Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients
Abstract
:1. Introduction
2. Results
2.1. Dilution Bank Preparation
2.2. Limit of Blank (LOB) and Limit of Detection (LOD) Establishment
2.3. PNA Clamp TaqMan Assay and ddPCR Results: Comparison
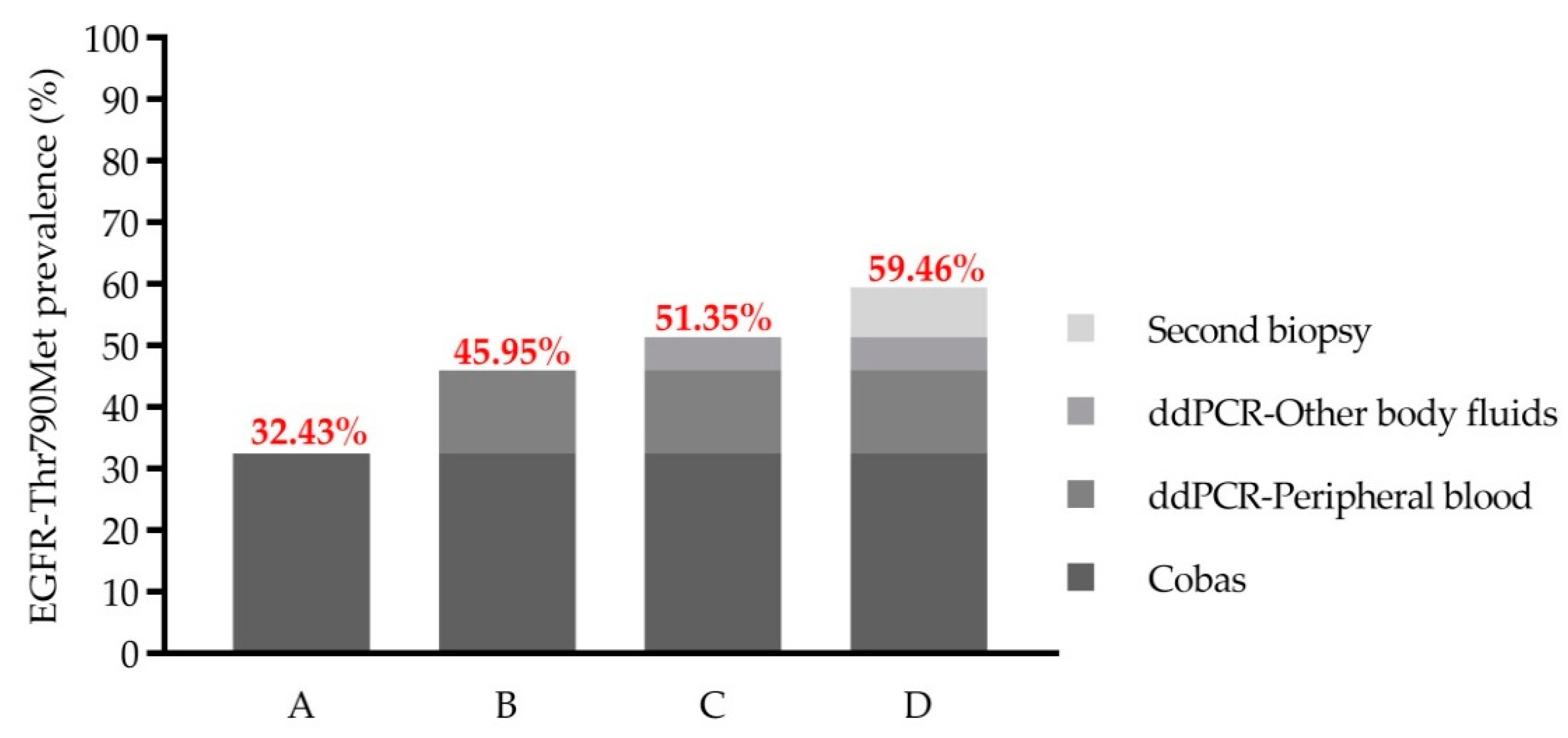
2.4. EGFR-Thr790Met Prevalence in Pretreated Stage IV NSCLC Association with Patients’ Characteristics
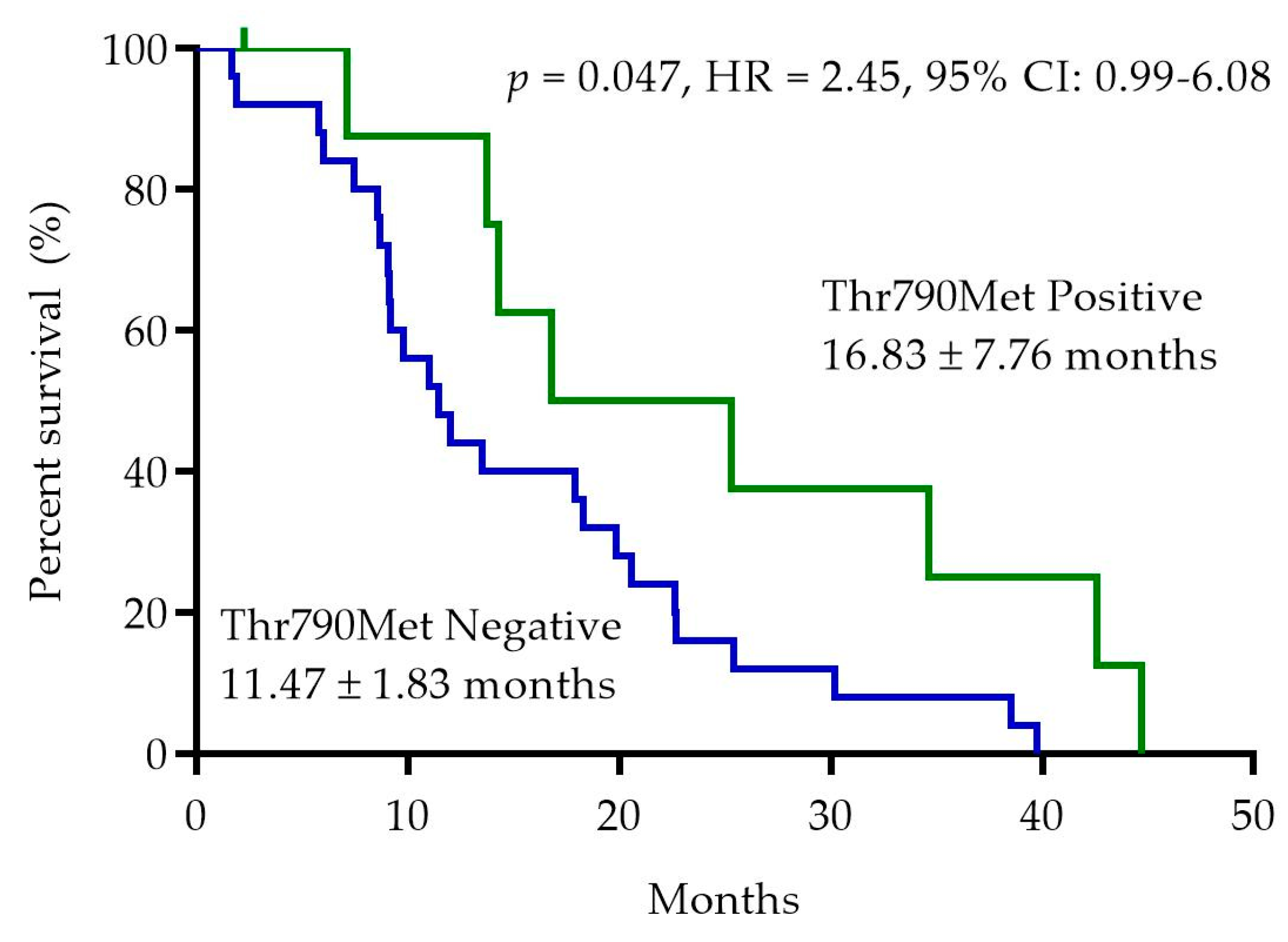
2.5. EGFR-Thr790Met Pretreatment Status and Progression-Free Survival (PFS)
2.6. EGFR-Thr790Met Detection in LB at the Time of Disease Progression
2.7. Early Detection of EGFR-Thr790Met in LB by ddPCR during Treatment Follow-Up
3. Discussion
4. Materials and Methods
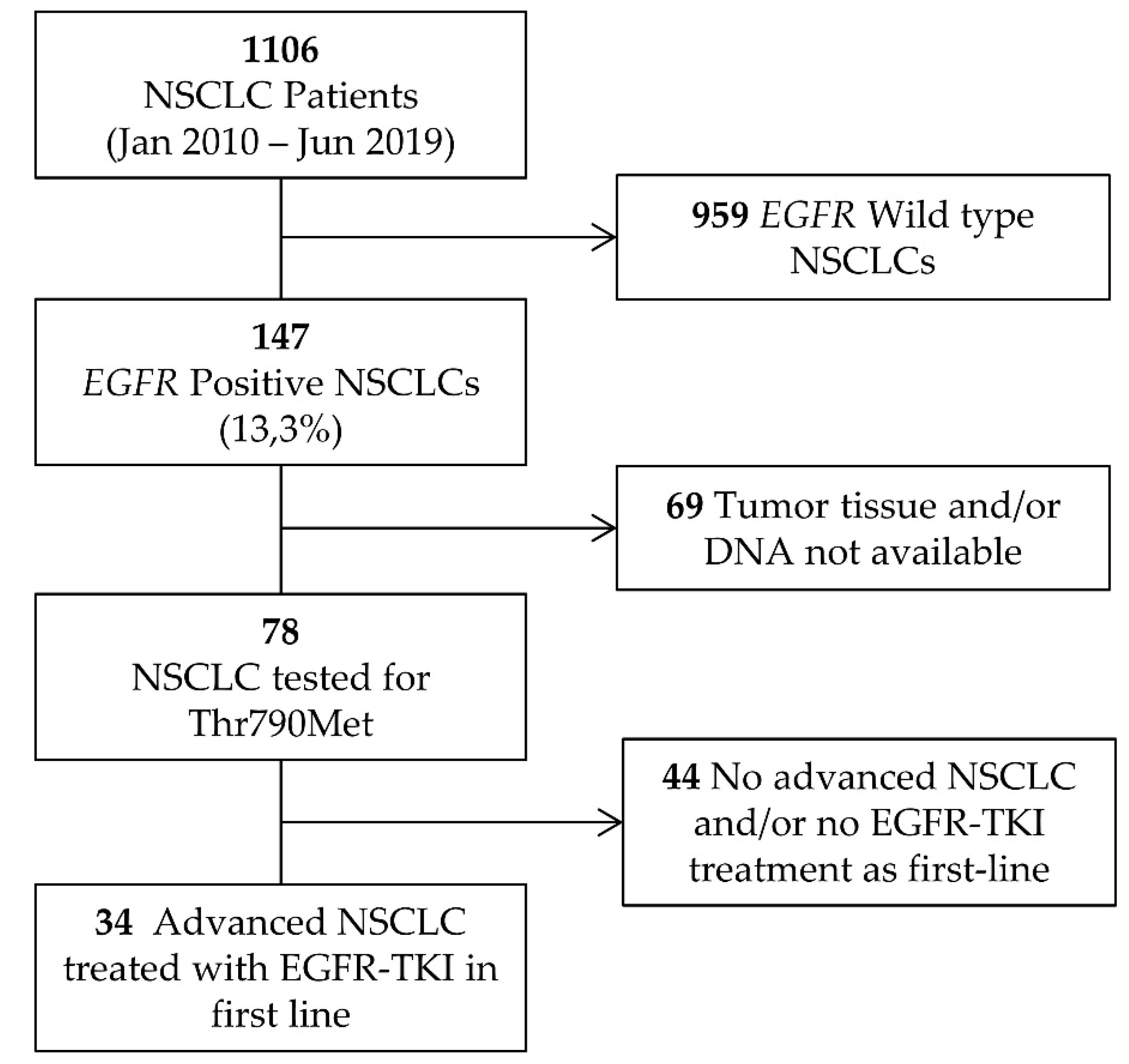
4.1. Patients
4.2. Reference Materials
4.3. DNA Extraction Plasma Isolation
4.4. EGFR Mutation Screening
4.5. PNA Clamp TaqMan Assay
4.6. Digital Droplet PCR (ddPCR)
4.7. Limit of Blank (LOB) and Limit of Detection (LOD)
4.8. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Graham, R.P.; Treece, A.L.; Lindeman, N.I.; Vasalos, P.; Shan, M.; Jennings, L.J.; Rimm, D.L. Worldwide Frequency of Commonly Detected EGFR Mutations. Arch. Pathol. Lab. Med. 2018, 142, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Takeda, M.; Nakagawa, K. First-and Second-Generation EGFR-TKIs Are All Replaced to Osimertinib in Chemo-Naive EGFR Mutation-Positive Non-Small Cell Lung Cancer? Int. J. Mol. Sci. 2019, 20, 146. [Google Scholar]
- Ohashi, K.; Maruvka, Y.E.; Michor, F.; Pao, W. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor-Resistant Disease. J. Clin. Oncol. 2013, 31, 1070. [Google Scholar]
- Nguyen, K.S.H.; Kobayashi, S.; Costa, D.B. Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancers Dependent on the Epidermal Growth Factor Receptor Pathway. Clin. Lung Cancer 2009, 10, 281–289. [Google Scholar]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005, 2, e73. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR Mutation and Resistance of Non-Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef]
- Yu, H.A.; Arcila, M.E.; Hellmann, M.D.; Kris, M.G.; Ladanyi, M.; Riely, G.J. Poor Response to Erlotinib in Patients with Tumors Containing Baseline EGFR T790M Mutations Found by Routine Clinical Molecular Testing. Ann. Oncol. 2014, 25, 423–428. [Google Scholar] [CrossRef]
- Kohsaka, S.; Petronczki, M.; Solca, F.; Maemondo, M. Tumor Clonality and Resistance Mechanisms in EGFR Mutation-Positive Non-Small-Cell Lung Cancer: Implications for Therapeutic Sequencing. Future Oncol. 2019, 15, 637–652. [Google Scholar]
- Takeda, M.; Sakai, K.; Hayashi, H.; Tanaka, K.; Haratani, K.; Takahama, T.; Kato, R.; Yonesaka, K.; Nishio, K.; Nakagawa, K. Impact of Coexisting Gene Mutations in EGFR-Mutated Non-Small Cell Lung Cancer before Treatment on EGFR T790M Mutation Status after EGFR-TKIs. Lung Cancer 2020, 139, 28–34. [Google Scholar] [CrossRef]
- Gibson, N.J. The Use of Real-Time PCR Methods in DNA Sequence Variation Analysis. Clin. Chim. Acta 2006, 363, 32–47. [Google Scholar] [CrossRef]
- D’Arcy, C.; Cummings, N.J.; Tra, T.; McLean, C. Assessing Minimal Tumour Cell Requirements and DNA Input for Accurate Molecular Analysis of FFPE Tissues Using MALDI-TOF MS. Pathology 2015, 47, S58. [Google Scholar] [CrossRef]
- Kobunai, T.; Watanabe, T.; Yamamoto, Y.; Eshima, K. The Frequency of KRAS Mutation Detection in Human Colon Carcinoma Is Influenced by the Sensitivity of Assay Methodology: A Comparison between Direct Sequencing and Real-Time PCR. Biochem. Biophys. Res. Commun. 2010, 395, 158–162. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Mamon, H.; Kulke, M.H.; Berbeco, R.; Makrigiorgos, G.M. Replacing PCR with COLD-PCR Enriches Variant DNA Sequences and Redefines the Sensitivity of Genetic Testing. Nat. Med. 2008, 14, 579–584. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, C.W.; Shao, Y.; Wang, H.T.; Wu, Y.F.; Song, Y.Y.; Li, X.B.; Zhang, Z.; Wang, W.J.; Li, L.Q.; et al. Comparison of Droplet Digital PCR and Conventional Quantitative PCR for Measuring EGFR Gene Mutation. Exp. Ther. Med. 2015, 9, 1383–1388. [Google Scholar] [CrossRef] [Green Version]
- Lettig, L.; Sahnane, N.; Pepe, F.; Cerutti, R.; Albeni, C.; Franzi, F.; Veronesi, G.; Ogliari, F.; Pastore, A.; Tuzi, A.; et al. EGFR T790M Detection Rate in Lung Adenocarcinomas at Baseline Using Droplet Digital PCR and Validation by Ultra-Deep next Generation Sequencing. Transl. Lung Cancer Res. 2019, 8, 584–592. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Mazieres, J.; Senal, R.; Rouquette, I.; Quantin, X.; Pujol, J.L.; Roch, B.; Bouidioua, A.; Godreuil, S.; Coyaud, E.; et al. Ultra-Sensitive EGFRT790M Detection as an Independent Prognostic Marker for Lung Cancer Patients Harboring EGFRdel19 Mutations and Treated with First-Generation TKIs. Clin. Cancer Res. 2019, 25, 4280–4289. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Kawaguchi, T.; Isa, S.I.; Ando, M.; Tamiya, A.; Kubo, A.; Saka, H.; Takeo, S.; Adachi, H.; Tagawa, T.; et al. Ultra-Sensitive Detection of the Pretreatment EGFR T790M Mutation in Non-Small Cell Lung Cancer Patients with an EGFR-Activating Mutation Using Droplet Digital PCR. Clin. Cancer Res. 2015, 21, 3552–3560. [Google Scholar] [CrossRef] [Green Version]
- Su, K.Y.; Chen, H.Y.; Li, K.C.; Kuo, M.L.; Yang, J.C.H.; Chan, W.K.; Ho, B.C.; Chang, G.C.; Shih, J.Y.; Yu, S.L.; et al. Pretreatment Epidermal Growth Factor Receptor (EGFR) T790M Mutation Predicts Shorter EGFR Tyrosine Kinase Inhibitor Response Duration in Patients with Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Lee, G.K.; Lee, Y.S.; Zhang, W.; Hwang, J.A.; Nam, B.H.; Kim, S.H.; Kim, J.H.; Yun, T.; Han, J.Y.; et al. Clinical Outcome According to the Level of Preexisting Epidermal Growth Factor Receptor T790M Mutation in Patients with Lung Cancer Harboring Sensitive Epidermal Growth Factor Receptor Mutations. Cancer 2014, 120, 2090–2098. [Google Scholar] [CrossRef]
- Rosell, R.; Molina, M.A.; Costa, C.; Simonetti, S.; Gimenez-Capitan, A.; Bertran-Alamillo, J.; Mayo, C.; Moran, T.; Mendez, P.; Cardenal, F.; et al. Pretreatment EGFR T790M Mutation and BRCA1 MRNA Expression in Erlotinib-Treated Advanced Non-Small-Cell Lung Cancer Patients with EGFR Mutations. Clin. Cancer Res. 2011, 17, 1160–1168. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.; Molina, M.A.; Drozdowskyj, A.; Gimeńez-Capitán, A.; Bertran-Alamillo, J.; Karachaliou, N.; Gervais, R.; Massuti, B.; Wei, J.; Moran, T.; et al. The Impact of EGFR T790M Mutations and BIM MRNA Expression on Outcome in Patients with EGFR-Mutant NSCLC Treated with Erlotinib or Chemotherapy in the Randomized Phase III EURTAC Trial. Clin. Cancer Res. 2014, 20, 2001–2010. [Google Scholar] [CrossRef] [Green Version]
- Tatematsu, T.; Okuda, K.; Suzuki, A.; Oda, R.; Sakane, T.; Kawano, O.; Haneda, H.; Moriyama, S.; Sasaki, H.; Nakanishi, R. The Detectability of the Pretreatment EGFR T790M Mutations in Lung Adenocarcinoma Using CAST-PCR and Digital PCR. J. Thorac. Dis. 2017, 9, 2397–2403. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.S.; Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.M.E.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef] [Green Version]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar]
- Takahama, T.; Sakai, K.; Takeda, M.; Azuma, K.; Hida, T.; Hirabayashi, M.; Oguri, T.; Tanaka, H.; Ebi, N.; Sawa, T.; et al. Detection of the T790M Mutation of EGFR in Plasma of Advanced Non-Small Cell Lung Cancer Patients with Acquired Resistance to Tyrosine Kinase Inhibitors (West Japan Oncology Group 8014LTR Study). Oncotarget 2016, 7, 58492–58499. [Google Scholar] [CrossRef] [Green Version]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using Circulating Cell-Free DNA to Monitor Personalized Cancer Therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive Detection of Response and Resistance in Egfrmutant Lung Cancer Using Quantitative Next-Generation Genotyping of Cell-Free Plasma DNA. Clin. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef] [Green Version]
- Remon, J.; Menis, J.; Hasan, B.; Peric, A.; de Maio, E.; Novello, S.; Reck, M.; Berghmans, T.; Wasag, B.; Besse, B.; et al. The APPLE Trial: Feasibility and Activity of AZD9291 (Osimertinib) Treatment on Positive PLasma T790M in EGFR-Mutant NSCLC Patients. EORTC 1613. Clin. Lung Cancer 2017, 18, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Beau-Faller, M.; Pencreach, E.; Leduc, C.; Blons, H.; Merlio, J.P.; Bringuier, P.P.; de Fraipont, F.; Escande, F.; Lemoine, A.; Ouafik, L.H.; et al. Independent Prognostic Value of Ultra-Sensitive Quantification of Tumor Pre-Treatment T790M Subclones in EGFR Mutated Non-Small Cell Lung Cancer (NSCLC) Treated by First/Second Generation TKI, Depends on Variant Allele Frequency (VAF): Results of the French Cooperative Thoracic Intergroup (IFCT) Biomarkers France Project. Lung Cancer 2020, 140, 19–26. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Sawa, K.; Fukui, M.; Oyanagi, J.; Yoshimoto, N.; Suzumura, T.; Watanabe, T.; Kaneda, H.; Mitsuoka, S.; Asai, K.; et al. Predictive Impact of Low-Frequency Pretreatment T790M Mutation in Patients with EGFR-Mutated Non-Small Cell Lung Cancer Treated with EGFR Tyrosine Kinase Inhibitors. Lung Cancer 2020, 139, 80–88. [Google Scholar] [CrossRef]
- Ye, X.; Zhu, Z.Z.; Zhong, L.; Lu, Y.; Sun, Y.; Yin, X.; Yang, Z.; Zhu, G.; Ji, Q. High T790M Detection Rate in TKI-Naive NSCLC with EGFR Sensitive Mutation: Truth or Artifact? J. Thorac. Oncol. 2013, 8, 1118–1120. [Google Scholar]
- Kim, S.; Park, C.; Ji, Y.; Kim, D.G.; Bae, H.; van Vrancken, M.; Kim, D.H.; Kim, K.M. Deamination Effects in Formalin-Fixed, Paraffin-Embedded Tissue Samples in the Era of Precision Medicine. J. Mol. Diagn. 2017, 19, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Suda, K.; Kimura, H.; Matsumoto, K.; Arao, T.; Nagai, T.; Saijo, N.; Yatabe, Y.; Mitsudomi, T.; Nishio, K. Highly Sensitive Detection of EGFR T790M Mutation Using Colony Hybridization Predicts Favorable Prognosis of Patients with Lung Cancer Harboring Activating EGFR Mutation. J. Thorac. Oncol. 2012, 7, 1640–1644. [Google Scholar] [CrossRef] [Green Version]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef] [Green Version]
- Ma, G.; Zhang, J.; Yin, L.; Jiang, H.; Zhang, W.; Song, Y.; Liu, M. The Prognostic Role of Pretreatment Epidermal Growth Factor Receptor T790M Mutation in Advanced Non-Small Cell Lung Cancer Patients Treated with EGFR Tyrosine Kinase Inhibitors. Oncotarget 2017, 8, 50941–50948. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.; Yu, Y.; Li, Z.; Niu, X.; Lu, S. The Predictive Role of Pretreatment Epidermal Growth Factor Receptor T790M Mutation on the Progression-Free Survival of Tyrosine-Kinase Inhibitor-Treated Non-Small Cell Lung Cancer Patients: A Meta-Analysis. OncoTargets Ther. 2014, 7, 387–393. [Google Scholar]
- Chmielecki, J.; Foo, J.; Oxnard, G.R.; Hutchinson, K.; Ohashi, K.; Somwar, R.; Wang, L.; Amato, K.R.; Arcila, M.; Sos, M.L.; et al. Optimization of Dosing for EGFR-Mutant Non-Small Cell Lung Cancer with Evolutionary Cancer Modeling. Sci. Transl. Med. 2011, 3, 90ra59. [Google Scholar] [CrossRef] [Green Version]
- Oxnard, G.R.; Arcila, M.E.; Sima, C.S.; Riely, G.J.; Chmielecki, J.; Kris, M.G.; Pao, W.; Ladanyi, M.; Miller, V.A. Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Lung Cancer: Distinct Natural History of Patients with Tumors Harboring the T790M Mutation. Clin. Cancer Res. 2011, 17, 1616–1622. [Google Scholar] [CrossRef] [Green Version]
- Hata, A.; Katakami, N.; Yoshioka, H.; Takeshita, J.; Tanaka, K.; Nanjo, S.; Fujita, S.; Kaji, R.; Imai, Y.; Monden, K.; et al. Rebiopsy of Non-Small Cell Lung Cancer Patients with Acquired Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor: Comparison between T790M Mutation-Positive and Mutation-Negative Populations. Cancer 2013, 119, 4325–4332. [Google Scholar] [CrossRef]
- Ko, B.; Paucar, D.; Halmos, B. EGFR T790M: Revealing the Secrets of a Gatekeeper. Lung Cancer Targets Ther. 2017, 8, 147–159. [Google Scholar]
- Hata, A.N.; Niederst, M.J.; Archibald, H.L.; Gomez-Caraballo, M.; Siddiqui, F.M.; Mulvey, H.E.; Maruvka, Y.E.; Ji, F.; Bhang, H.E.C.; Radhakrishna, V.K.; et al. Tumor Cells Can Follow Distinct Evolutionary Paths to Become Resistant to Epidermal Growth Factor Receptor Inhibition. Nat. Med. 2016, 22, 262–269. [Google Scholar] [CrossRef]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Lianidou, E.; Pantel, K. Liquid Biopsies. Genes Chromosomes Cancer 2019, 58, 219–232. [Google Scholar]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. S1), S49–S52. [Google Scholar]



| Characteristics | T790M Negative (n = 25) n (%) | T790M Positive (n = 9) n (%) | p-Value |
|---|---|---|---|
| Sex | 0.139 | ||
| Male | 9 (36.0%) | 6 (66.7%) | |
| Female | 16 (64.0%) | 3 (33.3%) | |
| Age (years) | 0.240 | ||
| ˂65 | 13 (52.0%) | 2 (22.2%) | |
| ≥65 | 12 (48.0%) | 7 (77.8%) | |
| Smoking status | 0.254 | ||
| Never smoker | 17 (68.0%) | 4 (44.4%) | |
| (Former) smoker | 8 (32.0%) | 5 (55.6%) | |
| CNS metastasis | 1.000 | ||
| No | 18 (72.0%) | 7 (77.8%) | |
| Yes | 7 (28.0%) | 2 (22.2%) | |
| Bone metastasis | 0.214 | ||
| No | 15 (60.0%) | 8 (88.9%) | |
| Yes | 10 (40.0%) | 1 (11.1%) | |
| Type of EGFR mutation * | 1.000 | ||
| Deletion exon 19 | 15 (62.5%) | 5 (55.6%) | |
| Leu858Arg | 9 (37.5%) | 4 (44.4%) | |
| EGFR-TKI Treatment | 0.216 | ||
| Erlotinib | 15 (60.0%) | 3 (33.3%) | |
| Gefitinib | 4 (16.0%) | 4 (44.4%) | |
| Afatinib | 3 (12.0%) | 0 (0.0%) | |
| Dacomitinib | 1 (4.0%) | 0 (0.0%) | |
| Erlotinib–Gefitinib | 1 (4.0%) | 1 (11.1%) | |
| Erlotinib + Ramucirumab | 1 (4.0%) | 0 (0.0%) | |
| Erlotinib + Bevacizumab | 0 (0.0%) | 1 (11.1%) |
| Characteristics | n | % |
|---|---|---|
| Sex | ||
| Male | 15 | 44.12% |
| Female | 19 | 55.88% |
| Age (years) | ||
| Mean | 65.47 | |
| Range | 32–85 | |
| Smoking status | ||
| Never smoker | 21 | 61.76% |
| Former smoker | 6 | 17.65% |
| Current smoker | 7 | 20.59% |
| Histologic type | ||
| Adenocarcinoma | 33 | 97.06% |
| Squamous | 1 | 2.94% |
| CNS metastasis | ||
| Yes | 9 | 26.47% |
| No | 25 | 73.53% |
| Bone metastasis | ||
| Yes | 11 | 32.35% |
| No | 23 | 67.65% |
| Type of EGFR mutation | ||
| Deletion 19 | 20 | 58.82% |
| Leu858Arg | 13 | 38.24% |
| Leu858Arg/Ser768Ile | 1 | 2.94% |
| EGFR-TKI Treatment | ||
| Erlotinib | 18 | 52.94% |
| Gefitinib | 8 | 23.53% |
| Afatinib | 3 | 8.82% |
| Dacomitinib | 1 | 2.94% |
| Erlotinib–Gefitinib | 2 | 5.88% |
| Erlotinib + Ramucirumab | 1 | 2.94% |
| Erlotinib + Bevacizumab | 1 | 2.94% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simarro, J.; Pérez-Simó, G.; Mancheño, N.; Ansotegui, E.; Muñoz-Núñez, C.F.; Gómez-Codina, J.; Juan, Ó.; Palanca, S. Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients. Int. J. Mol. Sci. 2022, 23, 8526. https://doi.org/10.3390/ijms23158526
Simarro J, Pérez-Simó G, Mancheño N, Ansotegui E, Muñoz-Núñez CF, Gómez-Codina J, Juan Ó, Palanca S. Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients. International Journal of Molecular Sciences. 2022; 23(15):8526. https://doi.org/10.3390/ijms23158526
Chicago/Turabian StyleSimarro, Javier, Gema Pérez-Simó, Nuria Mancheño, Emilio Ansotegui, Carlos Francisco Muñoz-Núñez, José Gómez-Codina, Óscar Juan, and Sarai Palanca. 2022. "Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients" International Journal of Molecular Sciences 23, no. 15: 8526. https://doi.org/10.3390/ijms23158526
APA StyleSimarro, J., Pérez-Simó, G., Mancheño, N., Ansotegui, E., Muñoz-Núñez, C. F., Gómez-Codina, J., Juan, Ó., & Palanca, S. (2022). Technical Validation and Clinical Implications of Ultrasensitive PCR Approaches for EGFR-Thr790Met Mutation Detection in Pretreatment FFPE Samples and in Liquid Biopsies from Non-Small Cell Lung Cancer Patients. International Journal of Molecular Sciences, 23(15), 8526. https://doi.org/10.3390/ijms23158526






