Key Residues Affecting Binding Affinity of Sirex noctilio Fabricius Odorant-Binding Protein (SnocOBP9) to Aggregation Pheromone
Abstract
:1. Introduction
2. Results
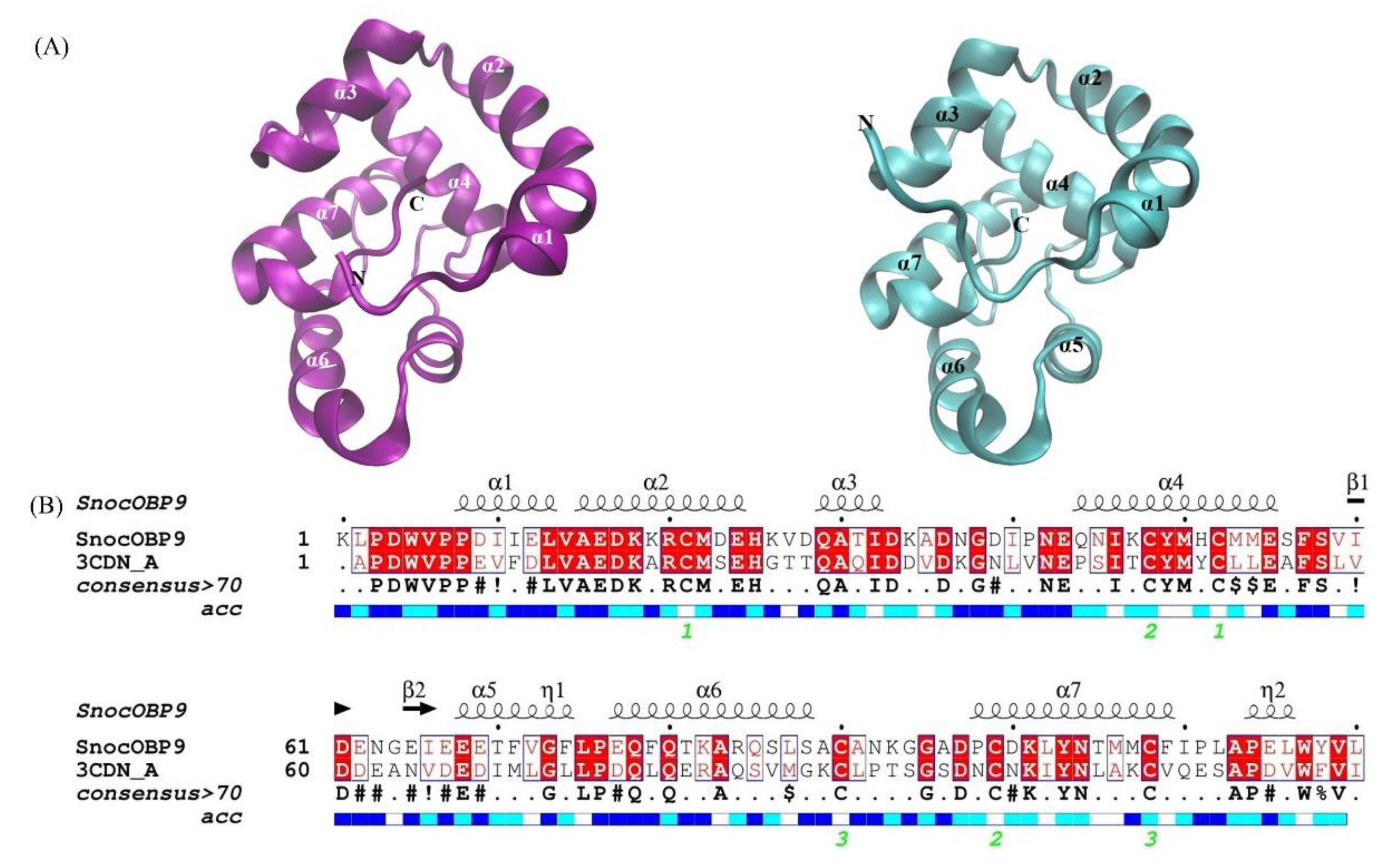
2.1. Sequence Analysis of SnocOBP9
2.2. Expression and Purification of Recombinant SnocOBP9WT and SnocOBP9MT
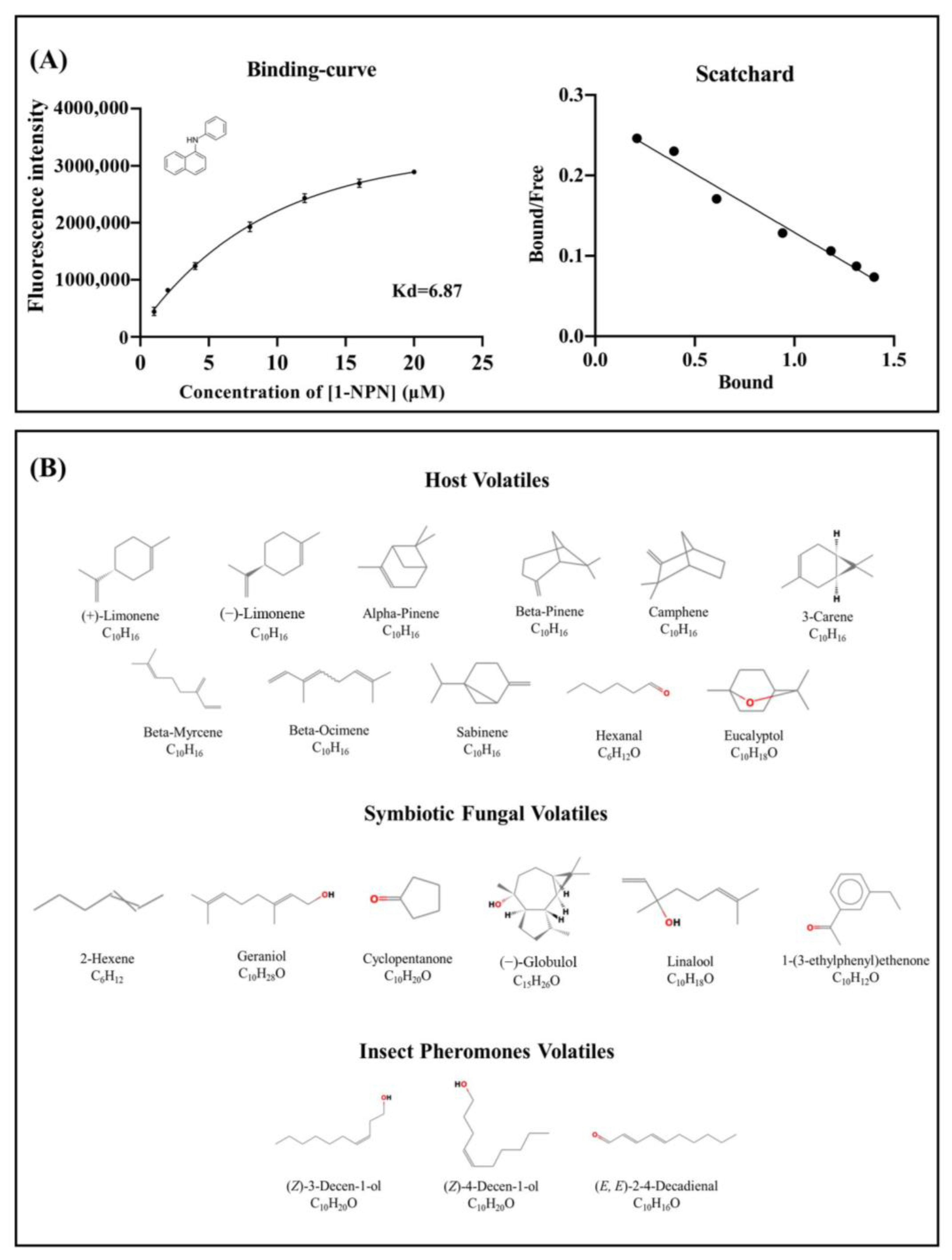
2.3. Fluorescence Competitive Binding Assay
2.4. Protein Homology Modeling and Molecular Docking
2.5. Molecular Dynamics Simulation of SnocOBP9-Z3D Complexes
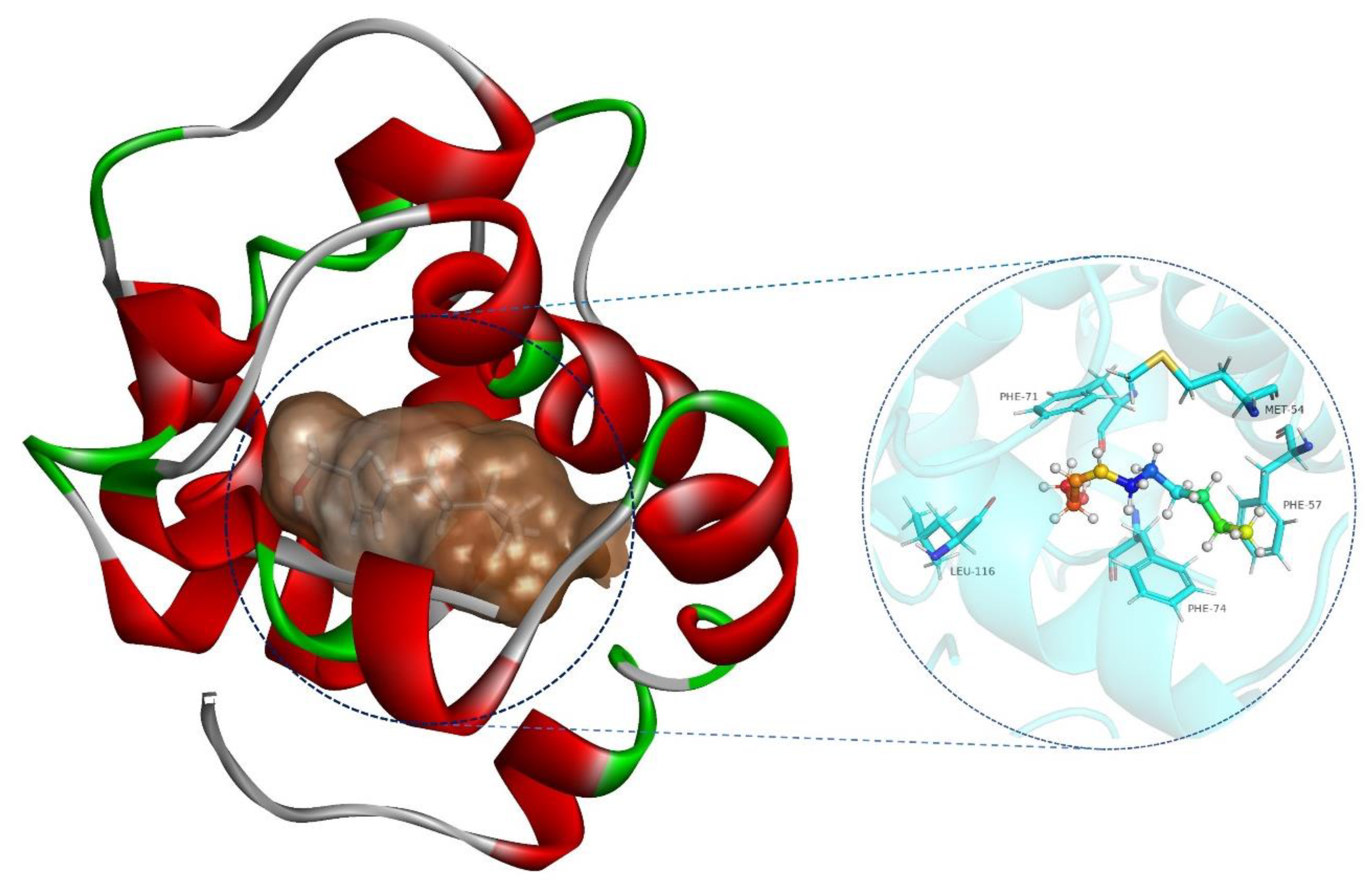
2.6. SnocOBP9-Z3D Binding Mode Analysis
2.7. SnocOBP9-Z3D Binding Energy Calculation
2.8. Per Residue Binding Free Energy Decomposition
2.9. Computational Alanine Scanning
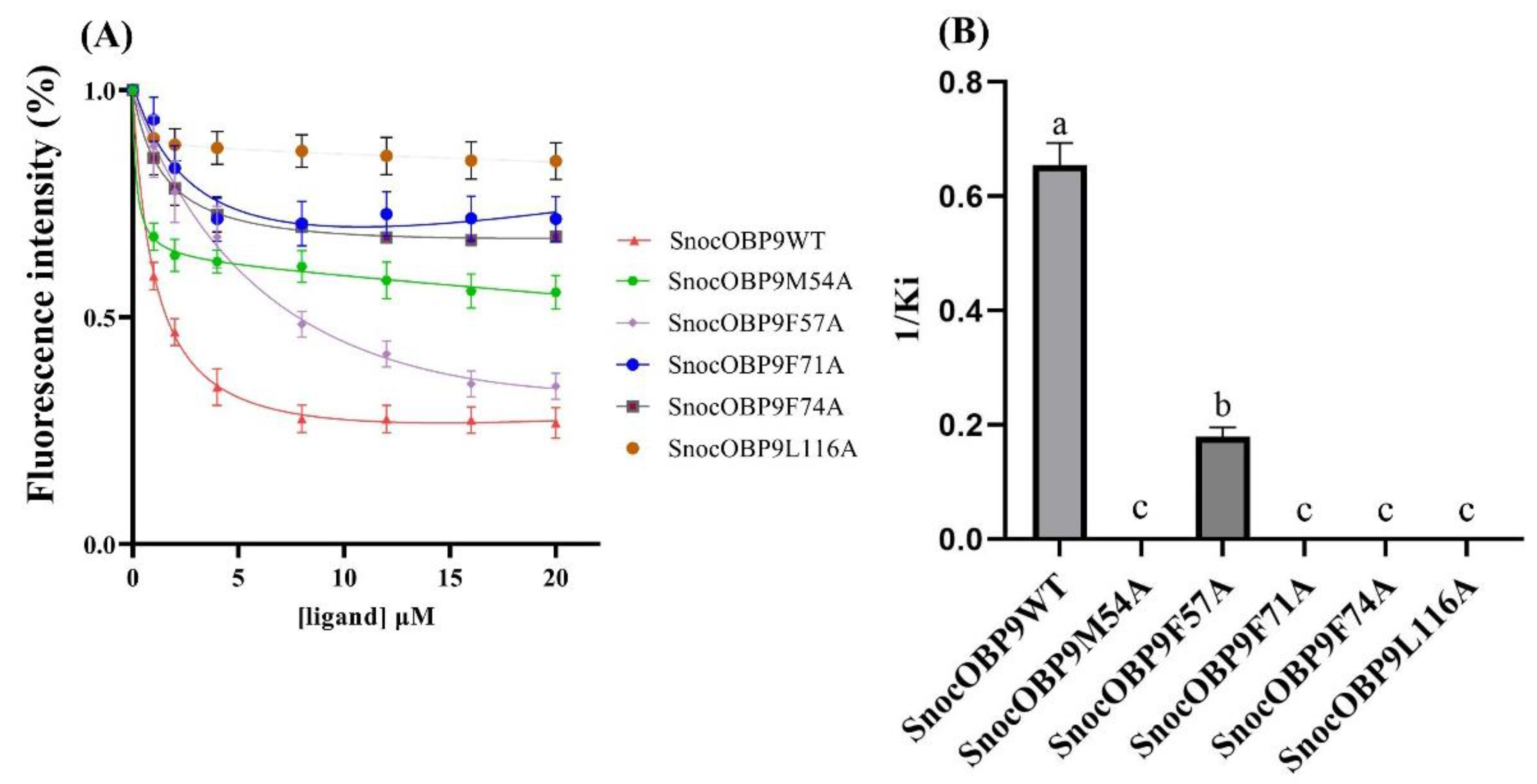
2.10. Site-Directed Mutation and Binding Affinity
3. Discussion
4. Materials and Methods
4.1. Insect Rearing and Tissue Collection
4.2. Total RNA Isolation, cDNA Synthesis and Sequence Analysis
4.3. Cloning, Expression and Purification of Recombinant Wild-Type SnocOBP9 Protein
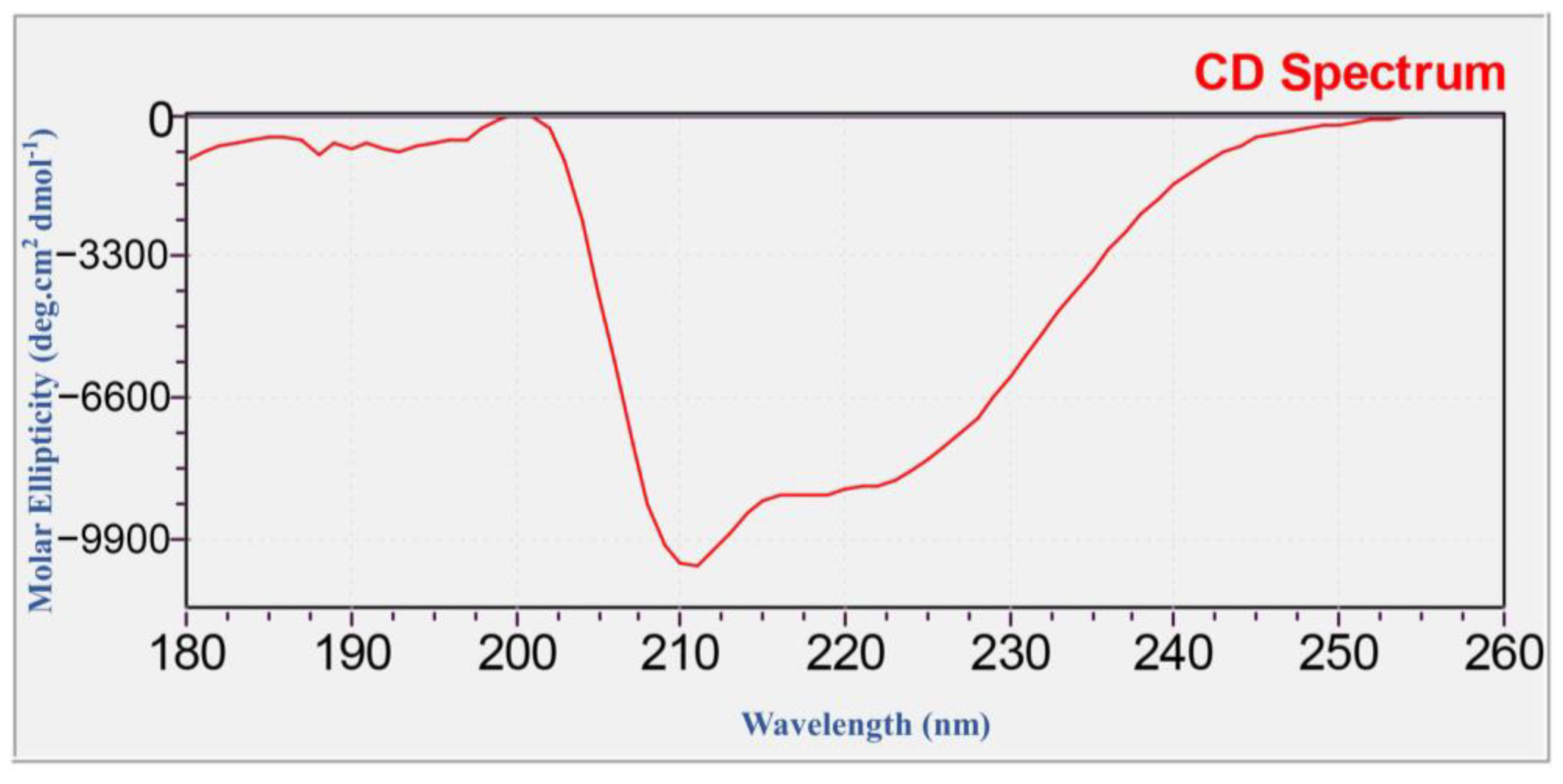
4.4. Identification of Recombinant SnocOBP9 (Western Blot, LC-MS/MS and CD-Spectrum)
4.5. Fluorescence Competitive Binding Assay
4.6. Homology Modeling and Molecular Docking
4.7. Molecular Dynamics Simulations
4.8. Binding Free Energy Calculation
4.9. Computational Alanine Scanning and Site-Directed Mutation
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| SnocOBP9 | Sirex noctilio odorant-binding protein 9 |
| 1-NPN | N-phenyl-1-naphthylamine |
| Z3D | (Z)-3-decen-ol |
| CAS. | Computational Alanine Scanning |
| SnoOBP9WT | Sirex noctilio odorant-binding protein 9 wild-type |
| SnoOBP9MT | Sirex noctilio odorant-binding protein 9 mutant-type |
References
- Zwiebel, L.; Takken, W. Olfactory regulation of mosquito-host interactions. Insect Biochem. Mol. Biol. 2004, 34, 645–652. [Google Scholar] [CrossRef] [Green Version]
- Brito, N.F.; Moreira, M.F.; Melo, A.C. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef]
- Piesik, D.; Rochat, D.; Van der Pers, J.; Marion-Poll, F. Pulsed Odors from Maize or Spinach Elicit Orientation in European Corn Borer Neonate Larvae. J. Chem. Ecol. 2009, 35, 1032–1042. [Google Scholar] [CrossRef]
- Skoczek, A.; Piesik, D.; Wenda-Piesik, A.; Buszewski, B.; Bocianowski, J.; Wawrzyniak, M. Volatile organic compounds released by maize following herbivory or insect extract application and communication between plants. J. Appl. Entomol. 2017, 141, 630–643. [Google Scholar] [CrossRef]
- Piesik, D.; Rochat, D.; Delaney, K.; Marion-Poll, F. Orientation of European corn borer first instar larvae to synthetic green leaf volatiles. J. Appl. Entomol. 2013, 137, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Piesik, D.; Wenda-Piesik, A. Sitophilus granarius responses to blends of five groups of cereal kernels and one group of plant volatiles. J. Stored Prod. Res. 2015, 62, 36–39. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein–bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Scaloni, A.; Monti, M.; Angeli, S.; Pelosi, P. Structural Analysis and Disulfide-Bridge Pairing of Two Odorant-Binding Proteins from Bombyx mori. Biochem. Biophys. Res. Commun. 1999, 266, 386–391. [Google Scholar] [CrossRef]
- Peng, G.; Leal, W. Identification and Cloning of a Pheromone-Binding Protein from the Oriental Beetle, Exomala orientalis. J. Chem. Ecol. 2001, 27, 2183–2192. [Google Scholar] [CrossRef]
- Guo, X.; Xuan, N.; Liu, G.; Xie, H.; Lou, Q.; Arnaud, P.; Offmann, B.; Picimbon, J.-F. An Expanded Survey of the Moth PBP/GOBP Clade in Bombyx mori: New Insight into Expression and Functional Roles. Front. Physiol. 2021, 1701, 712593. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Winberg, G.; Ren, H.; Zhang, S. Expression, purification and functional analysis of an odorant binding protein AaegOBP22 from Aedes aegypti. Protein Expr. Purif. 2011, 75, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Picimbon, J.-F.; Ji, S.; Kan, Y.; Chuanling, Q.; Zhou, J.-J.; Pelosi, P. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 2008, 372, 464–468. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Genta, F.A.; Sorgine, M.H.F.; Logullo, R.; Mesquita, R.D.; Paiva-Silva, G.O.; Majerowicz, D.; Medeiros, M.; Koerich, L.; Terra, W.R.; et al. An Insight into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius prolixus. PLoS Negl. Trop. Dis. 2014, 8, e2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.S.; Xiao, S.; Carlson, J.R. The diverse small proteins called odorant-binding proteins. R. Soc. Open Biol. 2018, 8, 180208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rihani, K.; Ferveur, J.-F.; Briand, L. The 40-Year Mystery of Insect Odorant-Binding Proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.-Z.; Liu, J.-T.; Zhou, J.-J.; Wang, Q.; Dong, J.-Z.; Zhang, Y.-J.; Li, X.-C.; Li, J.; Gu, S.-H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. Biol. 2018, 98, 34–47. [Google Scholar] [CrossRef]
- Jing, D.; Zhang, T.; Bai, S.; Prabu, S.; He, K.; Dewer, Y.; Wang, Z. GOBP1 Plays a Key Role in Sex Pheromones and Plant Volatiles Recognition in Yellow Peach Moth, Conogethes punctiferalis (Lepidoptera: Crambidae). Insects 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.-Y.; Liu, C.-C.; Dong, S.-L. Functional differentiation of pheromone-binding proteins in the common cutworm Spodoptera litura. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 165, 254–262. [Google Scholar] [CrossRef]
- Hu, P.; Gao, C.; Zong, S.; Luo, Y.; Tao, J. Pheromone Binding Protein EhipPBP1 Is Highly Enriched in the Male Antennae of the Seabuckthorn Carpenterworm and Is Binding to Sex Pheromone Components. Front. Physiol. 2018, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Tai, S.; Dai, W.; Zhang, C. Expression and Functional Analysis of Two Odorant-Binding Proteins from Bradysia odoriphaga (Diptera: Sciaridae). J. Agric. Food Chem. 2019, 67, 3565–3574. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Pan, C.; Huang, Y.; Li, Y.; Li, H.; Wu, C.; Chen, T.; Ma, D.; Li, Z. Functional characteristic analysis of three odorant-binding proteins from the sweet potato weevil (Cylas formicarius) in the perception of sex pheromones and host plant volatiles. Pest Manag. Sci. 2021, 77, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhong, Y.; Tang, R.; Rebijith, K.B.; Li, F.; Chen, G.; Zhang, F. Olfactory Reception of Host Alarm Pheromone Component by the Odorant-Binding Proteins in the Samurai Wasp, Trissolcus japonicus (Hymenoptera: Scelionidae). Front. Physiol. 2020, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Maung, K.L.; Jing, D.; Zhang, T.; Prabu, S.; He, K.; Bai, S.; Wang, Z. Molecular identification and functional analysis of Niemann-Pick type C2 protein in Macrocentrus cingulum Brischke (Hymenoptera: Braconidae). J. Asia-Pac. Entomol. 2021, 24, 7–14. [Google Scholar] [CrossRef]
- Du, Y.; Xu, K.; Zhao, H.; Jiang, Y.; Li, H. Identification and functional characterization of AcerOBP15 from Apis cerana cerana (Hymenoptera: Apidae). Apidologie 2021, 52, 668–683. [Google Scholar] [CrossRef]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S. Molecular Dynamics Simulation of Nanocomposites Using BIOVIA Materials Studio. In Lammps and Gromacs; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Yang, Z.; Qin, Y.; Wang, S.; Duan, H.; Yang, X. Computational investigation of the molecular conformation-dependent binding mode of (E)-β-farnesene analogs with a heterocycle to aphid odorant-binding proteins. J. Mol. Model. 2018, 24, 70. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, C.; Liu, D. Analyses of structural dynamics revealed flexible binding mechanism for the Agrilus mali odorant binding protein 8 towards plant volatiles. Pest Manag. Sci. 2020, 77, 1642–1653. [Google Scholar] [CrossRef]
- Zhu, J.; Zaremska, V.; D’Onofrio, C.; Knoll, W.; Pelosi, P. Site-directed mutagenesis of odorant-binding proteins. Methods Enzymol. 2020, 642, 301–324. [Google Scholar] [CrossRef]
- Parkin, E.A. Symbiosis in Larval Siricidæ (Hymenoptera). Nature 1941, 147, 329. [Google Scholar] [CrossRef]
- Rawlings, G. Recent observations on the Sirex noctilio population in Pinus radiata forest in New Zealand. N. Z. J. For. 1948, 5, 411–421. [Google Scholar]
- Gilbert, J.M.; Miller, L.W. An outbreak of Sirex noctilio F. in Tasmania. Aust. For. 1952, 16, 63–69. [Google Scholar] [CrossRef]
- Madden, J.L. Some treatments which render Monterey pine (Pinus radiata) attractive to the wood wasp Sirex noctilio F. Bull. Entomol. Res. 1971, 60, 467–472. [Google Scholar] [CrossRef]
- Haugen, D.; Bedding, R.; Underdown, M.; Neumann, F. National strategy for control of Sirex noctilio in Australia. Aust. For. Grow. 1990, 13. [Google Scholar]
- De Groot, P.; Nystrom, K.; Scarr, T. Discovery of Sirex noctilio (Hymenoptera: Siricidae) in Ontario, Canada. Great Lakes Entomol. 2006, 39, 5. [Google Scholar]
- Li, D.; Shi, J.; Lu, M.; Ren, L.; Zhen, C.; Luo, Y. Detection and Identification of the Invasive Sirex noctilio (Hymenoptera: Siricidae) Fungal Symbiont, Amylostereum areolatum (Russulales: Amylostereacea), in China and the Stimulating Effect of Insect Venom on Laccase Production by A. areolatum YQL03. J. Econ. Entomol. 2015, 108, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Tao, J.; Ren, L.; Shi, J.; Luo, Y. Identification of Sirex noctilio (Hymenoptera: Siricidae) Using a Species-Specific Cytochrome C Oxidase Subunit I PCR Assay. J. Econ. Entomol. 2016, 109, 1424–1430. [Google Scholar] [CrossRef] [Green Version]
- Martinson, S.; Ajó, A.F.; Martínez, A.; Krivak-Tetley, F.; Villacide, J.M.; Ayres, M.; Corley, J.C. Attack rates of Sirex noctilio and patterns of pine tree defenses and mortality in northern Patagonia. Bull. Entomol. Res. 2019, 109, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Cooperband, M.F.; Böröczky, K.; Hartness, A.; Jones, T.H.; Zylstra, K.E.; Tumlinson, J.H.; Mastro, V.C. Male-Produced Pheromone in the European Woodwasp, Sirex noctilio. J. Chem. Ecol. 2012, 38, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Böröczky, K.; Zylstra, K.E.; Mastro, V.C.; Tumlinson, J.H.; Böröczky, K.; Zylstra, K.E.; Mastro, V.C.; Tumlinson, J.H. Volatile profiles and trap catches of two pine-host species of Sirex noctilio. In Proceedings of the 20th U.S. Department of Agriculture Interagency Research Forum on Invasive Species, Annapolis, MD, USA, 13–16 January 2009. [Google Scholar]
- Böröczky, K.; Crook, D.J.; Jones, T.H.; Kenny, J.C.; Zylstra, K.E.; Mastro, V.C.; Tumlinson, J.H. Monoalkenes as Contact Sex Pheromone Components of the Woodwasp Sirex noctilio. J. Chem. Ecol. 2009, 35, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Qiao, H.-L.; Shi, J.; Luo, Y.; Lu, P.-F. Research Progress in Reproductive Behavior and Chemical Ecological Regulation of the European Woodwasp (Sirex noctilio), a Severe invasive pest. Sci. Silvae Sin. 2020, 56, 127–141. [Google Scholar]
- Liu, R. Identification of Pheromone Components of Sirex noctilio Fabricius and Trapping Technology in Forest. Master’s Thesis, Beijing Forestry University, Beijing, China, 2019. [Google Scholar]
- Gao, C.Y. Study on the Associated Semiochemicals and Trapping Techniquesin Forest of Sirex noctilio Fabricius and S Nitobei. Master’s Thesis, Beijing Forestry University, Beijing, China, 2019. [Google Scholar]
- Guignard, Q.; Bouwer, M.; Slippers, B.; Allison, J. Biology of a putative male aggregation-sex pheromone in Sirex noctilio (Hymenoptera: Siricidae). PLoS ONE 2020, 15, e0244943. [Google Scholar] [CrossRef]
- Dodds, K.J.; Zylstra, K.E.; Dubois, G.D.; Hoebeke, E.R. Arboreal Insects Associated with Herbicide-Stressed Pinus resinosa and Pinus sylvestris Used as Sirex noctilio Trap Trees in New York. Environ. Entomol. 2012, 41, 1350–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boroczky, K.; Crook, D.J.; Francese, J.A.; Mastro, V.C.; Tumlinson, J.H. Chemical ecology of Sirex noctilio. In Proceedings of the 19th U.S. Department of Agriculture Interagency Research Forum on Invasive Species, Annapolis, MD, USA, 8–11 January 2008. [Google Scholar]
- Xu, Q.; Sun, X.-T.; Lu, P.-F.; Luo, Y.-Q.; Shi, J. Volatile profiles of three tree species in the northeastern China and associated effects on Sirex noctilio activity. J. Plant Interact. 2019, 14, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Hurley, B.P.; Garnas, J.; Cooperband, M.F. Assessing trap and lure effectiveness for the monitoring of Sirex noctilio. Agric. For. Entomol. 2015, 17, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Batista, E.S.P.; Redak, R.A.; Busoli, A.C.; Camargo, M.B.; Allison, J.D. Trapping for Sirex Woodwasp in Brazilian Pine Plantations: Lure, Trap Type and Height of Deployment. J. Insect Behav. 2018, 31, 210–221. [Google Scholar] [CrossRef] [Green Version]
- Coutts, M. The mechanism of pathogenicity of Sirex noctilio on Pinus radiata I. Effects of the symbiotic fungus Amylostereum sp. (Thelophoraceae). Aust. J. Biol. Sci. 1969, 22, 915–924. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.-X.; Ren, L.-L.; Liu, X.-B.; Shi, J.; Wang, J.-Z.; Luo, Y.-Q. Effects of endophytic fungi in Mongolian pine on the selection behavior of woodwasp (Sirex noctilio) and the growth of its fungal symbiont. Pest Manag. Sci. 2018, 75, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Hao, E.; Qiao, H.; Wang, J.; Wu, W.; Zhou, J.; Lu, P. Antennal transcriptome analysis of olfactory genes and characterizations of odorant binding proteins in two woodwasps, Sirex noctilio and Sirex nitobei (Hymenoptera: Siricidae). BMC Genom. 2021, 22, 172. [Google Scholar] [CrossRef]
- Guo, B.; Hao, E.; Wang, J.; Lu, P.; Qiao, H. Molecular docking of odorant binding proteins and its related semiochemicals of sirex woodwasp Sirex noctilio, an invasive insect pest. J. Plant Prot. 2019, 46, 65–78. [Google Scholar]
- Danty, E.; Michard-Vanhee, C.; Huet, J.-C.; Genecque, E.; Pernollet, J.-C.; Masson, C. Biochemical characterization, molecular cloning and localization of a putative odorant-binding protein in the honey bee Apis mellifera L. (Hymenoptera: Apidea). Febs Lett. 1997, 414, 595–598. [Google Scholar] [CrossRef] [Green Version]
- Danty, E.; Arnold, G.; Huet, J.-C.; Huet, D.; Masson, C.; Pernollet, J.-C. Separation, Characterization and Sexual Heterogeneity of Multiple Putative Odorant-binding Proteins in the Honeybee Apis mellifera L. (Hymenoptera: Apidea). Chem. Senses 1998, 23, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Billen, J. Occurrence and structural organization of the exocrine glands in the legs of ants. Arthropod Struct. Dev. 2009, 38, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Kainoh, Y.; Oishi, Y. Source of sex pheromone of the egg-larval parasitoid, Ascogaster reticulatus Watanabe (Hymenoptera: Braconidae). J. Chem. Ecol. 1993, 19, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Beani, L.; Calloni, C. Leg tegumental glands and male rubbing behavior at leks in Polistes dominulus (Hymenoptera: Vespidae). J. Insect Behav. 1991, 4, 449–462. [Google Scholar] [CrossRef]
- Jarau, S.; Hrncir, M.; Ayasse, M.; Schulz, C.; Francke, W.; Zucchi, R.; Barth, F.G. A Stingless Bee (Melipona seminigra) Marks Food Sources with a Pheromone from Its Claw Retractor Tendons. J. Chem. Ecol. 2004, 30, 793–804. [Google Scholar] [CrossRef] [PubMed]
- González-González, A.; Rubio-Meléndez, M.E.; Ballesteros, G.I.; Ramírez, C.C.; Palma-Millanao, R. Sex- and tissue-specific expression of odorant-binding proteins and chemosensory proteins in adults of the scarab beetle Hylamorpha elegans (Burmeister) (Coleoptera: Scarabaeidae). PeerJ 2019, 7, e7054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.-M.; Zhang, L.-Y.; Fu, X.-B.; Wu, F.; Tan, J.; Li, H.-L. Various Bee Pheromones Binding Affinity, Exclusive Chemosensillar Localization, and Key Amino Acid Sites Reveal the Distinctive Characteristics of Odorant-Binding Protein 11 in the Eastern Honey Bee, Apis cerana. Front. Physiol. 2018, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J. Odorant-binding proteins in insects. In Vitamins & Hormones; Elsevier: Amsterdam, The Netherlands, 2010; Volume 83, pp. 241–272. [Google Scholar] [CrossRef]
- Liu, N.; Yang, F.; Yang, K.; He, P.; Niu, X.H.; Xu, W.; Anderson, A.; Dong, S.L. Two subclasses of odorant-binding proteins in Spodoptera exigua display structural conservation and functional divergence. Insect Mol. Biol. 2015, 24, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-Y.; Yang, K.; Liu, Y.; Xu, W.; Anderson, A.; Dong, S.-L. Two general-odorant binding proteins in Spodoptera litura are differentially tuned to sex pheromones and plant odorants. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 180, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Khuhro, S.A.; Liao, H.; Dong, X.-T.; Yu, Q.; Yan, Q.; Dong, S.-L. Two general odorant binding proteins display high bindings to both host plant volatiles and sex pheromones in a pyralid moth Chilo suppressalis (Lepidoptera: Pyralidae). J. Asia-Pacific Entomol. 2017, 20, 521–528. [Google Scholar] [CrossRef]
- Sun, M.; Liu, Y.; Wang, G. Expression patterns and binding properties of three pheromone binding proteins in the diamondback moth, Plutella xyllotella. J. Insect Physiol. 2013, 59, 46–55. [Google Scholar] [CrossRef]
- Campanacci, V.; Krieger, J.; Bette, S.; Sturgis, J.N.; Lartigue, A.; Cambillau, C.; Breer, H.; Tegoni, M. Revisiting the Specificity of Mamestra brassicaeand Antheraea polyphemus Pheromone-binding Proteins with a Fluorescence Binding Assay. J. Biol. Chem. 2001, 276, 20078–20084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, M.; Li, E.; Li, J.; Chen, Q.; Zhou, H.; Zhang, S.; Li, K.; Ren, B.; Wang, Y.; Yin, J. Molecular Characterization and Key Binding Sites of Sex Pheromone-Binding Proteins from the Meadow Moth, Loxostege sticticalis. J. Agric. Food Chem. 2019, 67, 12685–12695. [Google Scholar] [CrossRef]
- Shen, J.; Hu, L.; Dai, J.; Chen, B.; Zhong, G.; Zhou, X. Mutations in pheromone-binding protein3 contribute to pheromone response variations in Plutella xylostella (L.) (Lepidoptera: Plutellidae). Pest Manag. Sci. 2019, 75, 2034–2042. [Google Scholar] [CrossRef]
- Hurley, B.P.; Slippers, B.; Wingfield, M.J. A comparison of control results for the alien invasive woodwasp, Sirex noctilio, in the southern hemisphere. Agric. For. Entomol. 2010, 9, 159–171. [Google Scholar] [CrossRef] [Green Version]
- Sarvary, M.A.; Cooperband, M.F.; Hajek, A.E. The importance of olfactory and visual cues in developing better monitoring tools for Sirex noctilio(Hymenoptera: Siricidae). Agric. For. Entomol. 2015, 17, 29–35. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, J.; Zhang, Y. Key Residues Involved in the Interaction between Cydia pomonella Pheromone Binding Protein 1 (CpomPBP1) and Codlemone. J. Agric. Food Chem. 2016, 64, 7994–8001. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Zhang, X.-Q.; Xu, J.-W.; Li, L.-L.; Zhu, X.-Y.; Wang, J.-J.; Wei, J.-Y.; Mang, D.-Z.; Zhang, F.; Yuan, X.; et al. Key Amino Acid Residues Influencing Binding Affinities of Pheromone-Binding Protein from Athetis lepigone to Two Sex Pheromones. J. Agric. Food Chem. 2020, 68, 6092–6103. [Google Scholar] [CrossRef]
- Fu, N.; Li, J.; Wang, M.; Ren, L.; Luo, Y. Genes Identification, Molecular Docking and Dynamics Simulation Analysis of Laccases from Amylostereum areolatum Provides Molecular Basis of Laccase Bound to Lignin. Int. J. Mol. Sci. 2020, 21, 8845. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Froeyen, M.; Herdewijn, P. Computational alanine scanning and free energy decomposition for E. coli type I signal peptidase with lipopeptide inhibitor complex. J. Mol. Graph. Model. 2008, 26, 813–823. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Significance of Root-Mean-Square Deviation in Comparing Three-dimensional Structures of Globular Proteins. J. Mol. Biol. 1994, 235, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.-C.; Tegoni, M.; Cambillau, C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Xu, J.-W.; Zhang, X.-C.; Li, L.-L.; Yuan, X.; Mang, D.-Z.; Zhu, X.-Y.; Zhang, F.; Dewer, Y.; Xu, L.; et al. Organophosphorus insecticide interacts with the pheromone-binding proteins of Athetis lepigone: Implication for olfactory dysfunction. J. Hazard. Mater. 2020, 397, 122777. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Li, Y.; Xing, Y.; Li, R.; Liu, J. Structural Insights into Two Representative Conformations of the Complex Formed by Grapholita molesta (Busck) Pheromone Binding Protein 2 and Z-8-Dodecenyl Acetate. J. Agric. Food Chem. 2019, 67, 4425–4434. [Google Scholar] [CrossRef] [PubMed]
- Kortemme, T.; Kim, D.E.; Baker, D. Computational Alanine Scanning of Protein-Protein Interfaces. Sci. STKE 2004, 2004, pl2. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.-L.; Huang, L.-Q.; Pelosi, P.; Wang, C.-Z. A Lysine at the C-Terminus of an Odorant-Binding Protein is Involved in Binding Aldehyde Pheromone Components in Two Helicoverpa Species. PLoS ONE 2013, 8, e55132. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ban, L.; Song, L.-M.; Liu, Y.; Pelosi, P.; Wang, G. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem. Mol. Biol. 2016, 72, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Gu, S.H.; Han, L.; Guo, Y.Y.; Zhang, Y.J. Specific involvement of two amino acid residues in cis-nerolidol binding to odorant-binding protein 5 AlinOBP5 in the alfalfa plant bug, Adelphocoris lineolatus (Goeze). Insect Mol. Biol. 2013, 22, 172–182. [Google Scholar] [CrossRef]
- Liu, H.; Duan, H.; Wang, Q.; Xiao, Y.; Wang, Q.; Xiao, Q.; Sun, L.; Zhang, Y. Key Amino Residues Determining Binding Activities of the Odorant Binding Protein AlucOBP22 to Two Host Plant Terpenoids of Apolygus lucorum. J. Agric. Food Chem. 2019, 67, 5949–5956. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, Z.; Zhang, Y. Structure-based discovery of potentially active semiochemicals for Cydia pomonella (L.). Sci. Rep. 2016, 6, 34600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, B.; Sali, A. Comparative protein structure modeling using modeller. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [Green Version]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar]
- Fiser, A.; Do, R.K.G.; Šali, A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Schrodinger, Inc. The PyMOL Molecular Graphics System, Version 1.8; Schrodinger, Inc.: New York, NY, USA, 2015.
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.W.S.; Vranken, W.F. ACPYPE-Antechamber python parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [Green Version]
- Kumari, R.; Kumar, R.; Consortium, O.S.D.D.; Lynn, A. G_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-F.; Wang, F.; Chen, Y.-Z.; Hao, G.-F.; Yang, G.-F. LARMD: Integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform. 2020, 21, 2206–2218. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Lin, S.-J.; Ren, J.-Y.; Liu, T.; Wang, Y.-M.; Li, C.-M.; Xu, W.-W.; He, Y.-W.; Zheng, W.-H.; Zhao, J.; et al. Molecular Docking and Molecular Dynamics (MD) Simulation of Human Anti-Complement Factor H (CFH) Antibody Ab42 and CFH Polypeptide. Int. J. Mol. Sci. 2019, 20, 2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Protein ID | Majority Protein ID | Sequence Coverage | Unique Sequence Coverage | Unique Peptides | Weight a | Score b | Intensity c |
|---|---|---|---|---|---|---|---|
| SnocOBP9 | SnocOBP9; A0A857N3E7 | 19.1% | 19.1% | 3 | 16.162 | 323.31 | 3.5801 × 109 |
| Peptides Sequence | Length a | Mass c | Proteins | Start Position a | End Position a | Score b |
|---|---|---|---|---|---|---|
| ADNGDIPNEQNIK | 13 | 1426.6688 | SnocOBP9;A0A857N3E7 | 56 | 68 | 112.93 |
| CMDEHKVDQATIDKADNGDIPNEQNIK | 27 | 3097.4081 | SnocOBP9;A0A857N3E7 | 42 | 68 | 118.41 |
| VDQATIDKADNGDIPNEQNIK | 21 | 2297.1135 | SnocOBP9;A0A857N3E7 | 48 | 68 | 107.33 |
| Ligand Name | CAS Number | IC50 a | Ki a |
|---|---|---|---|
| Host Volatiles | |||
| (+)-Limonene | 5989-27-5 | 6.09 ± 1.43 | 4.87 ± 0.94 |
| (−)-Limonene | 5989-54-8 | 6.95 ± 0.61 | 5.50 ± 0.49 |
| Alpha-Pinene | 80-56-8 | >20 b | — c |
| Beta-Pinene | 127-91-3 | >20 | — |
| 3-Carene | 13466-78-9 | >20 | — |
| Camphene | 79-92-5 | >20 | — |
| Beta-Myrcene | 123-35-3 | >20 | — |
| Beta-Ocimene | 13877-91-3 | >20 | — |
| Sabinene | 3387-41-5 | >20 | — |
| Hexanal | 66-25-1 | >20 | — |
| Eucalyptol | 470-82-6 | >20 | — |
| Symbiotic Fungal Volatiles | |||
| Geraniol | 106-24-1 | >20 | — |
| 2-Hexene | 592-43-8 | >20 | — |
| Cyclopentanone | 120-92-3 | >20 | — |
| (−)-Globulol | 489-41-8 | >20 | — |
| Linalool | 78-70-6 | >20 | — |
| 1-(3-ethylphenyl)ethenone | 22699-70-3 | >20 | — |
| Insect Pheromones Volatiles | |||
| (Z)-3-Decen-ol | 10340-22-4 | 1.92 ± 0.14 | 1.53 ± 0.09 |
| (Z)-4-Decen-ol | 57074-37-0 | >20 | — |
| (E, E)-2,4-Decadienal | 25152-84-5 | >20 | — |
| System | Clusters | Occurrence [%] |
|---|---|---|
| SnocOBP9-Z3D | I | 66.02 |
| II | 26.86 | |
| III | 2.09 | |
| IV | 1.52 | |
| V | 0.95 | |
| VI | 0.88 | |
| VII | 0.67 | |
| VIII | 0.50 | |
| IX | 0.31 | |
| X | 0.19 |
| Complex | Sampling Conformations | Cluster b | Van Der Waal Energy a (ΔGvdw) | Electrostatic Energy a (ΔGele) | Polar Solvation Energy a (ΔGPB) | Apolar Solvation Energy a (ΔGSA) | Total Energy a (ΔGbind) |
|---|---|---|---|---|---|---|---|
| SnocOBP9-Z3D | 50,000 | 100–200 | 32.146 ± 0.045 | −4.595 ± 0.031 | 12.773 ± 0.036 | −3.163 ± 0.003 | −27.13 ± 0.043 |
| Complex | Residue | MM Energy a (ΔGMM) | Polar Energy a (ΔGPB) | Apolar Energy a (ΔGSA) | Total Energy a (ΔGbind) | Average Distance b | Standard Deviation b |
|---|---|---|---|---|---|---|---|
| SnocOBP9-Z3D | MET-54 | −1.412 | 0.332 | −0.073 | −1.153 | 6.3592 | 0.5401 |
| PHE-57 | −1.328 | 0.318 | −0.097 | −1.107 | 5.8364 | 0.8025 | |
| PHE-71 | −1.776 | 0.582 | −0.109 | −1.303 | 6.9383 | 0.7486 | |
| PHE-74 | −2.094 | 0.644 | −0.134 | −1.583 | 7.1332 | 1.7607 | |
| LEU-116 | −1.326 | 0.329 | −0.033 | −1.030 | 9.2189 | 0.7678 |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| SnocOBP9WT-F | CCGGAATCCAAGCTTCCCG ATTGGGTACC |
| SnocOBP9WT-R | CCGCTCGAGTTACAACACG TACCATAATT CTGGAGC |
| SnocOBP9M54A-F | GTTACATGCATTGCATGGCGGAGTCGTTCAGTGTG |
| SnocOBP9M54A-R | CACACTGAACGACTCCGCCATGCAATGCATGTAAC |
| SnocOBP9F57A-F | CATTGCATGATGGAGTCGGCCAGTGTGATCGACGAGAAC |
| SnocOBP9F57A-R | GTTCTCGTCGATCACACTGGCCGACTCCATCATGCAATG |
| SnocOBP9F71A-F | GTGAAATCGAAGAAGAAACTGCTGTGGGATTCCTACCAGAAC |
| SnocOBP9F71A-R | GTTCTGGTAGGAATCCCACAGCAGTTTCTTCTTCGATTTCAC |
| SnocOBP9F74A-F | GAAGAAGAAACTTTTGTGGGAGCCCTACCAGAACAGTTTCAGAC |
| SnocOBP9F74A-R | GTCTGAAACTGTTCTGGTAGGGCTCCCACAAAAGTTTCTTCTTC |
| SnocOBP9L116A-F | CCACTTGCTCCAGAATTAGCGTACGTGTTG |
| SnocOBP9L116A-R | CAACACGTACGCTAATTCTGGAGCAAGTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, E.; Li, Y.; Guo, B.; Yang, X.; Lu, P.; Qiao, H. Key Residues Affecting Binding Affinity of Sirex noctilio Fabricius Odorant-Binding Protein (SnocOBP9) to Aggregation Pheromone. Int. J. Mol. Sci. 2022, 23, 8456. https://doi.org/10.3390/ijms23158456
Hao E, Li Y, Guo B, Yang X, Lu P, Qiao H. Key Residues Affecting Binding Affinity of Sirex noctilio Fabricius Odorant-Binding Protein (SnocOBP9) to Aggregation Pheromone. International Journal of Molecular Sciences. 2022; 23(15):8456. https://doi.org/10.3390/ijms23158456
Chicago/Turabian StyleHao, Enhua, Yini Li, Bing Guo, Xi Yang, Pengfei Lu, and Haili Qiao. 2022. "Key Residues Affecting Binding Affinity of Sirex noctilio Fabricius Odorant-Binding Protein (SnocOBP9) to Aggregation Pheromone" International Journal of Molecular Sciences 23, no. 15: 8456. https://doi.org/10.3390/ijms23158456
APA StyleHao, E., Li, Y., Guo, B., Yang, X., Lu, P., & Qiao, H. (2022). Key Residues Affecting Binding Affinity of Sirex noctilio Fabricius Odorant-Binding Protein (SnocOBP9) to Aggregation Pheromone. International Journal of Molecular Sciences, 23(15), 8456. https://doi.org/10.3390/ijms23158456






