A Systematic Study on the Optimal Nucleotide Analogue Concentration and Rate Limiting Nucleotide of the SARS-CoV-2 RNA-Dependent RNA Polymerase
Abstract
:1. Introduction
2. Results
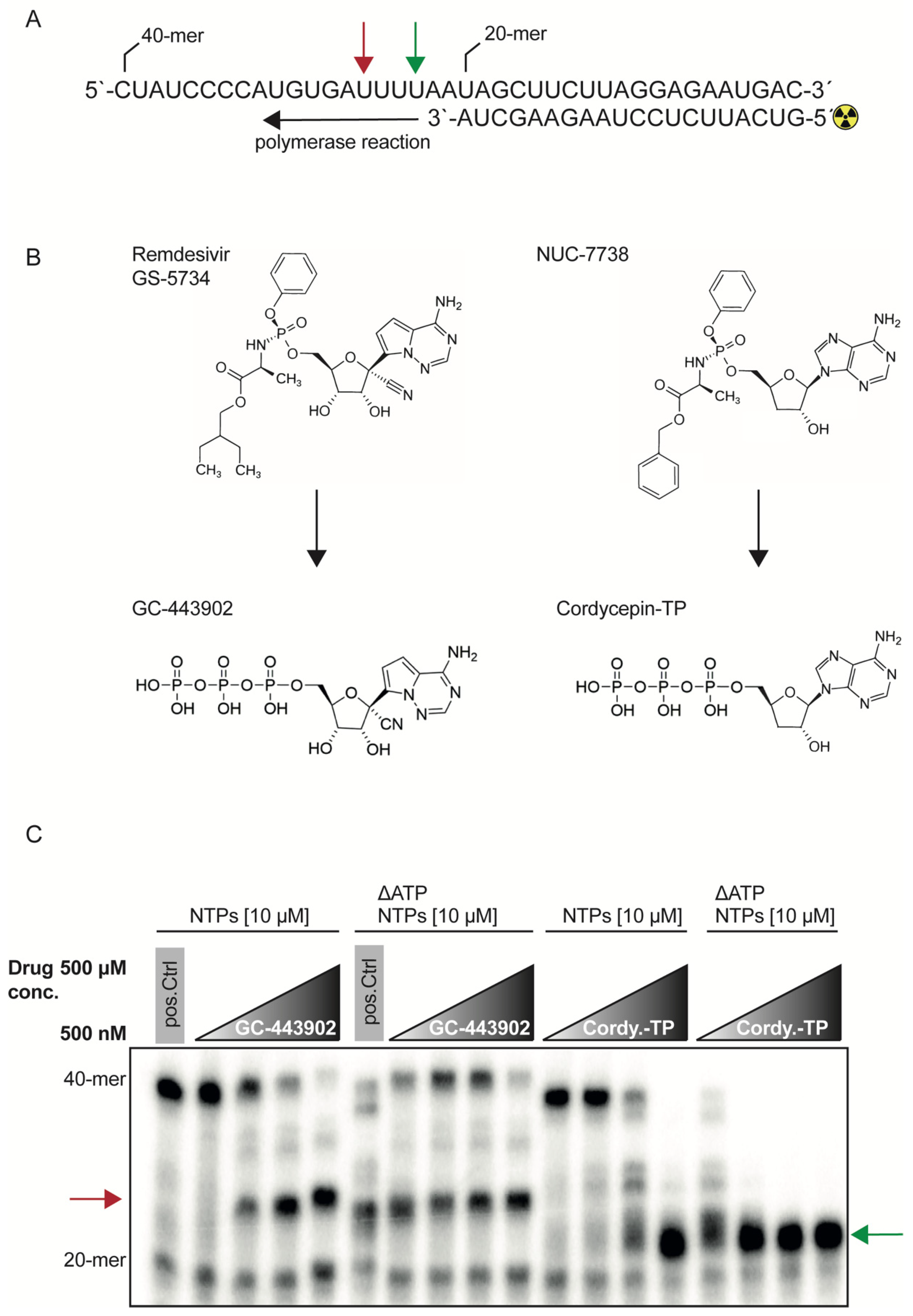
2.1. RdRp Inhibition with Remdesivir-TP and Cordycepin-TP Is NTP Dependent
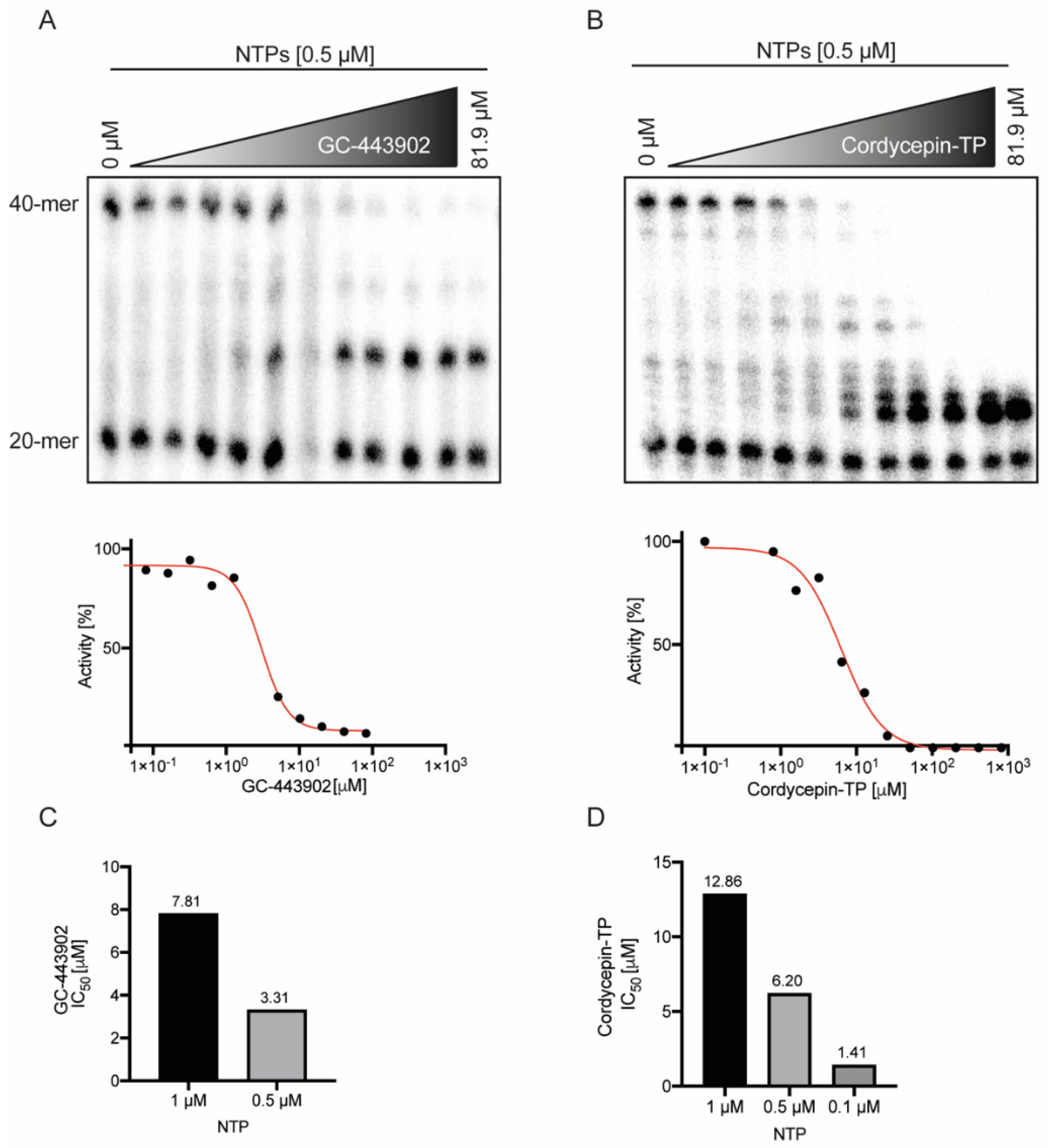
2.2. Remdesivir-TP and Cordycepin-TP Have Similar IC50
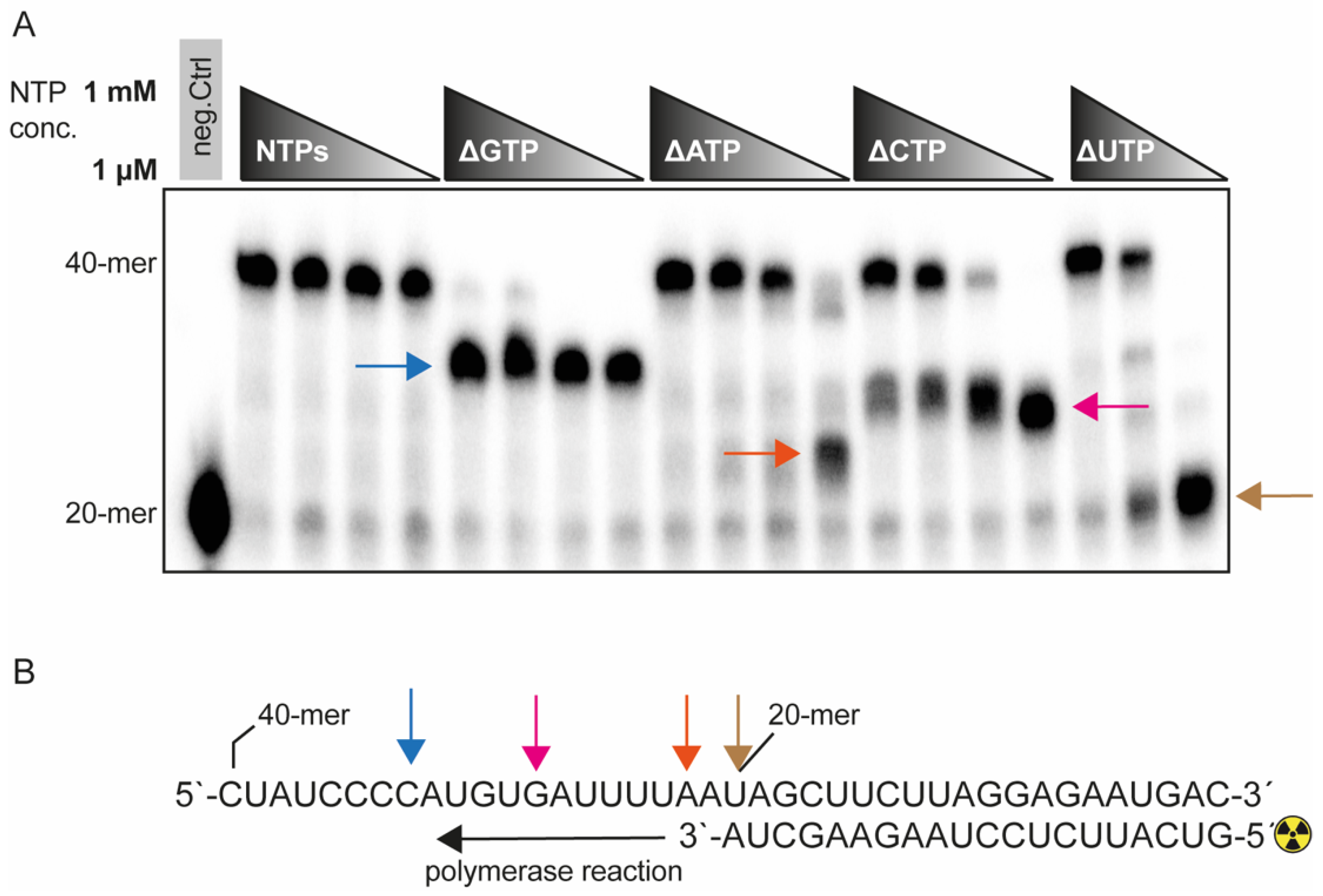
2.3. The Fidelity of the SARS-CoV-2 RdRp Complex Is Sensitive to GTP
3. Discussion
4. Materials and Methods
4.1. Expression Constructs and Recombinant Protein Expression
4.2. RNA Elongation Assay
4.3. IC50 Calculations
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hilgenfeld, R.; Peiris, M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013, 100, 286–295. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 2020, 580, E7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, R.; Gilby, N.B.; Wei, G.W. Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance. J. Chem. Inf. Model. 2022; Preprint. [Google Scholar] [CrossRef]
- Tsai, C.H.; Lee, P.Y.; Stollar, V.; Li, M.L. Antiviral therapy targeting viral polymerase. Curr. Pharm. Des. 2006, 12, 1339–1355. [Google Scholar] [CrossRef] [PubMed]
- Buonaguro, L.; Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Med. 2020, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Drozdzal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Kotfis, K.; Ghavami, S.; Los, M.J. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist. Updates 2020, 53, 100719. [Google Scholar] [CrossRef] [PubMed]
- Pradere, U.; Garnier-Amblard, E.C.; Coats, S.J.; Amblard, F.; Schinazi, R.F. Synthesis of nucleoside phosphate and phosphonate prodrugs. Chem. Rev. 2014, 114, 9154–9218. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Choy, K.T.; Wong, A.Y.; Kaewpreedee, P.; Sia, S.F.; Chen, D.; Hui, K.P.Y.; Chu, D.K.W.; Chan, M.C.W.; Cheung, P.P.; Huang, X.; et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020, 178, 104786. [Google Scholar] [CrossRef]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Abeldano Zuniga, R.A.; Coca, S.M.; Abeldano, G.F.; Gonzalez-Villoria, R.A.M. Clinical effectiveness of drugs in hospitalized patients with COVID-19: A systematic review and meta-analysis. Ther. Adv. Respir. Dis. 2021, 15, 17534666211007214. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Gotte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.C.; Johnson, R.M.; Ayyanathan, K.; Miller, J.; Whig, K.; Kamalia, B.; Dittmar, M.; Weston, S.; Hammond, H.L.; Dillen, C.; et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature 2022, 604, 134–140. [Google Scholar] [CrossRef]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573.e13. [Google Scholar] [CrossRef] [PubMed]
- Schwenzer, H.; De Zan, E.; Elshani, M.; van Stiphout, R.; Kudsy, M.; Morris, J.; Ferrari, V.; Um, I.H.; Chettle, J.; Kazmi, F.; et al. The Novel Nucleoside Analogue ProTide NUC-7738 Overcomes Cancer Resistance Mechanisms In Vitro and in a First-In-Human Phase I Clinical Trial. Clin. Cancer Res. 2021, 27, 6500–6513. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Hobartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.P.; Fan, H.; Keown, J.R.; Knight, M.L.; Grimes, J.M.; Fodor, E. The SARS-CoV-2 RNA polymerase is a viral RNA capping enzyme. Nucleic Acids Res. 2021, 49, 13019–13030. [Google Scholar] [CrossRef] [PubMed]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. Adenosine as an endogenous immunoregulator in cancer pathogenesis: Where to go? Purinergic Signal 2013, 9, 145–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burnstock, G.; Boeynaems, J.M. Purinergic signalling and immune cells. Purinergic Signal 2014, 10, 529–564. [Google Scholar] [CrossRef] [Green Version]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 16, 177–192. [Google Scholar] [CrossRef]
- Rosenke, K.; Hansen, F.; Schwarz, B.; Feldmann, F.; Haddock, E.; Rosenke, R.; Barbian, K.; Meade-White, K.; Okumura, A.; Leventhal, S.; et al. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat. Commun. 2021, 12, 2295. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.J.; Su, M.I.; Wu, J.L.; Kumar, S.; Lim, L.H.; Wang, C.W.; Nelissen, F.H.; Chen, M.C.; Doreleijers, J.F.; Wijmenga, S.S.; et al. How a low-fidelity DNA polymerase chooses non-Watson-Crick from Watson-Crick incorporation. J. Am. Chem. Soc. 2014, 136, 4927–4937. [Google Scholar] [CrossRef] [PubMed]
- Ganai, R.A.; Johansson, E. DNA Replication-A Matter of Fidelity. Mol. Cell 2016, 62, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Sanjuan, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [Green Version]
- Sydow, J.F.; Cramer, P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009, 19, 732–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006, 439, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [Green Version]
- Malone, B.; Chen, J.; Wang, Q.; Llewellyn, E.; Choi, Y.J.; Olinares, P.D.B.; Cao, X.; Hernandez, C.; Eng, E.T.; Chait, B.T.; et al. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2102516118. [Google Scholar] [CrossRef]
- Ogando, N.S.; Ferron, F.; Decroly, E.; Canard, B.; Posthuma, C.C.; Snijder, E.J. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front. Microbiol. 2019, 10, 1813. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vatandaslar, H. A Systematic Study on the Optimal Nucleotide Analogue Concentration and Rate Limiting Nucleotide of the SARS-CoV-2 RNA-Dependent RNA Polymerase. Int. J. Mol. Sci. 2022, 23, 8302. https://doi.org/10.3390/ijms23158302
Vatandaslar H. A Systematic Study on the Optimal Nucleotide Analogue Concentration and Rate Limiting Nucleotide of the SARS-CoV-2 RNA-Dependent RNA Polymerase. International Journal of Molecular Sciences. 2022; 23(15):8302. https://doi.org/10.3390/ijms23158302
Chicago/Turabian StyleVatandaslar, Hasan. 2022. "A Systematic Study on the Optimal Nucleotide Analogue Concentration and Rate Limiting Nucleotide of the SARS-CoV-2 RNA-Dependent RNA Polymerase" International Journal of Molecular Sciences 23, no. 15: 8302. https://doi.org/10.3390/ijms23158302
APA StyleVatandaslar, H. (2022). A Systematic Study on the Optimal Nucleotide Analogue Concentration and Rate Limiting Nucleotide of the SARS-CoV-2 RNA-Dependent RNA Polymerase. International Journal of Molecular Sciences, 23(15), 8302. https://doi.org/10.3390/ijms23158302





