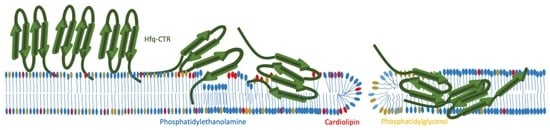
Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq
Abstract
1. Introduction
2. Results
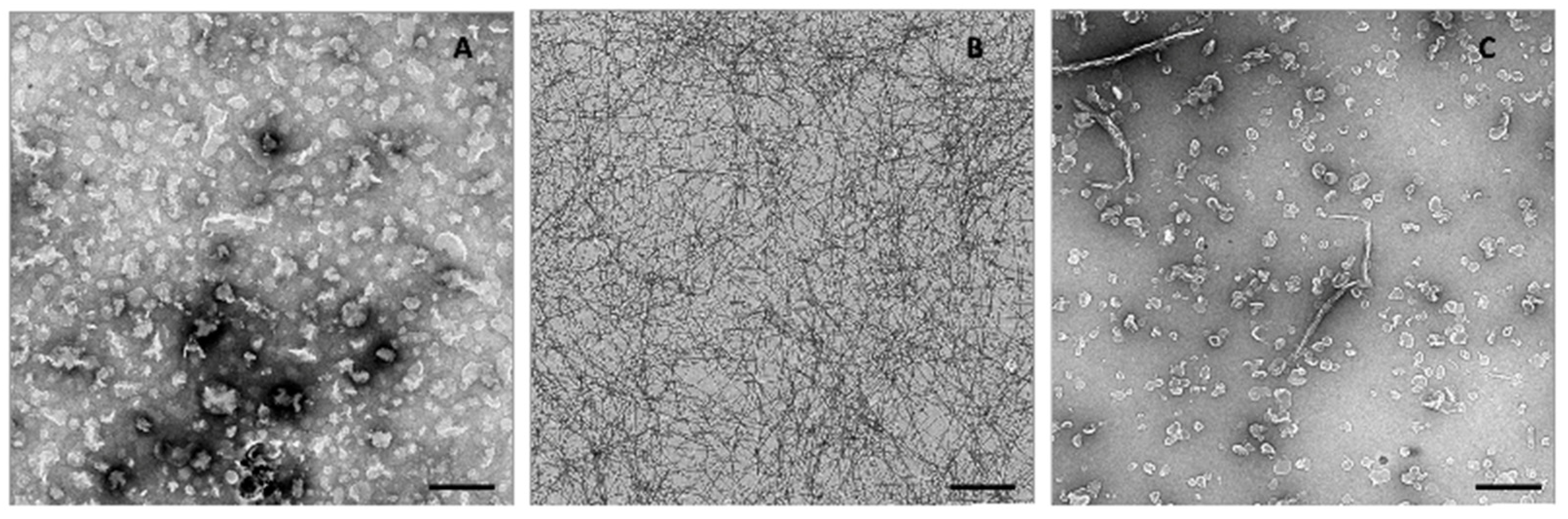
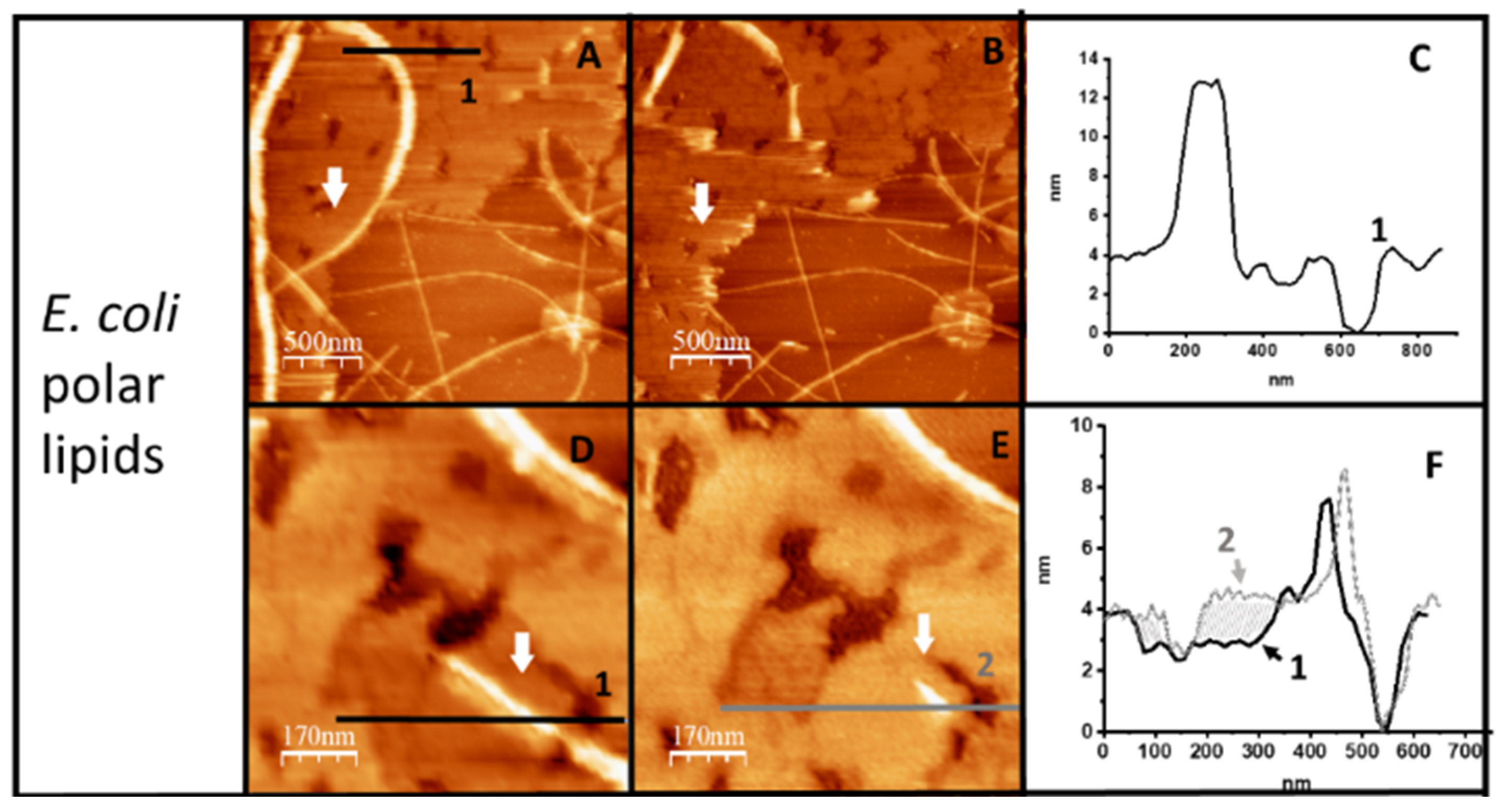
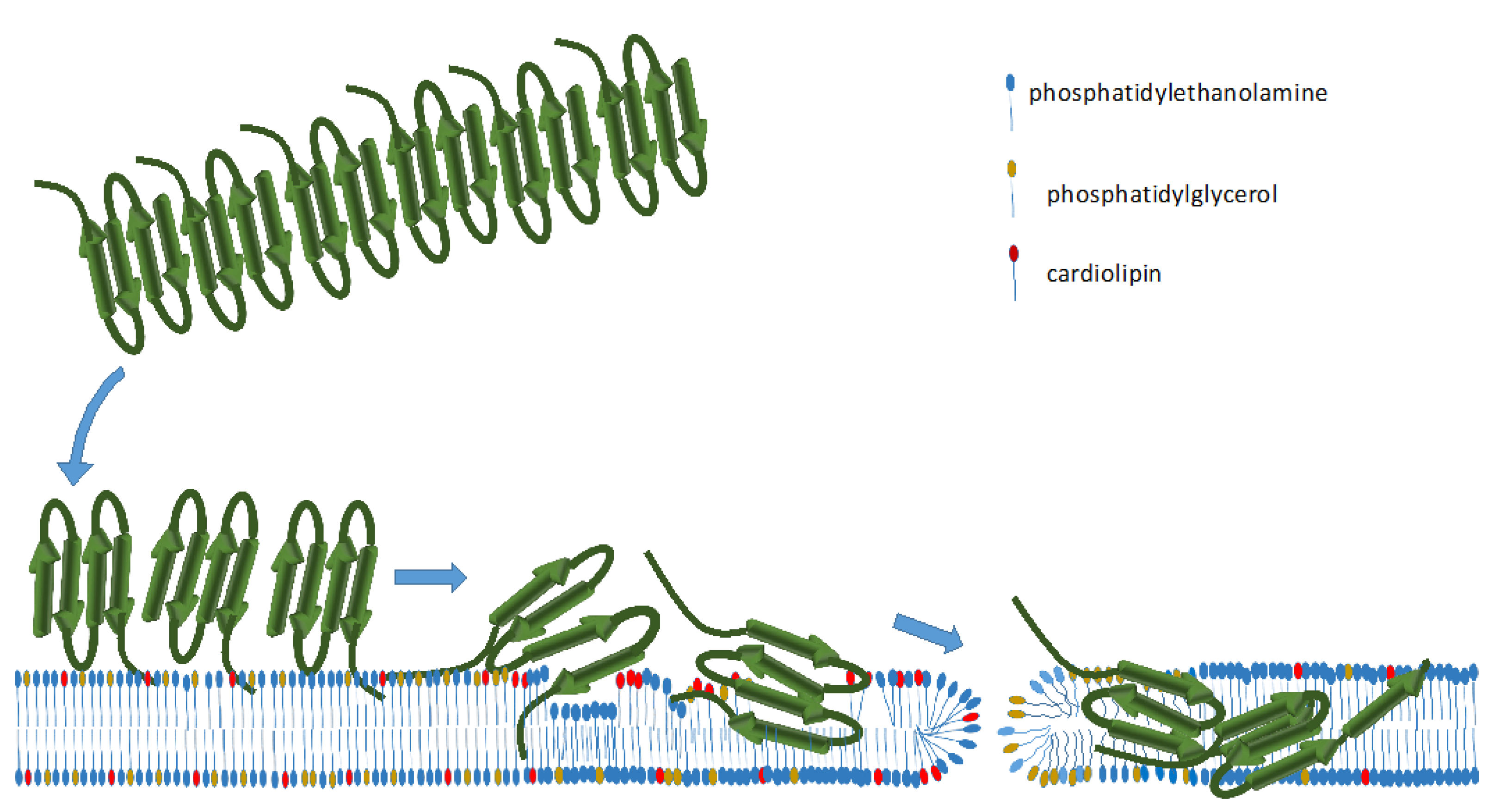
2.1. Hfq-CTR Filaments Disrupt Membranes while, Conversely, Membranes Disrupt Hfq-CTR Filaments
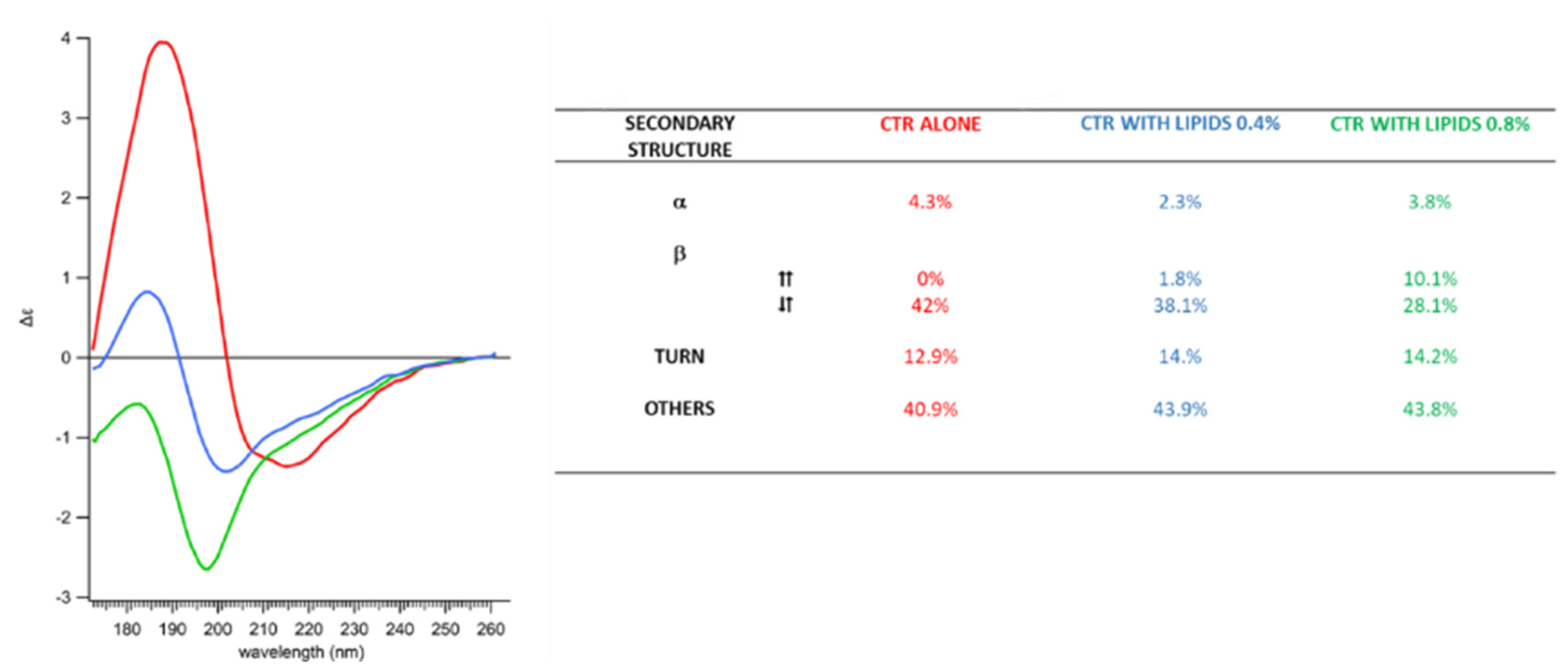
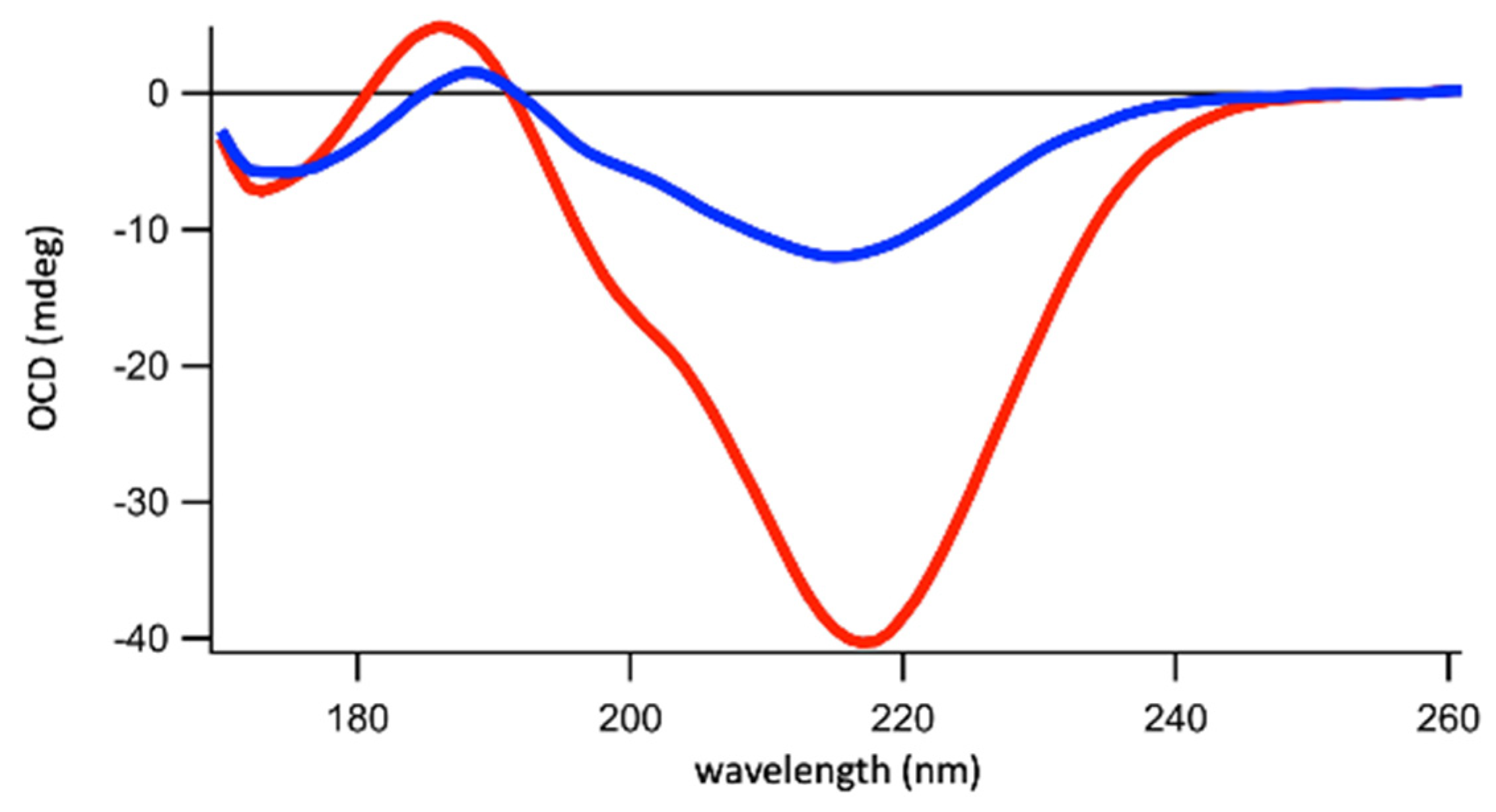
2.2. Membranes Break Hfq-CTR Amyloid-like Structure
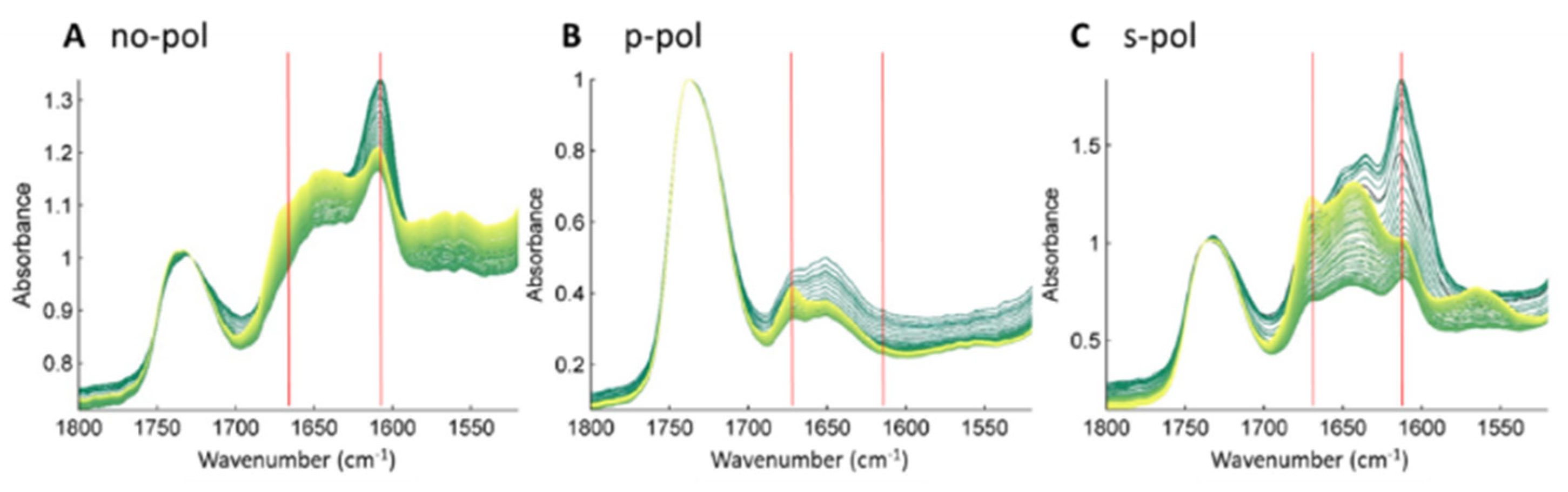
2.3. Membrane-Induced Fibril-Disruption Results in the Insertion of β-Sheets
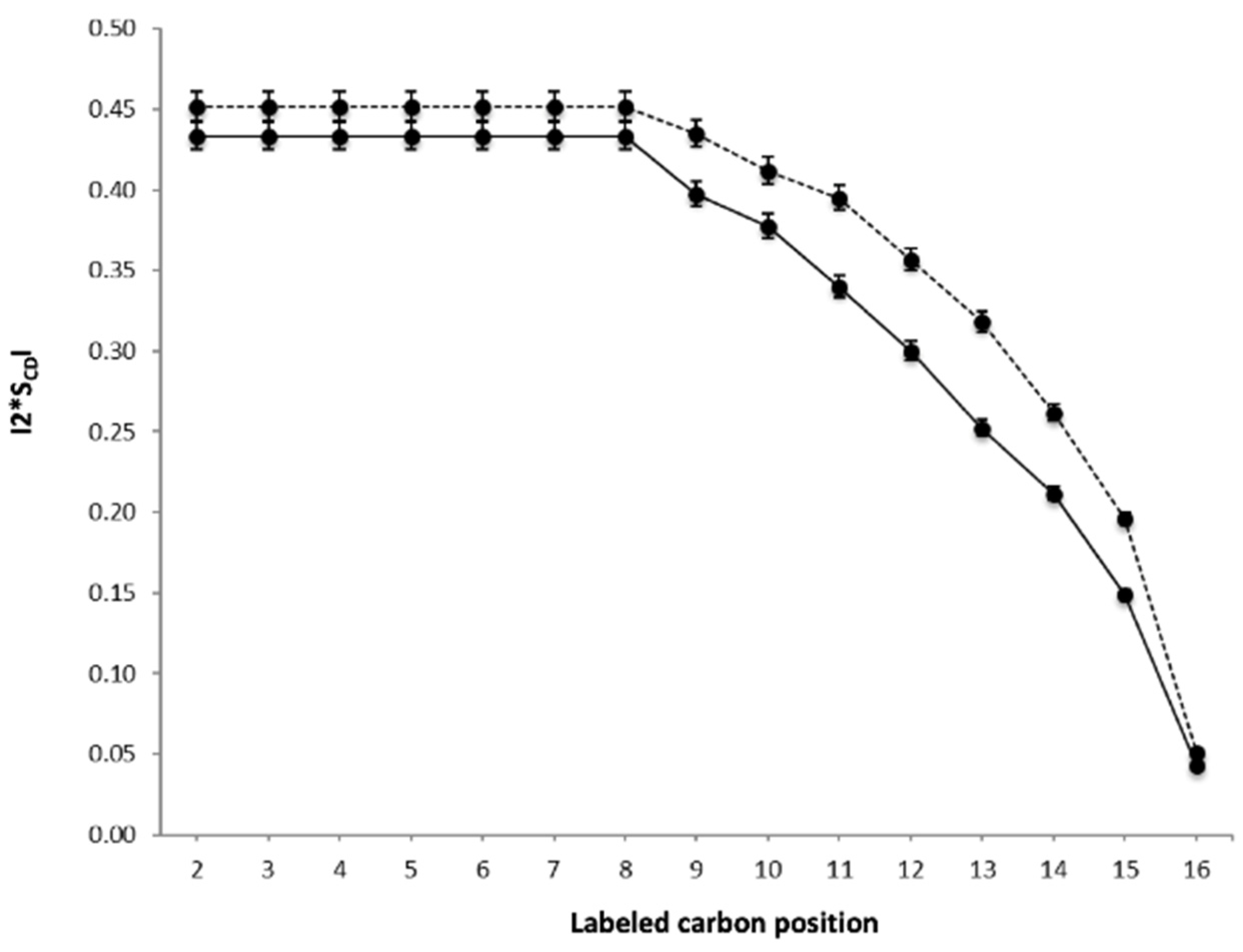
2.4. Dynamics of the Lipid Membrane Is Affected by Hfq-CTR
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Hfq Proteins
4.2. Preparation of Small Unilamellar Vesicles (SUVs)
4.3. AFM Imaging in Solution
4.4. FTIR
4.5. Transmission Electron Microscopy (TEM)
4.6. Synchrotron Radiation Circular Dichroism (SRCD) and Orientated Circular Dichroism (OCD)
4.7. Solid-State NMR Spectroscopy
4.8. Sample Preparation for Solid-State NMR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brennan, R.G.; Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007, 10, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Wassarman, K.M.; Rosenow, C.; Tjaden, B.C.; Storz, G.; Gottesman, S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003, 50, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef]
- Storz, G.; Opdyke, J.A.; Zhang, A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004, 7, 140–144. [Google Scholar] [CrossRef]
- Zambrano, N.; Guichard, P.P.; Bi, Y.; Cayrol, B.; Marco, S.; Arluison, V. Involvement of HFq protein in the post-transcriptional regulation of E. coli bacterial cytoskeleton and cell division proteins. Cell Cycle 2009, 8, 2470–2472. [Google Scholar] [CrossRef]
- Bardill, J.P.; Zhao, X.; Hammer, B.K. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 2011, 80, 1381–1394. [Google Scholar] [CrossRef]
- Gripenland, J.; Netterling, S.; Loh, E.; Tiensuu, T.; Toledo-Arana, A.; Johansson, J. RNAs: Regulators of bacterial virulence. Nat. Rev. Microbiol. 2010, 8, 857–866. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S.; Storz, G. Modulating the outer membrane with small RNAs. Genes Dev. 2006, 20, 2338–2348. [Google Scholar] [CrossRef]
- Vogt, S.L.; Raivio, T.L. Hfq reduces envelope stress by controlling expression of envelope-localized proteins and protein complexes in enteropathogenic Escherichia coli. Mol. Microbiol. 2014, 92, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Orans, J.; Kovach, A.R.; Hoff, K.E.; Horstmann, N.M.; Brennan, R.G. Crystal structure of an Escherichia coli Hfq Core (residues 2-69)-DNA complex reveals multifunctional nucleic acid binding sites. Nucleic Acids Res. 2020, 48, 3987–3997. [Google Scholar] [CrossRef] [PubMed]
- Seraphin, B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. Embo J. 1995, 14, 2089–2098. [Google Scholar] [CrossRef]
- Wilusz, C.J.; Wilusz, J. Lsm proteins and Hfq: Life at the 3’ end. RNA Biol. 2013, 10, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Pearson, R.F.; Moller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: A bacterial Sm-like protein. Embo J. 2002, 21, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef] [PubMed]
- Updegrove, T.B.; Correia, J.J.; Galletto, R.; Bujalowski, W.; Wartell, R.M. E. coli DNA associated with isolated Hfq interacts with Hfq’s distal surface and C-terminal domain. Biochim. Biophys. Acta 2010, 1799, 588–596. [Google Scholar] [CrossRef]
- Dimastrogiovanni, D.; Frohlich, K.S.; Bandyra, K.J.; Bruce, H.A.; Hohensee, S.; Vogel, J.; Luisi, B.F. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife 2014, 3, e05375. [Google Scholar] [CrossRef]
- Arluison, V.; Folichon, M.; Marco, S.; Derreumaux, P.; Pellegrini, O.; Seguin, J.; Hajnsdorf, E.; Regnier, P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004, 271, 1258–1265. [Google Scholar] [CrossRef]
- Wen, B.; Wang, W.; Zhang, J.; Gong, Q.; Shi, Y.; Wu, J.; Zhang, Z. Structural and dynamic properties of the C-terminal region of the Escherichia coli RNA chaperone Hfq: Integrative experimental and computational studies. Phys. Chem. Chem. Phys. 2017, 19, 21152–21164. [Google Scholar] [CrossRef]
- Nikulin, A.; Stolboushkina, E.; Perederina, A.; Vassilieva, I.; Blaesi, U.; Moll, I.; Kachalova, G.; Yokoyama, S.; Vassylyev, D.; Garber, M.; et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Sauer, E.; Weichenrieder, O. Structural basis for RNA 3’-end recognition by Hfq. Proc. Natl. Acad. Sci. USA 2011, 108, 13065–13070. [Google Scholar] [CrossRef] [PubMed]
- El Hamoui, O.; Yadav, I.; Radiom, M.; Wien, F.; Berret, J.-F.; van der Maarel, J.R.C.; Arluison, V. Interactions between DNA and the Hfq Amyloid-like Region Trigger a Viscoelastic Response. Biomacromolecules 2020, 21, 3668–3677. [Google Scholar] [CrossRef] [PubMed]
- Wien, F.; Martinez, D.; Le Brun, E.; Jones, N.C.; Vronning Hoffmann, S.; Waeytens, J.; Berbon, M.; Habenstein, B.; Arluison, V. The Bacterial Amyloid-Like Hfq Promotes In Vitro DNA Alignment. Microorganisms 2019, 7, 639. [Google Scholar] [CrossRef]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef]
- Jiang, K.; Zhang, C.; Guttula, D.; Liu, F.; van Kan, J.A.; Lavelle, C.; Kubiak, K.; Malabirade, A.; Lapp, A.; Arluison, V.; et al. Effects of Hfq on the conformation and compaction of DNA. Nucleic Acids Res. 2015, 43, 4332–4341. [Google Scholar] [CrossRef]
- Santiago-Frangos, A.; Kavita, K.; Schu, D.J.; Gottesman, S.; Woodson, S.A. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. USA 2016, 113, E6089–E6096. [Google Scholar] [CrossRef]
- Kavita, K.; Zhang, A.; Tai, C.H.; Majdalani, N.; Storz, G.; Gottesman, S. Multiple in vivo roles for the C-terminal domain of the RNA chaperone Hfq. Nucleic Acids Res. 2022, 50, 1718–1733. [Google Scholar] [CrossRef]
- Turbant, F.; Wu, P.; Wien, F.; Arluison, V. The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology 2021, 10, 900. [Google Scholar] [CrossRef]
- Beich-Frandsen, M.; Vecerek, B.; Konarev, P.V.; Sjoblom, B.; Kloiber, K.; Hammerle, H.; Rajkowitsch, L.; Miles, A.J.; Kontaxis, G.; Wallace, B.A.; et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011, 39, 4900–4915. [Google Scholar] [CrossRef] [PubMed]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trepout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35, e00190. [Google Scholar] [CrossRef]
- Arluison, V.; Mura, C.; Guzman, M.R.; Liquier, J.; Pellegrini, O.; Gingery, M.; Regnier, P.; Marco, S. Three-dimensional Structures of Fibrillar Sm Proteins: Hfq and Other Sm-like Proteins. J. Mol. Biol. 2006, 356, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Partouche, D.; Militello, V.; Gomez-Zavaglia, A.; Wien, F.; Sandt, C.; Arluison, V. In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study. Pathogens 2019, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Partouche, D.; Mathurin, J.; Malabirade, A.; Marco, S.; Sandt, C.; Arluison, V.; Deniset, A.; Trepout, S. Correlative infrared nanospectroscopy and transmission electron microscopy to investigate nanometric amyloid fibrils: Prospects and challenges. J. Microsc. 2019, 274, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Taghbalout, A.; Yang, Q.; Arluison, V. The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J. 2014, 458, 11–22. [Google Scholar] [CrossRef]
- Mura, C.; Kozhukhovsky, A.; Gingery, M.; Phillips, M.; Eisenberg, D. The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs). Protein Sci. 2003, 12, 832–847. [Google Scholar] [CrossRef]
- Malabirade, A.; Morgado-Brajones, J.; Trepout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 10724. [Google Scholar] [CrossRef]
- Arluison, V.; Taghbalout, A. Cellular localization of RNA degradation and processing components in Escherichia coli. Methods Mol. Biol. 2015, 1259, 87–101. [Google Scholar]
- Taghbalout, A.; Rothfield, L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. USA 2007, 104, 1667–1672. [Google Scholar] [CrossRef]
- Park, S.; Prevost, K.; Heideman, E.M.; Carrier, M.C.; Azam, M.S.; Reyer, M.A.; Liu, W.; Masse, E.; Fei, J. Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. Elife 2021, 10, e64207. [Google Scholar] [CrossRef] [PubMed]
- Iosub, I.A.; van Nues, R.W.; McKellar, S.W.; Nieken, K.J.; Marchioretto, M.; Sy, B.; Tree, J.J.; Viero, G.; Granneman, S. Hfq CLASH uncovers sRNA-target interaction networks linked to nutrient availability adaptation. Elife 2020, 9, e54655. [Google Scholar] [CrossRef] [PubMed]
- Kannaiah, S.; Livny, J.; Amster-Choder, O. Spatiotemporal Organization of the E. coli Transcriptome: Translation Independence and Engagement in Regulation. Mol. Cell 2019, 76, 574–589.e7. [Google Scholar] [CrossRef]
- Eason, I.R.; Kaur, H.P.; Alexander, K.A.; Sukhodolets, M.V. Growth phase-specific changes in the composition of E. coli transcription complexes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1109, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, K.S.; Gottesman, S. Small Regulatory RNAs in the Enterobacterial Response to Envelope Damage and Oxidative Stress. Microbiol. Spectr. 2018, 6, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M. Bacterial communication through membrane vesicles. Biosci. Biotechnol. Biochem. 2019, 83, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, A.; Upadhyaya, B.B.; Fritz, J.V.; Heintz-Buschart, A.; Desai, M.S.; Yusuf, D.; Huang, D.; Baumuratov, A.; Wang, K.; Galas, D.; et al. The extracellular RNA complement of Escherichia coli. Microbiologyopen 2015, 4, 252–266. [Google Scholar] [CrossRef]
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A.; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016, 12, e1005672. [Google Scholar] [CrossRef]
- Partouche, D.; Turbant, F.; El Hamoui, O.; Campidelli, C.; Bombled, M.; Trepout, S.; Wien, F.; Arluison, V. Epigallocatechin Gallate Remodelling of Hfq Amyloid-Like Region Affects Escherichia coli Survival. Pathogens 2018, 7, 95. [Google Scholar] [CrossRef]
- Turbant, F.; Partouche, D.; El Hamoui, O.; Trepout, S.; Legoubey, T.; Wien, F.; Arluison, V. Apomorphine Targets the Pleiotropic Bacterial Regulator Hfq. Antibiotics 2021, 10, 10030257. [Google Scholar] [CrossRef]
- Goormaghtigh, E.; Cabiaux, V.; Ruysschaert, J.M. Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy. II. Experimental aspects, side chain structure, and H/D exchange. Sub-Cell. Biochem. 1994, 23, 363–403. [Google Scholar]
- Ruysschaert, J.-M.; Raussens, V. ATR-FTIR Analysis of Amyloid Proteins. Methods Mol. Biol. 2018, 1777, 69–81. [Google Scholar] [PubMed]
- Waeytens, J.; Mathurin, J.; Deniset-Besseau, A.; Arluison, V.; Bousset, L.; Rezaei, H.; Raussens, V.; Dazzi, A. Probing amyloid fibril secondary structures by infrared nanospectroscopy: Experimental and theoretical considerations. Analyst 2020, 146, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Sarroukh, R.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta 2013, 1828, 2328–2338. [Google Scholar] [CrossRef]
- Burck, J.; Wadhwani, P.; Fanghanel, S.; Ulrich, A.S. Oriented Circular Dichroism: A Method to Characterize Membrane-Active Peptides in Oriented Lipid Bilayers. Acc. Chem. Res. 2016, 49, 184–192. [Google Scholar] [CrossRef]
- Yao, H.; Hong, M. Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR. J. Am. Chem. Soc. 2014, 136, 2611–2624. [Google Scholar] [CrossRef]
- Davis, J.H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta 1983, 737, 117–171. [Google Scholar] [CrossRef]
- Vahdat Shariat Panahi, A.; Hultman, P.; Ollinger, K.; Westermark, G.T.; Lundmark, K. Lipid membranes accelerate amyloid formation in the mouse model of AA amyloidosis. Amyloid 2019, 26, 34–44. [Google Scholar] [CrossRef]
- Banerjee, S.; Hashemi, M.; Zagorski, K.; Lyubchenko, Y.L. Interaction of Abeta42 with Membranes Triggers the Self-Assembly into Oligomers. Int. J. Mol. Sci. 2020, 21, 1129. [Google Scholar] [CrossRef]
- Canale, C.; Oropesa-Nunez, R.; Diaspro, A.; Dante, S. Amyloid and membrane complexity: The toxic interplay revealed by AFM. Semin. Cell Dev. Biol. 2018, 73, 82–94. [Google Scholar] [CrossRef]
- Gal, N.; Morag, A.; Kolusheva, S.; Winter, R.; Landau, M.; Jelinek, R. Lipid bilayers significantly modulate cross-fibrillation of two distinct amyloidogenic peptides. J. Am. Chem. Soc. 2013, 135, 13582–13589. [Google Scholar] [CrossRef] [PubMed]
- Feuillie, C.; Lambert, E.; Ewald, M.; Azouz, M.; Henry, S.; Marsaudon, S.; Cullin, C.; Lecomte, S.; Molinari, M. High Speed AFM and NanoInfrared Spectroscopy Investigation of Abeta1-42 Peptide Variants and Their Interaction With POPC/SM/Chol/GM1 Model Membranes. Front. Mol. Biosci. 2020, 7, 571696. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Almaguer, S.; Mell, M.; Almendro-Vedia, V.G.; Calero, M.; Robledo-Sanchez, K.C.M.; Ruiz-Suarez, C.; Alarcon, T.; Barrio, R.A.; Hernandez-Machado, A.; Monroy, F. Membrane rigidity regulates E. coli proliferation rates. Sci. Rep. 2022, 12, 933. [Google Scholar] [CrossRef]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef]
- Wang, G.; Galli, T. Reciprocal link between cell biomechanics and exocytosis. Traffic 2018, 19, 741–749. [Google Scholar] [CrossRef]
- Munishkina, L.A.; Cooper, E.M.; Uversky, V.N.; Fink, A.L. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J. Mol. Recognit. 2004, 17, 456–464. [Google Scholar] [CrossRef]
- Dauros-Singorenko, P.; Blenkiron, C.; Phillips, A.; Swift, S. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 2018, 365, fny023. [Google Scholar] [CrossRef]
- Masse, E.; Escorcia, F.E.; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003, 17, 2374–2383. [Google Scholar] [CrossRef]
- Schu, D.J.; Zhang, A.; Gottesman, S.; Storz, G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015, 34, 2557–2573. [Google Scholar] [CrossRef]
- Bianco, C.M.; Frohlich, K.S.; Vanderpool, C.K. Bacterial Cyclopropane Fatty Acid Synthase mRNA Is Targeted by Activating and Repressing Small RNAs. J. Bacteriol. 2019, 201, e00461-19. [Google Scholar] [CrossRef] [PubMed]
- Horcas, I.; Fernandez, R.; Gómez-Rodríguez, M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef] [PubMed]
- Refregiers, M.; Wien, F.; Ta, H.P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron Radiat. 2012, 19, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Wien, F.; Wallace, B.A. Calcium fluoride micro cells for synchrotron radiation circular dichroism spectroscopy. Appl. Spectrosc. 2005, 59, 1109–1113. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. CDtoolX, a downloadable software package for processing and analyses of circular dichroism spectroscopic data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef]
- Wien, F.; Paternostre, M.; Gobeaux, F.; Artzner, F.; Refregiers, M. Calibration and quality assurance procedures at the far UV linear and circular dichroism experimental station DISCO. J. Phys. Conf. Ser. 2013, 425, 122014. [Google Scholar] [CrossRef]
- Chen, F.Y.; Lee, M.T.; Huang, H.W. Sigmoidal concentration dependence of antimicrobial peptide activities: A case study on alamethicin. Biophys. J. 2002, 82, 908–914. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turbant, F.; Waeytens, J.; Campidelli, C.; Bombled, M.; Martinez, D.; Grélard, A.; Habenstein, B.; Raussens, V.; Velez, M.; Wien, F.; et al. Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq. Int. J. Mol. Sci. 2022, 23, 8739. https://doi.org/10.3390/ijms23158739
Turbant F, Waeytens J, Campidelli C, Bombled M, Martinez D, Grélard A, Habenstein B, Raussens V, Velez M, Wien F, et al. Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq. International Journal of Molecular Sciences. 2022; 23(15):8739. https://doi.org/10.3390/ijms23158739
Chicago/Turabian StyleTurbant, Florian, Jehan Waeytens, Camille Campidelli, Marianne Bombled, Denis Martinez, Axelle Grélard, Birgit Habenstein, Vincent Raussens, Marisela Velez, Frank Wien, and et al. 2022. "Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq" International Journal of Molecular Sciences 23, no. 15: 8739. https://doi.org/10.3390/ijms23158739
APA StyleTurbant, F., Waeytens, J., Campidelli, C., Bombled, M., Martinez, D., Grélard, A., Habenstein, B., Raussens, V., Velez, M., Wien, F., & Arluison, V. (2022). Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq. International Journal of Molecular Sciences, 23(15), 8739. https://doi.org/10.3390/ijms23158739