Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis
Abstract
:1. Introduction
2. Results
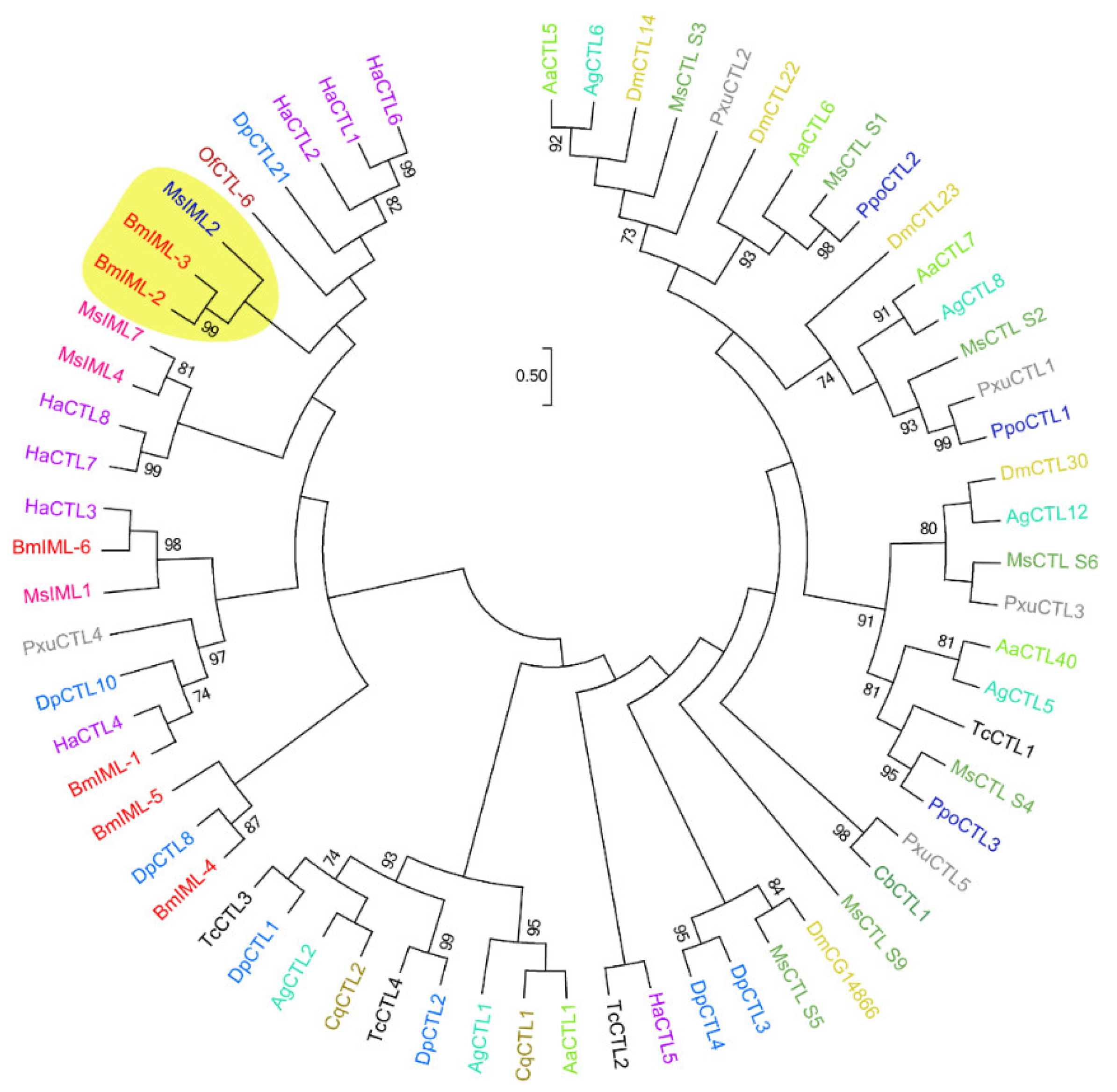
2.1. Sequence Characteristics and Bioinformatics Analysis of BmIML-2
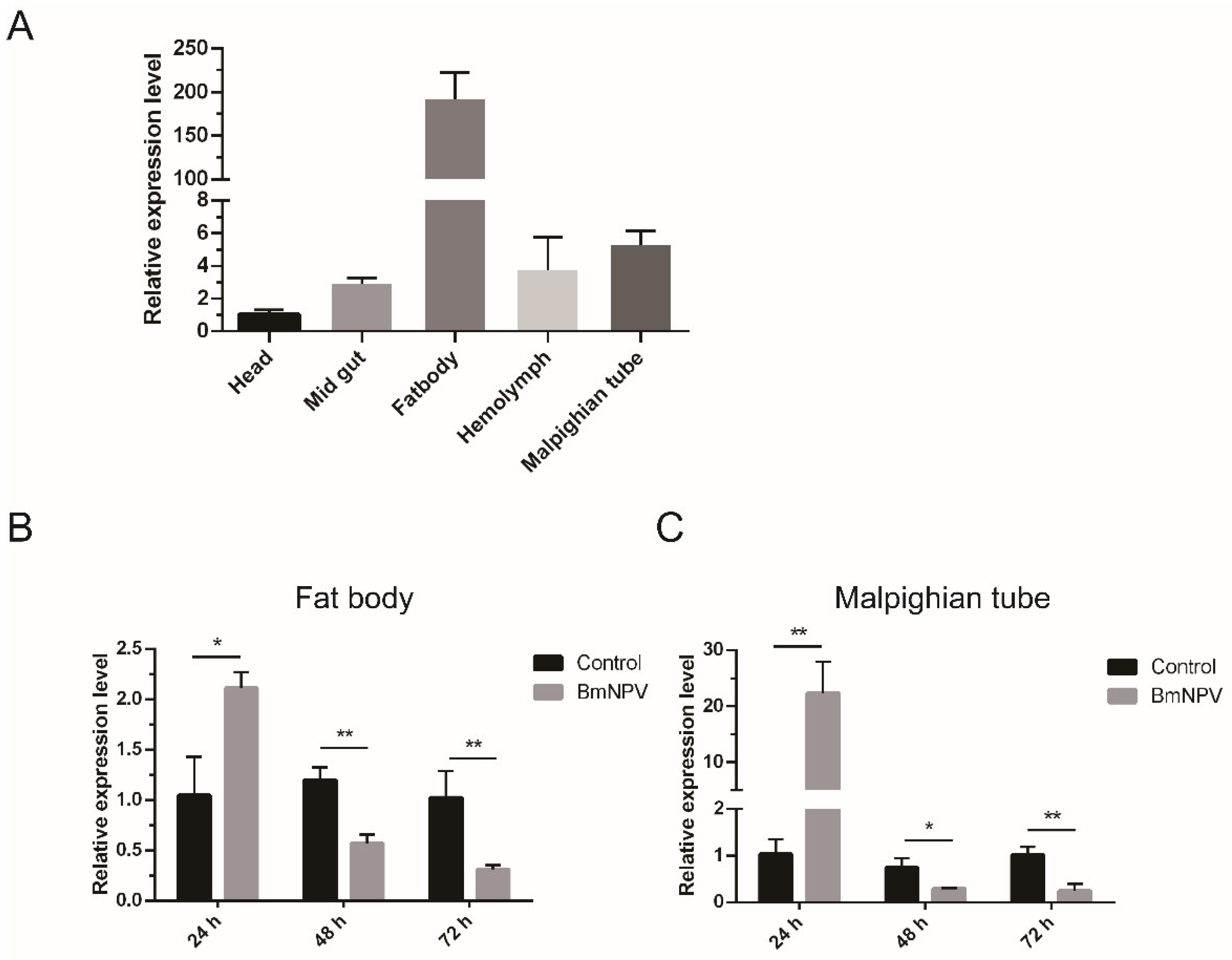
2.2. Tissue and Induced Expression Pattern of BmIML-2
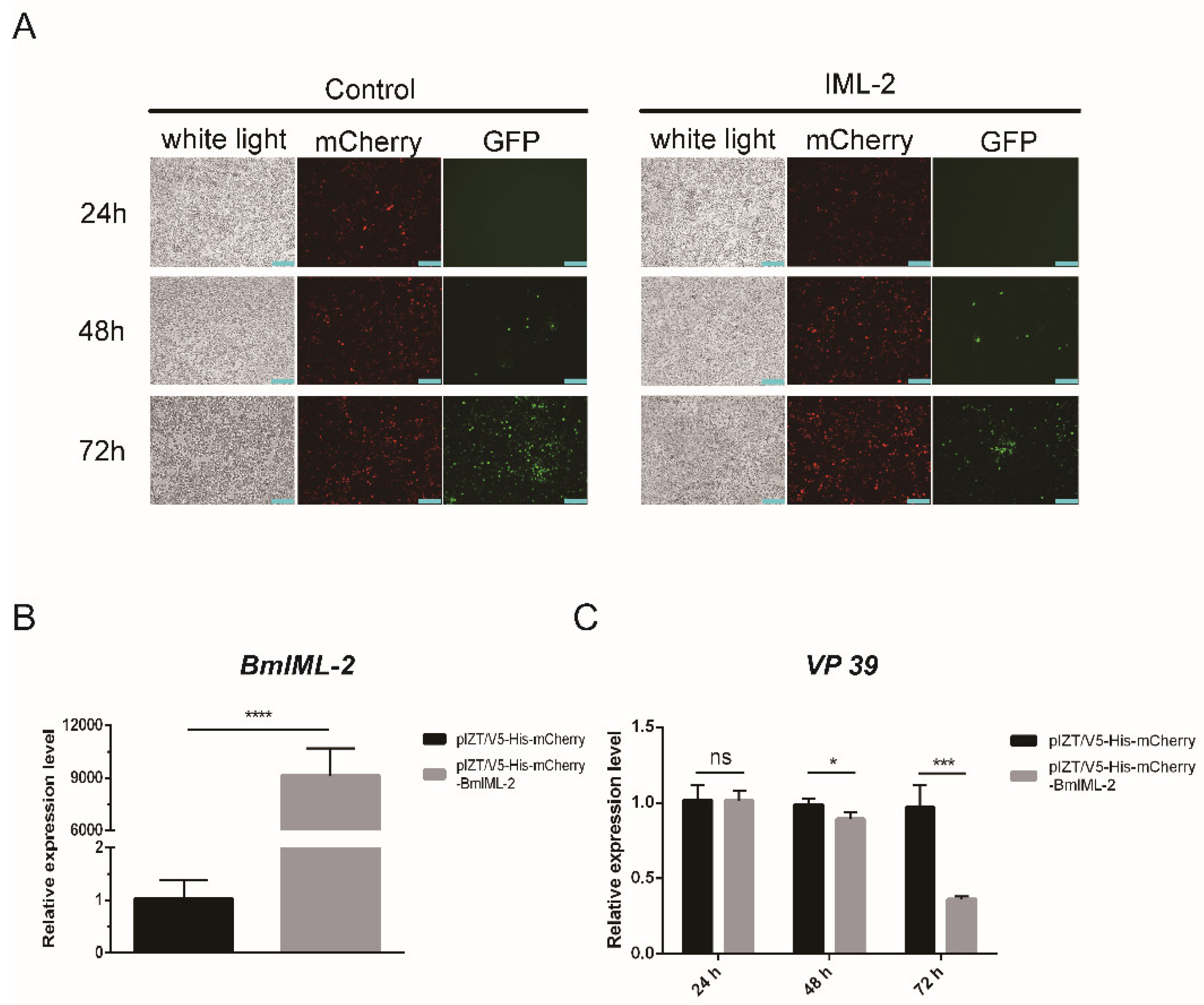
2.3. Overexpression of BmIML-2 Inhibited BmNPV Infection and Replication
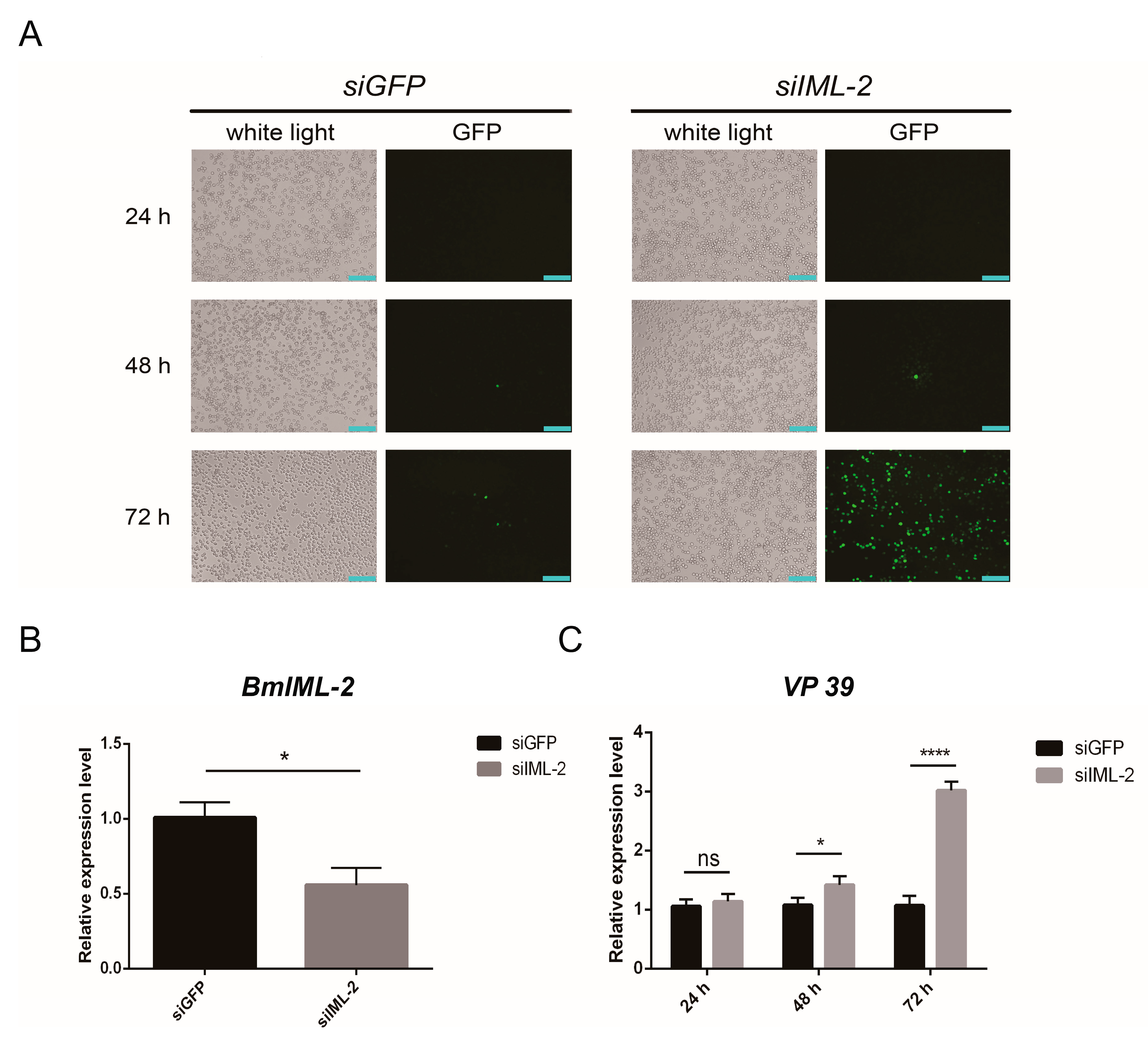
2.4. Knockdown of BmIML-2 Promoted BmNPV Infection and Replication
2.5. Overexpression of BmIML-2 Affected the Expression of Apoptosis-Related Genes
2.6. Overexpression of BmIML-2 Facilitated BmNPV-Induced Apoptosis
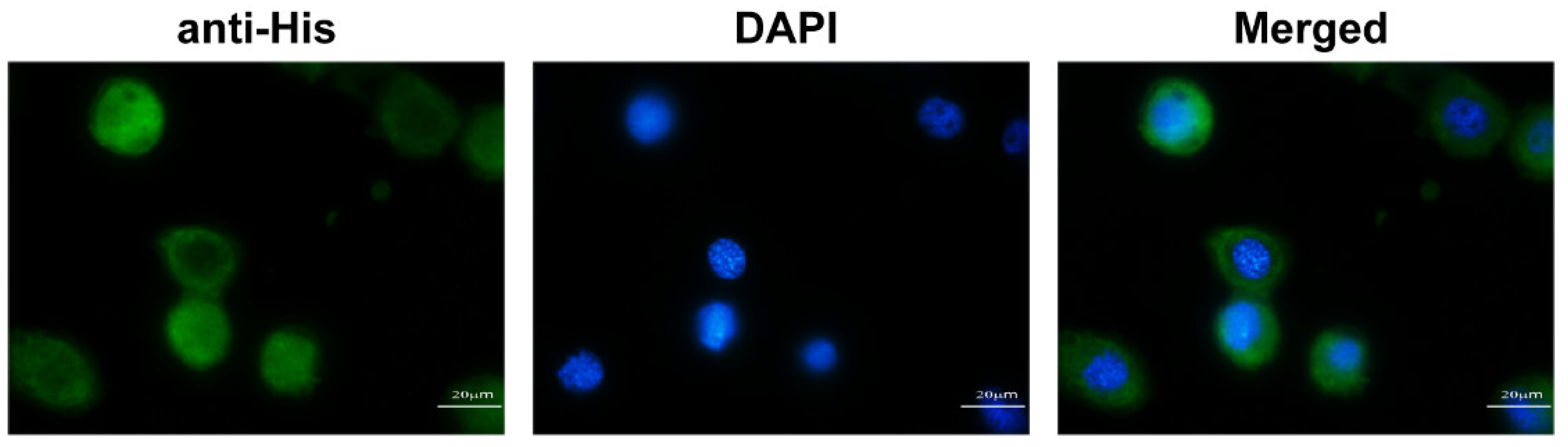
2.7. Immunofluorescence Localization Analysis of BmIML-2
3. Discussion
4. Materials and Methods
4.1. Preparation of Silkworm, BmN Cells, and Virus
4.2. Bioinformatics Analysis of BmIML-2
4.3. RNA Extraction and cDNA Synthesis
4.4. Overexpression Vector Construction and Small Interfering RNA (siRNA) Synthesis
4.5. Cell Transient Transfection and BmNPV Infection
4.6. Genomic DNA Extraction
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Observation of Apoptosis Morphology of Cells after BmIML-2 Overexpression
4.9. Immunofluorescence Analysis of BmIML-2
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [CrossRef] [PubMed]
- Vepari, C.; Kaplan, D.L. Silk as a Biomaterial. Prog. Polym. Sci. 2007, 32, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y. Facilitating Drug Discovery in Human Disease Models Using Insects. Biol. Pharm. Bull. 2020, 43, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Panthee, S.; Paudel, A.; Hamamoto, H.; Sekimizu, K. Advantages of the Silkworm As an Animal Model for Developing Novel Antimicrobial Agents. Front. Microbiol. 2017, 8, 373. [Google Scholar] [CrossRef]
- Hu, X.; Shen, Y.; Zheng, Q.; Wang, G.; Wu, X.; Gong, C. Bm59 is an early gene, but is unessential for the propagation and assembly of Bombyx mori nucleopolyhedrovirus. Mol. Genet. Genom. 2016, 291, 145–154. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yu, H.Z.; Geng, L.; Xu, J.P.; Yu, D.; Zhang, S.Z.; Ma, Y.; Fei, D.Q. Comparative Transcriptome Analysis of Bombyx mori (Lepidoptera) Larval Midgut Response to BmNPV in Susceptible and Near-Isogenic Resistant Strains. PLoS ONE 2016, 11, e155341. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, D.; Zhao, Q.; Zhang, G.; Zhang, Y.; Qiu, Z.; Shen, D.; Lu, C. Label-free proteomic analysis of silkworm midgut infected by Bombyx mori nuclear polyhedrosis virus. J. Proteom. 2019, 200, 40–50. [Google Scholar] [CrossRef]
- Feng, M.; Kong, X.; Zhang, J.; Xu, W.; Wu, X. Identification of a novel host protein SINAL10 interacting with GP64 and its role in Bombyx mori nucleopolyhedrovirus infection. Virus Res. 2018, 247, 102–110. [Google Scholar] [CrossRef]
- Dong, X.; Liu, T.; Wang, W.; Pan, C.; Wu, Y.; Du, G.; Chen, P.; Lu, C.; Pan, M. BmREEPa Is a Novel Gene that Facilitates BmNPV Entry into Silkworm Cells. PLoS ONE 2015, 10, e144575. [Google Scholar] [CrossRef]
- Cherry, S.; Silverman, N. Host-pathogen interactions in Drosophila: New tricks from an old friend. Nat. Immunol. 2006, 7, 911–917. [Google Scholar] [CrossRef]
- Hughes, A.L. Evolution of the betaGRP/GNBP/beta-1,3-glucanase family of insects. Immunogenetics 2012, 64, 549–558. [Google Scholar] [CrossRef]
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in lepidopteran insects. Adv. Exp. Med. Biol. 2010, 708, 181–204. [Google Scholar] [PubMed]
- Costa, A.; Jan, E.; Sarnow, P.; Schneider, D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 2009, 4, e7436. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, W.; Guo, H.; Dang, Y.; Cheng, T.; Yang, W.; Sun, Q.; Wang, B.; Wang, Y.; Xie, E.; et al. Distinct Functions of Bombyx mori Peptidoglycan Recognition Protein 2 in Immune Responses to Bacteria and Viruses. Front Immunol. 2019, 10, 776. [Google Scholar] [CrossRef]
- Drickamer, K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 1992, 360, 183–186. [Google Scholar]
- Robinson, M.J.; Sancho, D.; Slack, E.C.; LeibundGut-Landmann, S.; Reis, E.S.C. Myeloid C-type lectins in innate immunity. Nat. Immunol. 2006, 7, 1258–1265. [Google Scholar] [CrossRef]
- Drickamer, K.; Taylor, M.E. Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 2015, 34, 26–34. [Google Scholar] [CrossRef]
- Brown, G.D. C-type lectins in innate antifungal immunity: A key to the therapeutic future? Mycoses 2015, 583, 1. [Google Scholar]
- Geng, T.; Lu, F.; Wu, H.; Wang, Y.; Lou, D.; Tu, N.; Zhu, F.; Wang, S. C-type lectin 5, a novel pattern recognition receptor for the JAK/STAT signaling pathway in Bombyx mori. J. Invertebr. Pathol. 2021, 179, 107473. [Google Scholar] [CrossRef]
- Liu, K.; Qian, Y.; Jung, Y.S.; Zhou, B.; Cao, R.; Shen, T.; Shao, D.; Wei, J.; Ma, Z.; Chen, P.; et al. mosGCTL-7, a C-Type Lectin Protein, Mediates Japanese Encephalitis Virus Infection in Mosquitoes. J. Virol. 2017, 91, e01348-16. [Google Scholar] [CrossRef]
- Cheng, G.; Cox, J.; Wang, P.; Krishnan, M.N.; Dai, J.; Qian, F.; Anderson, J.F.; Fikrig, E. A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell 2010, 142, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fang, N.N.; Zheng, Y.; Liu, K.Y.; Mao, B.; Kong, L.N.; Chen, Y.; Ai, H. Identification and characterization of two novel C-type lectins from the larvae of housefly, Musca domestica L. Arch. Insect Biochem. Physiol. 2018, 98, e21467. [Google Scholar] [CrossRef]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Jiravanichpaisal, P.; Lee, B.L.; Soderhall, K. Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology 2006, 211, 213–236. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, L.B.; Ma, Y.; Liu, Y.X.; Cao, H.H.; Wang, Y.L.; Kong, X.; Huang, Z.H.; Zhu, H.D.; Wang, Y.X.; et al. Bombyx mori beta-1,3-Glucan Recognition Protein 4 (BmbetaGRP4) Could Inhibit the Proliferation of B. mori Nucleopolyhedrovirus through Promoting Apoptosis. Insects 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Clem, R.J. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top Microbiol. Immunol. 2005, 289, 113–129. [Google Scholar] [CrossRef]
- Clarke, T.E.; Clem, R.J. Insect defenses against virus infection: The role of apoptosis. Int. Rev. Immunol. 2003, 22, 401–424. [Google Scholar] [CrossRef]
- Clem, R.J. Baculoviruses and apoptosis: A diversity of genes and responses. Curr. Drug Targets 2007, 8, 1069–1074. [Google Scholar] [CrossRef]
- Ikeda, M.; Yamada, H.; Hamajima, R.; Kobayashi, M. Baculovirus genes modulating intracellular innate antiviral immunity of lepidopteran insect cells. Virology 2013, 435, 1–13. [Google Scholar] [CrossRef]
- Yamada, H.; Shibuya, M.; Kobayashi, M.; Ikeda, M. Identification of a novel apoptosis suppressor gene from the baculovirus Lymantria dispar multicapsid nucleopolyhedrovirus. J. Virol. 2011, 85, 5237–5242. [Google Scholar] [CrossRef]
- Suganuma, I.; Ushiyama, T.; Yamada, H.; Iwamoto, A.; Kobayashi, M.; Ikeda, M. Cloning and characterization of a dronc homologue in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2011, 41, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Kitaguchi, K.; Hamajima, R.; Kobayashi, M.; Ikeda, M. Novel apoptosis suppressor Apsup from the baculovirus Lymantria dispar multiple nucleopolyhedrovirus precludes apoptosis by preventing proteolytic processing of initiator caspase Dronc. J. Virol. 2013, 87, 12925–12934. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Hamajima, R.; Saito, A.; Tomizaki, M.; Iwamoto, A.; Kobayashi, M.; Yamada, H.; Ikeda, M. Bombyx mori homolog of tumor suppressor p53 is involved in apoptosis-mediated antiviral immunity of B. mori cells infected with nucleopolyhedrovirus. Dev. Comp. Immunol. 2018, 84, 133–141. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, X.; Li, C.; Peng, P.; Wang, J.; He, E.; Qiu, Z.; Xia, D.; Zhao, Q.; Shen, D. Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis. Int. J. Mol. Sci. 2022, 23, 8369. https://doi.org/10.3390/ijms23158369
Mei X, Li C, Peng P, Wang J, He E, Qiu Z, Xia D, Zhao Q, Shen D. Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis. International Journal of Molecular Sciences. 2022; 23(15):8369. https://doi.org/10.3390/ijms23158369
Chicago/Turabian StyleMei, Xianghan, Chun Li, Peilin Peng, Jue Wang, Enxi He, Zhiyong Qiu, Dingguo Xia, Qiaoling Zhao, and Dongxu Shen. 2022. "Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis" International Journal of Molecular Sciences 23, no. 15: 8369. https://doi.org/10.3390/ijms23158369
APA StyleMei, X., Li, C., Peng, P., Wang, J., He, E., Qiu, Z., Xia, D., Zhao, Q., & Shen, D. (2022). Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis. International Journal of Molecular Sciences, 23(15), 8369. https://doi.org/10.3390/ijms23158369





