T.; Schwarz, A. Decreased Expression of G-Protein-Coupled Receptors GPR43 and GPR109a in Psoriatic Skin Can Be Restored by Topical Application of Sodium Butyrate. Arch. Dermatol. Res. 2018, 310, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Rutting, S.; Xenaki, D.; Malouf, M.; Horvat, J.C.; Wood, L.G.; Hansbro, P.M.; Oliver, B.G. Short-Chain Fatty Acids Increase TNFα-Induced Inflammation in Primary Human Lung Mesenchymal Cells through the Activation of P38 MAPK. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 316, L157–L174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanford, J.A.; O’Neill, A.M.; Zouboulis, C.C.; Gallo, R.L. Short-Chain Fatty Acids from Cutibacterium Acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J. Immunol. 2019, 202, 1767–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Schlatterer, K.; Peschel, A.; Kretschmer, D. Short-Chain Fatty Acid and FFAR2 Activation-A New Option for Treating Infections? Front. Cell. Infect. Microbiol. 2021, 11, 785833. [Google Scholar] [CrossRef]
- Chiu, C.-Y.; Lin, G.; Wang, C.-J.; Hung, S.-I.; Chung, W.-H. Metabolomics Reveals Microbial-Derived Metabolites Associated with Immunoglobulin E Responses in Filaggrin-Related Atopic Dermatitis. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2021, 32, 1709–1717. [Google Scholar] [CrossRef]
- Hotze, M.; Baurecht, H.; Rodríguez, E.; Chapman-Rothe, N.; Ollert, M.; Fölster-Holst, R.; Adamski, J.; Illig, T.; Ring, J.; Weidinger, S. Increased Efficacy of Omalizumab in Atopic Dermatitis Patients with Wild-Type Filaggrin Status and Higher Serum Levels of Phosphatidylcholines. Allergy 2014, 69, 132–135. [Google Scholar] [CrossRef]
- Matthias, J.; Maul, J.; Noster, R.; Meinl, H.; Chao, Y.-Y.; Gerstenberg, H.; Jeschke, F.; Gasparoni, G.; Welle, A.; Walter, J.; et al. Sodium Chloride Is an Ionic Checkpoint for Human TH2 Cells and Shapes the Atopic Skin Microenvironment. Sci. Transl. Med. 2019, 11, eaau0683. [Google Scholar] [CrossRef]
- Ottas, A.; Fishman, D.; Okas, T.-L.; Püssa, T.; Toomik, P.; Märtson, A.; Kingo, K.; Soomets, U. Blood Serum Metabolome of Atopic Dermatitis: Altered Energy Cycle and the Markers of Systemic Inflammation. PLoS ONE 2017, 12, e0188580. [Google Scholar] [CrossRef] [Green Version]
- Jacob, M.; Gu, X.; Luo, X.; Al-Mousa, H.; Arnaout, R.; Al-Saud, B.; Lopata, A.L.; Li, L.; Dasouki, M.; Rahman, A.M.A. Metabolomics Distinguishes DOCK8 Deficiency from Atopic Dermatitis: Towards a Biomarker Discovery. Metabolites 2019, 9, 274. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, K.; Sivamani, R.K.; Newman, J.W. Noninvasive Profiling of Sweat-Derived Lipid Mediators for Cutaneous Research. Ski. Res. Technol. 2019, 25, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihály, J.; Gericke, J.; Törőcsik, D.; Gáspár, K.; Szegedi, A.; Rühl, R. Reduced Lipoxygenase and Cyclooxygenase Mediated Signaling in PBMC of Atopic Dermatitis Patients. Prostaglandins Other Lipid Mediat. 2013, 107, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Lee, S.Y.; Kang, M.J.; Kim, B.S.; Lee, M.J.; Jung, S.S.; Yoon, J.S.; Cho, H.J.; Lee, E.; Yang, S.I.; et al. Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant. Allergy Asthma Immunol. Res. 2020, 12, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Ta, L.D.H.; Chan, J.C.Y.; Yap, G.C.; Purbojati, R.W.; Drautz-Moses, D.I.; Koh, Y.M.; Tay, C.J.X.; Huang, C.-H.; Kioh, D.Y.Q.; Woon, J.Y.; et al. A Compromised Developmental Trajectory of the Infant Gut Microbiome and Metabolome in Atopic Eczema. Gut Microbes 2020, 12, 1801964. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.S.; Tsuyama, N.; Inoue, H.; Guo, Y.; Mokuda, S.; Nobukiyo, A.; Nakatani, N.; Yamaide, F.; Nakano, T.; Kohno, Y.; et al. Long-Chain Saturated Fatty Acids in Breast Milk Are Associated with the Pathogenesis of Atopic Dermatitis via Induction of Inflammatory ILC3s. Sci. Rep. 2021, 11, 13109. [Google Scholar] [CrossRef]
- Herberth, G.; Offenberg, K.; Rolle-Kampczyk, U.; Bauer, M.; Otto, W.; Röder, S.; Grützmann, K.; Sack, U.; Simon, J.-C.; Borte, M.; et al. Endogenous Metabolites and Inflammasome Activity in Early Childhood and Links to Respiratory Diseases. J. Allergy Clin. Immunol. 2015, 136, 495–497. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Lee, E.; Yon, D.K.; Jee, H.M.; Baek, H.S.; Lee, S.W.; Cho, J.-Y.; Han, M.Y. The Potential Pathways Underlying the Association of Propyl-Paraben Exposure with Aeroallergen Sensitization and EASI Score Using Metabolomics Analysis. Sci. Rep. 2021, 11, 3772. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, G.; Liu, X.; Shao, Y.; Gao, P.; Xin, C.; Cui, Z.; Zhao, X.; Xu, G. Serum Metabolomics Study and Eicosanoid Analysis of Childhood Atopic Dermatitis Based on Liquid Chromatography-Mass Spectrometry. J. Proteome Res. 2014, 13, 5715–5723. [Google Scholar] [CrossRef]
- Assfalg, M.; Bortoletti, E.; D’Onofrio, M.; Pigozzi, R.; Molinari, H.; Boner, A.L.; Peroni, D.G.; Piacentini, G.L. An Exploratory (1) H-Nuclear Magnetic Resonance Metabolomics Study Reveals Altered Urine Spectral Profiles in Infants with Atopic Dermatitis. Br. J. Dermatol. 2012, 166, 1123–1125. [Google Scholar] [CrossRef]
- Wang, H.; Cui, L.; Jia, Y.; Gao, Y.; Zhang, G.; He, C. Application of Lipidomics to Reveal Differences of Facial Skin Surface Lipids between Atopic Dermatitis and Healthy Infants. J. Cosmet. Dermatol. 2020, 19, 1528–1534. [Google Scholar] [CrossRef]
- Yang, L.; Sato, M.; Saito-Abe, M.; Nishizato, M.; Mezawa, H.; Yamamoto-Hanada, K.; Ohya, Y. Serum 25-Hydroxyvitamin D Concentrations and Atopic Dermatitis in Early Childhood: Findings from the Japan Environment and Children’s Study. Nutrients 2021, 13, 2761. [Google Scholar] [CrossRef] [PubMed]
- Brüggen, M.-C.; Stingl, G. Subcutaneous White Adipose Tissue: The Deepest Layer of the Cutaneous Immune Barrier. JDDG J. Dtsch. Dermatol. Ges. 2020, 18, 1225–1227. [Google Scholar] [CrossRef] [PubMed]
- Proksch, E. PH in Nature, Humans and Skin. J. Dermatol. 2018, 45, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Yousaf, M.; Ayasse, M.; Ahmed, A.; Gwillim, E.C.; Janmohamed, S.R.; Yousaf, A.; Patel, K.R.; Thyssen, J.P.; Silverberg, J.I. Association between Atopic Dermatitis and Hypertension: A Systematic Review and Meta-Analysis. Br. J. Dermatol. 2022, 186, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Selvarajah, V.; Connolly, K.; McEniery, C.; Wilkinson, I. Skin Sodium and Hypertension: A Paradigm Shift? Curr. Hypertens. Rep. 2018, 20, 94. [Google Scholar] [CrossRef] [Green Version]
- Selvarajah, V.; Mäki-Petäjä, K.M.; Pedro, L.; Bruggraber, S.F.A.; Burling, K.; Goodhart, A.K.; Brown, M.J.; McEniery, C.M.; Wilkinson, I.B. Novel Mechanism for Buffering Dietary Salt in Humans: Effects of Salt Loading on Skin Sodium, Vascular Endothelial Growth Factor C, and Blood Pressure. Hypertension 2017, 70, 930–937. [Google Scholar] [CrossRef]
- Hon, K.L.; Kung, J.S.C.; Ng, W.G.; Tsang, K.Y.C.; Cheng, N.; Leung, T.F. Are Skin Equipment for Assessing Childhood Eczema Any Good? J. Dermatol. Treat. 2021, 32, 45–48. [Google Scholar] [CrossRef]
- Reilly, D.M.; Parslew, R.; Sharpe, G.R.; Powell, S.; Green, M.R. Inflammatory Mediators in Normal, Sensitive and Diseased Skin Types. Acta Derm. Venereol. 2000, 80, 171–174. [Google Scholar] [CrossRef]
- Behrendt, H.; Ring, J. Histamine, Antihistamines and Atopic Eczema. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 1990, 20, 25–30. [Google Scholar] [CrossRef]
- Masini, E.; Giannella, E.; Bani-Sacchi, T.; Fantozzi, R.; Palmerani, B.; Mannaioni, P.F. Histamine Release from Serosal Mast Cells by Intermediate Products of Arachidonic Acid Metabolism. Agents Actions 1987, 20, 202–205. [Google Scholar] [CrossRef]
| References | Matrices | Target | Sample Size | Findings |
|---|---|---|---|---|
| [70] | Skin biopsy | Metabolites in Biocrates Absolute IDQ® P180 kit | 15 AE 17 HE | ↑ Putrescine in AE LS as compared to AE NL and HE ↑ ADMA in AE LS as compared to AE NL ↑ Amino acids in AE LS as compared to AE NL and HE ↑ Sphingolipids in AE LS as compared to AE NL and HE |
| [71] | Tape strips | Metabolites in Waters Co. TrueMass® Stratum corneum metabolon lipid panel | 10 AE 10 HE | FLG is not associated with changing lipid composition of AE ↑ Short-chain ceramides in AE ↑ FFA in AE ↑ Cholesterol-sulphate in AE SCSFAs negatively correlated with Staphylococcus presence Several ceramide species positively correlate with Staphylococcus presence |
| [72] | Skin biopsy | FFA, eicosanoids, and docosanoids | 3 AE 6 HE | ↑ Arachidonic acid in AE LS and NL vs. HE ↑ PUFA-hydroxy metabolites in AE vs. HE ↑ 5-lipoxygenase-derived metabolites in AE LS ↑ in cyclooxygenases metabolites in AE vs. HE |
| [53] | Sweat | UT | 21 AE 6 Other dermatoses 10 HE | Glucose positively correlates with AE severity ↑ Glucose in AE with acute inflammation versus chronic inflammation and HE No difference in lactate between AE and HE No difference in sodium and salt content between AE and HE |
| [73] | Sweat | Lipid mediators | 11 AE 12 HE | ↑ C30-40 NS ceramides in AE ↑ C30-40 NS ceramides in AE ↑ C18:1 sphingosine in AE ↑ 10-nitrooleate in AE |
| [74] | Skin Biopsy | FFA, Eicosanoids, and Docosanoids | 18 AE 3 IV 14 HE | Stratum corneum lipid structure alteration is not related to the FLG genotype ↑ Arachidonic acid in AE FLG(+/−) compared to AE FLG(+/+), HE, and IV ↑ Hydroxy fatty acid HETE-12 in AE FLG(+/−) compared to other groups |
| [75] | Stratum corneum lipid extraction | Lipids, ceramides, cholesterols, FFA, and triglycerides | 27 AE 15 HE | ↓ FFA in AE with S. aureus growth ↓ Triglycerides in AE with S. aureus growth Cholesterol is not associated with S. aureus growth ↑ Short-chain ceramides in AE versus HE ↓ Long-chain ceramides in AE with S. aureus growth vs. AE without S. aureus Certain ceramides correlate with S. aureus presence |
| [76] | Skin biopsy | Lipids | 15 AE 9 HE | ↑ Arachidonic acid AE LS compared to AE NL ↑ SCSFAs in AE ↓ LCSFAs in AE n-6 FA inversely correlated with disease severity in AE NL ↑ Phospholipids in AE (NL and LS) compared to HE |
| [77] | Interstitial fluid | Arachidonic-acid-derived mediators | 16 AE 9 P 12 HE | ↑ LBT4 within AE LS compared to NL and HE, with similar results in P LBT4 has no correlation with AE disease severity No significant difference in PGE2 levels between groups |
| [78] | Tape strips | Ceramides | 10 AE 10 HE | ↑ C26 ceramide within non-lesional and lesional skin treated with dupilumab C26 ceramide did not correlate with reduction in AE severity in dupilumab-treated AE participants C26 ceramide did correlate with stratum corneum hydration |
| Endotype | Categories | Matrix | Metabolite Profile Relative to First Category |
|---|---|---|---|
| IgE | Mediated (extrinsic) vs. non-mediated (intrinsic) | Blood | ↑ Isopropanol [97] ↑ Threonine [97] ↑ Betanine [97] ↑ Creatinine [97] ↑ Dimethylamine [97] |
| FLG | Deficient vs. non-deficient | Skin | No broad differences in lipid composition [71,74] ↓ Arachidonic acid [74] ↓ Hydroxy fatty acid HETE-12 [74] |
| Blood | ↓ Glycerophospholipids [98] ↓ Sphingomyelin [98] ↓ Amino Acids [98] ↓ Acylcarnitines [98] ↑ Isopropanol [97] ↑ Iso-butyrate [97] ↑ Isoleucine [97] ↑ Tyramine [97] ↑ Histidine [97] ↑ Threonine [97] | ||
| Staphylococci | Present vs. absent | Skin | ↓ SCSFAs [71] ↑ Ceramide subspecies AS, ADS, NS and NDS [71] |
| S. aureus | Influenced vs. independent | Skin | ↓ FFA [75] ↓ Triglycerides [75] ↓ Long-chain ceramides [75] |
| Biological race | Asian vs. African vs. European | u.s. |
| References | Matrices | Target | Sample Size | Findings |
|---|---|---|---|---|
| [103] | PBMCs and plasma | PUFA | 20 AE 20 HE | ↓ n3-PUFA in AE vs. HE ↓ Linoleic acid in AE vs. HE ↓ 12-HETE in AE vs. HE ↑ Arachidonic acid in AE PBMCs vs. HE ↓ Arachidonic acid in AE plasma vs. HE |
| [100] | Serum | Metabolites in Biocrates Absolute IDQ® P180 kit and UT | 25 AE 24 HE 13 AE 15 HE | ↓ Total acylcarnitines in AE vs. HE ↓ Phosphatidylcholines in AE vs. HE No PCA separation between AE as compared to HE |
| [98] | Serum | Glycerophospholipids, acylcarnitines, sphingomyelins, amino acids, carbohydrates | 20 AE | Only FLG (+/+) had response to drug targeting of IgE ↓ Glycerophospholipids in FLG (+/+) vs. FLG mutant ↓ Sphingomyelin in FLG (+/+) vs. FLG mutant ↓ Amino acids in FLG (+/+) vs. FLG mutant ↓ Acylcarnitines in FLG (+/+) vs. FLG mutant |
| [97] | Plasma | UT | 58 AE 23 HE | Distinct metabolite differences for endotypes: FLG mutant and high IgE Iso-butyrate, isoleucine, tyramine, histidine, threonine, and isopropanol are associated with FLG mutation Isopropanol is associated with IgE levels |
| [102] | Sebum | Lipid mediators, non-esterified and total fatty acid forms | 11 AE 9 HE | Sweat and sebum highly overlap in detection but concentrations of metabolites were typically higher in sebum. |
| [72] | Serum | FFA, eicosanoids and docosanoids | 6 AE 6 HE | No change in arachidonic acid in AE vs. HE |
| [101] | Serum | Amine/phenol sub-metabolomes | 9 AE 10 Dock8-deficient 33 HE | AE and DOCK8-deficient individuals have unique metabolomics profiles ↑ 3-hydroxyanthranilic acid in DOCK8-deficient vs. HE and AE ↑ Aspartic acid in DOCK8-deficient vs. HE ↓ Hypo-taurine in DOCK8-deficient vs. AE ↓ Glycyl-phenylalanine in DOCK8-deficient vs. AE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afghani, J.; Traidl-Hoffmann, C.; Schmitt-Kopplin, P.; Reiger, M.; Mueller, C. An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study. Int. J. Mol. Sci. 2022, 23, 8791. https://doi.org/10.3390/ijms23158791
Afghani J, Traidl-Hoffmann C, Schmitt-Kopplin P, Reiger M, Mueller C. An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study. International Journal of Molecular Sciences. 2022; 23(15):8791. https://doi.org/10.3390/ijms23158791
Chicago/Turabian StyleAfghani, Jamie, Claudia Traidl-Hoffmann, Philippe Schmitt-Kopplin, Matthias Reiger, and Constanze Mueller. 2022. "An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study" International Journal of Molecular Sciences 23, no. 15: 8791. https://doi.org/10.3390/ijms23158791
APA StyleAfghani, J., Traidl-Hoffmann, C., Schmitt-Kopplin, P., Reiger, M., & Mueller, C. (2022). An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study. International Journal of Molecular Sciences, 23(15), 8791. https://doi.org/10.3390/ijms23158791







