Abstract
Non-coding RNAs (ncRNAs) represent a research hotspot by playing a key role in epigenetic and transcriptional regulation of diverse biological functions and due to their involvement in different diseases, including oral inflammatory diseases. Based on ncRNAs’ suitability for salivary biomarkers and their involvement in neuropathic pain and tissue regeneration signaling pathways, the present narrative review aims to highlight the potential clinical applications of ncRNAs in oral inflammatory diseases, with an emphasis on salivary diagnostics, regenerative dentistry, and precision medicine for neuropathic orofacial pain.
1. Introduction
Oral inflammatory diseases are commonly observed pathologies in everyday dental clinical practice and via activation of the systemic immune response can lead to the development of systemic inflammatory disorders and impairment of the individual’s general health [1]. In order to improve clinical outcomes and increase patient quality of life, there is a tendency to incorporate molecular profiling into clinical decision-making and molecular signature-guided therapies. Based on the understanding of non-coding RNAs (ncRNAs) in the regulation of inflammatory signaling, new avenues for ncRNA in diagnostics and therapeutic intervention in inflammatory diseases have opened.
2. Non-Coding RNA Transcriptome
It has been proposed that approximately 70% of the human genome is transcribed into mRNA, but only around 2% are protein-coding [2,3,4], suggesting that a very tiny proportion of the human genome sequences and elements translate into proteins. According to this, a vast majority of transcribed sequences of RNA molecules are regulatory elements. Non-coding RNAs are genome elements that do not encode for amino acids [4]. They are involved in various cellular processes and interfere with signaling pathways. Non-coding RNAs are localized in almost every cellular compartment and interact with nucleic acids and proteins, thus changing their conformation and functions. Some non-coding RNAs interact with chromatin, contributing to chromatin remodeling, and thus shaping gene activity. In the backbone of every pathological condition lies non-coding RNA transcriptome changes, and their interactome. The localization and abundance of non-coding RNA are tissue-specific [5] and can change in response to a wide span of stressogenic factors and cellular and environmental events. Non-coding RNA transcriptome changes should be more closely discovered in association with disease development and have the potential to be utilized as diagnostic, predictive, and prognostic biomarkers; parameters for the stratification of patients into more specific groups; and potential targets for therapeutics. The non-coding RNA transcriptome comprises RNA molecules of various structures, lengths, and functions [4]. The basic classification on long non-coding (lncRNA) and small non-coding RNA is based on their length.
Transfer RNA (tRNAs) and ribosomal rRNAs (rRNAs) were among the first RNAs found that do not translate into proteins. However, high-throughput “whole transcriptome” techniques discovered seemingly insignificant but now very important regulators of all cellular processes in the two additional classes of non-coding RNAs: long non-coding and small non-coding RNA molecules. [6]. Classification of non-coding RNAs is based mostly on their length, but there are other types of non-coding RNA based on their special features such as packing into microvesicles, including extracellular RNAs [7].
2.1. Long Non-Coding RNA
LncRNAs are endogenous cellular RNAs lacking an ORF (open reading frame) or with a shorter ORF than mRNA, which is used as one of the criteria for distinguishing lncRNAs from messenger RNAs [8,9]. LncRNAs are classified as more than 200-nucleotide-long RNA molecules [10]. There are more than 16,000 lncRNA genes, while estimations suggest more than 100,000 lncRNAs [11]. The expression and abundance of lncRNAs is tissue specific; they have specific cellular localization, and have the ability to alter signaling pathways by interacting with nucleic acids and proteins. They are involved in gene expression regulation at the pre-, post-, and transcriptional level via changes in the chromatin state, and transcriptional activity, splicing, and translation [11,12]. LncRNAs can be subclassified into the groups of very long intergenic RNAs longer than 10 kb, and macroRNAs, which are pathway specific [13]. Five additional subclasses of RNA transcripts can be distinguished: intronic, intergenic, sense, antisense, and bidirectional [8]. Regarding the functional significance of lncRNA, one should be cautious, and conclusions should rely on specific studies with genetic quantitative loss-of-function or gain-of-function models in each species due to the different levels of lncRNA evolutionary conservation [9]. Namely, it has been suggested that lncRNA conservation between species could be considered at four dimensions: at the sequence level, where lncRNAs from different species can have sequence homology and thus similar transcripts; at the structural level, where similar structures could be produced despite a lack of lncRNA sequence homology; at the functional level, where similar functions could be executed despite lncRNAs’ different sequences and structures; and at the transcriptional level, where the locus of transcription is conserved, thus mediating functions despite different lncRNA transcripts [9].
2.2. Small Non-Coding RNA
Small non-coding RNAs (sncRNAs) are 20–200-nucleotide (some around even 400 nts in length) RNA molecules involved in the regulation of gene expression, transcription, translation, splicing, RNA modification, and methylation by the interaction with target mRNA, either through full or partial complementarity [14]. Different types of snRNAs such as microRNAs (miRs), endogenous short interfering RNA (endo-siRNA), piwi-interacting RNA (pi-RNAs), transfer RNA (tRNAs), transfer RNA fragments (tRFs), small nuclear (snRNAs), small nucleolar (snoRNAs), small Cajal-body RNAs (scaRNAs), YRNAs (small stem-loop RNA structures), and short hairpin RNA (shRNA) are synthesized through various biogenesis and maturation pathways by various enzymes and by interaction with different proteins, such as Argonaute or Pi-wi like, combined into the ribonucleoproteins [14,15,16]. Ribonucleoprotein complexes guide non-coding RNA to the target and enable them to interact not only with RNA molecules but with proteins and DNA in some cases, such as microRNAs, thus forming a very complex coding-non-coding RNA-DNA-protein reactome [17,18,19].
So far, the vast number of studies have focused on microRNAs as critical molecules governing development and stress responses. Very recent studies have paid attention to other sncRNAs, tRNA and tRFs, that accumulate in the stressed cell [20]. Because both sncRNAs have a Dicer-dependent biogenesis pathway, it has been proposed that tRFs could function akin to an miR to inhibit the translation or cleave partial complementary target sites [21]. The canonical pathway of miR biogenesis requires two enzymes: Drosha and Dicer. Primary miR transcripts are first cleaved by the nuclear “microprocessor” complex, which contains the enzyme Drosha, and then exported to the cytoplasm, where they are further processed by another enzyme, Dicer, forming the mature miR. Dicer is critical for most miRNAs, but the 5p miRNAs appear to be produced to some extent even without Dicer. Moreover, unlike canonical miR-21-5p, the synthesis of noncanonical miRs, including miR-320a-3p and miR-484–3p, is Drosha independent [22].
In addition to those described above, it is noteworthy to mention extracellular RNAs (exRNAs) [7] and competing endogenous RNAs (ceRNAs) packed into lipid or protein particles, such as exosomes, microvesicles, or oncosomes, which are especially significant for cancer research, which bare and transfer very important genetic and biological information between cells and organ systems [7]. The competing RNA (ceRNA) term is associated more with their function and role rather than the length or another structural feature or localization. The competing RNA phenomenon is associated more with interaction with long and small non-coding RNA and other transcripts, thus acting as sponges by controlling the amounts of active (free to interact with) non-coding RNA transcripts [23].
2.3. Challenges Related to the Clinical Application of ncRNAs
The introduction of single-cell sequencing has enabled investigation of not only the landscape of ncRNAs but also their cellular function. Furthermore, bioinformatic tools are becoming more sophisticated and are used for analyzing ncRNA sequencing data. However, detection of ncRNAs still remains challenging due to the low expression and unique features of certain ncRNAs and the bias of RNA sequencing and bioinformatic methods, leading to erroneous identification of ncRNA species [24].
Currently, eleven RNA-based therapeutics have been approved by the US Food and drug administration and/or the European Medicines Agency involving antisense oligonucleotides (ASOs) or small interfering RNAs (siRNAs) while others, including miRNA mimics and antimiRs, are in phase II or III clinical investigations [25].
The use of miR-based therapeutics has several advantages [26]. Namely, miRs are naturally occurring molecules in human cells, contrary to synthetic chemotherapy compounds or ASOs, and all the mechanisms in cells for their processing and downstream gene target selection are available. Additionally, miRs, by targeting multiple genes within one pathway, show a broader yet specific response [26]. It is noteworthy that circulating miRs (in serum, saliva, and other body fluids), due to their stability and distinctive function, have the advantage of being biomarkers compared with other biomarkers such as proteins (cytokines). Moreover, as controllers of gene transcription, miRNAs and their expression have a higher probability of being related to clinical variables and since they reflect cellular disturbance that occurs years before the appearance of related clinical signs, they enable disease-preventing actions [27].
Clinical applications of all RNA-based therapeutics are hindered by several issues: specificity, related to undesired effects due to uptake in non-target cells or overdosing; delivery, mainly related to inefficient intracellular delivery and the lack of suitable delivery vehicles; and tolerability, caused by the recognition of RNA molecules by pathogen-associated molecular pattern (PAMP) receptors, causing strong immune effects [25]. Therefore, in order to achieve the application of ncRNA as therapeutics and biomarkers, further research should focus on immunogenicity screening, extensive pharmacodynamic and pharmacokinetic studies regarding delivery systems, and chemical modifications to improve specificity.
3. ncRNAs and Oral Inflammatory Diseases
Every disease is a consequence of the specific interactions among the genetic and epigenetic backgrounds of each individual and cellular and environmental conditions. During the last decade, it has been discovered that the non-coding elements of the genome underlie the onset and progression of inflammatory diseases [12].
Pulpitis is a chronic inflammatory condition of the dental pulp, mostly caused by cavities [28]. Periodontal disease is a result of the complex interaction among inflammatory and immune responses induced by pathogenic bacteria [29] and the individual’s genetic and epigenetic background. Periodontitis is mainly caused by bacterial infection affecting periodontal tissue, which can lead to the loss of alveolar bone attachment [30]. Peri-implantitis is an inflammatory process of the tissues around an osseointegrated implant and may result in supporting bone loss, and miRs, such as ncRNAs, could be engaged in the prognosis of peri-implant bone resorption [31].
Sjögren’s syndrome (SS) is a rheumatoid pathological condition predominantly affecting the salivary and lachrymal glands, leading to oral dryness and salivary gland swelling. Patients with SS show an increased risk of developing non-Hodgkin’s lymphoma [32]. Oral lichen planus (OLP) is a premalignant epithelial oral lesion, with the potential to malignantly transform [33]. Oral squamous cell carcinoma (OSCC) is a malignant epithelial tumor with low overall survival rates and poor prognosis, regardless of the advances in surgery, chemotherapy, and radiotherapy [34]. Growing evidence supports an association between oral squamous cell carcinoma (OSCC) and chronic inflammation and the involvement of long non-coding transcriptome alterations [34].
3.1. Long Non-Coding RNA and Oral Inflammatory Diseases, Premalignant States, and Oral Squamous Cell Carcinoma
LncRNAs were shown to be closely associated with oral inflammatory diseases and malignant transformation in the oral cavity epithelium, but the mechanisms underlying these pathological conditions are still unknown. Long non-coding RNAs were shown to be immunomodulatory [35]. Thus, antisense non-coding RNA in the INK4 locus (ANRIL) regulates the STAT1 pathway and thus the production of the proinflammatory cytokine interferon-gamma (IFN-γ). ANRIL is one of the regulators and a component of the NF-kB pathway, and after TNFα treatment, it induces IL-6 and IL-8 expression [36], indicating that it is an important regulator of inflammation underlying the pathology of this type of disease. Upregulation of another proinflammatory long non-coding RNA, of lncRNA MEG3 (lncRNA maternally expressed gene 3), was associated with pulpitis progression while its downregulation was associated with dental pulp regeneration [37]. The authors also found that inhibition of lncMEG3 lowered the secretion of proinflammatory cytokines in dental pulp cells treated with LPS, probably via p38/MAPK signaling pathway regulation [37]. LncRNA FGD5 antisense RNA1 was shown to be underexpressed in gingivae in patients with periodontitis by blocking miR-142 to silence NF-kB signaling and the inflammatory response [38]. LncRNA MALAT1 acts as a sponge to miR-20a and increases inflammatory processes in periodontal tissue via activation of the toll-like receptor 4 pathway [39]. LncRNA DQ786243 was shown to be overexpressed in the CD4+ lymphocytes of oral lichen planus patients compared with healthy individuals and associated with increased proinflammatory miR-146a through Foxp3 activation, which downregulates the NF-κB pathway and, in turn, affected the resulting LncRNA DQ786243 expression [40]. It has been shown that lncRNA TMEVPG1 was increased in CD4+ T helper cells in patients with Sjögren syndrome compared with healthy individuals [41], indicating its potential involvement in the development of SS. It was reviewed by Benedittis et al. that lncs LINC00426, NRIR, CYTOR, TPTEP1, BISPR, AC017002.1, n336161, LINC00426-003, NR_002712, LINC02384, TCONS_l2_00014794, lnc-UTS2D-1:1, and n340599 expression was altered in PBMCs or salivary glands in SS patients [42].
Jia et al. [43] showed the potential of four lncRNAs (ENST00000412740, NR_131012, ENST00000588803, and NR_038323) to distinguish early stage from advanced-stage OSCC. They also showed significant differences between OSCC and healthy controls, indicating that these four lncRNAs not only have prognostic potential but diagnostic potential as well [43]. Other lncRNAs frequently associated with oral cancers are lncRNA cancer susceptibility candidate 9 (lncRNA CASC9) [44] and lncRNA HOTAIR, which was described as an oncogene that boosts the invasive and metastatic potential of OSCC [45]. LncRNA CASC9 was overexpressed in OSCC tissue and SCC15, TSCCA, and CAL27 oral cancer cell lines compared with healthy matched tumorous tissue and HOMEC, a normal cell line derived from oral keratinocytes. Higher lncRNA CASC9 levels were also associated with an advanced T stage, positive lymph node status, and an advanced clinical stage [44]. LncRNA HOTAIR showed higher expression levels in OSCC and cell lines and advanced TNM stages, higher tumor grade, and positive lymph node status compared with healthy oral mucosa epithelium and a normal cell line derived from keratinocytes from the oral cavity. Furthermore, LncRNA HOTAIR acts as a sponge for miR-326, thus decreasing its ability to silence the translation of oncogenes, such as metastasis-associated gene 2, which was confirmed by Tao et al. [45].
3.2. Small Non-Coding and Oral Inflammatory Diseases, Premalignant States, and Oral Squamous Cell Carcinomas
In oral diseases and pathology, microRNAs are the most investigated small non-coding RNA with the highest potential to be utilized in clinical practice. MicroRNAs play a very important role in dental pulp pathology via their role in regulating the immune response and inflammation [46]. miR-21 was shown to mitigate the inflammatory signaling in LPS-stimulated dental pulp cells [47]. Bacterial infection with P. gingivalis lipopolysaccharides (LPS) was shown to induce miR-584 overexpression, which induced IL-8 production and inflammation in the gingival epithelium [48,49].
Kamal et al. [50] compared the differential expression of miRs from the saliva and plasma of patients with chronic periodontitis (CP) with miRs extracted from healthy controls. The authors pointed out that the arrays from saliva and plasma differed from each other and found that miR-let-7d/miR-103a-3p/126-3p/150-5p/199a-3p/4485-5p/6088/6821-5p were significantly lower in the saliva and plasma of the CP patients compared with the controls, indicating that these miRs might be used as diagnostic and prognostic factors of CP [50]. MicroRNA miR-146a and miR-155 deserve special attention because it has been shown that these miRs can modulate immune responses [51]. Their upregulation was observed in the crevicular fluid of patients with chronic periodontitis associated with diabetes mellitus type II [52]. Sipert et al. [51] investigated the differences in the miR expression levels in cultivated fibroblasts in dental pulp, gingival, and periodontal ligament fibroblasts from the same individual by microarray and RT-qPCR analysis. The authors showed a cell-type-specific miR expression pattern and an increase in proinflammatory miR-146a in gingival fibroblasts after stimulation with LPS, and that miR-155 in gingival fibroblasts was decreased after LPS addition [51]. By employing differential expression analysis, miR-517/525/624/3128/3658/3692/3912/3920/4683/4690 were identified as predictors of periimplantitis in the five years after implant surgery, which regulate critical processes in peri-implant tissues, such as inflammation or cellular proliferation [27].
Exosomes represent cellular particles that contain proteins, lipids, coding and non-coding RNAs, microRNA, and cytokines, thus having the ability to transfer information among neighboring and distant cells. It has been also shown that exosomal microRNA can modulate the immune response and inflammation [53,54]. Zheng et al. [55] showed that dental pulp stem cell-derived small extracellular vesicles (DPSCs-sEV) and their cargo, including 81 miRs, have immunomodulatory features in dental pulp cells. Especially, miR-125a-3p jumped was notable for its immunomodulatory potential by regulating the NF-κΒ and toll-like receptor (TLR) axis via silencing of the inhibitor of nuclear factor-kappa B kinase subunit beta (IKBKB) [55]. Exosome-derived microRNA molecules can alter the translation of mRNAs in immune cells [54]. MicroRNA miR-142-3p, which was found in exosomes from T cells, was associated with exocrine gland malfunction in SS [56]. Some other miRs from exosomes such as miR-124a/192-5p and miR-150-5p were associated with an impaired immune response in rheumatoid arthritis [54].
In OLP, miRs are the most studied small non-coding RNA. Gassling et al. [57] identified 16 altered miRNAs by microarray analysis in 7 patients with OLP. Oncogenic miRs, such as miR-21, were shown to be significantly upregulated, meaning that OLP might be a precursor state for malignant transformation. Additionally, miR-31/132/143/155/15a/342-3p were also differentially expressed in OLP compared with healthy controls [57]. Scapoli et al. [58] found that miR-21/23/25/146b/489/129/338/212, among others, were overexpressed in OSCC and that tumor-suppressive miR-34a/520h/197/378/135b, and miR-224 were lower than in healthy control samples. Furthermore, miR-let-7i, miR-155, and miR-146a downregulation was associated with OSCC metastasis [58].
A study investigated circulating small non-coding RNA derived from seven male patients with oral cancer and found a significant difference in the distribution of small non-coding RNA between the cancer and healthy control groups [59]. According to their results, a vast majority of investigated small non-coding RNA (50%) were miRs (miR-103-3p and miR-107 emerged as the most important, and associated with the tumor size), 38% were YRNAs, and 10% were tRNAs while snRNAs, rRNA, snRNA, and snoRNA together contributed only 1% of the small-noncoding RNA transcriptome [59]. These findings indicate the importance of microRNA in future clinical practice for oral disease diagnosis, prognosis, and treatment. The list of lncRNA and sncRNA associated with oral inflammatory diseases was presented in Table 1.

Table 1.
LncRNA and sncRNA association with oral inflammatory diseases in clinical settings.
4. Clinical Perspectives of ncRNAs in Oral Inflammatory Diseases
Due to their involvement in DNA translational control, their regulation of mRNA and protein expression levels, and their ability to reprogram cellular signaling pathways in oral inflammatory diseases, ncRNAs could be used to diagnose and predict disease and to improve patient-tailored treatments as an integral part of precision medicine for oral inflammatory diseases (Figure 1).
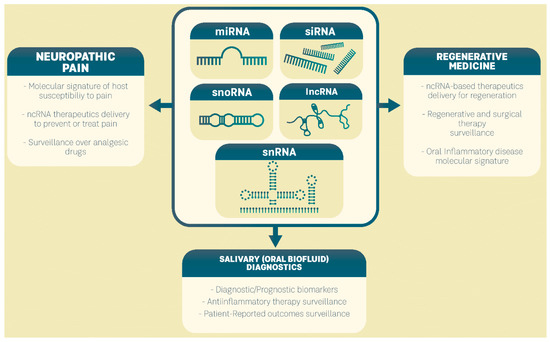
Figure 1.
Clinical perspectives of ncRNAs in precision medicine of oral inflammatory diseases.
4.1. ncRNAs as Salivary Diagnostic Markers of Oral Inflammatory Diseases
Saliva sampling represents a cost-effective and non-invasive procedure while ncRNAs, due to their short size, body fluid stability, and main location inside exosomes, represent very suitable salivary biomarkers. Prominently investigated, microRNAs are engaged in the regulation of cytokine expression and have been established as significant in the pathogenesis of oral inflammatory diseases. As a result, their evaluation in body fluids may be helpful in assessing disease status and progression and in the evaluation of the treatment process.
Salivary miR-21 can be used as a diagnostic marker for oral potentially malignant disorders, showing a specificity of 66% and sensitivity of 69% and an area under the receiver operating characteristic (ROC) curve (AUC) of 0.82 [60]. Regarding OSCC, ROC curve analysis of salivary miR-424, miR-31, and miR-345 showed that each miR had limited power individually to differentiate between OSCC and healthy controls, with miR-345 having the largest AUC of 0.77. However, their combination could differentiate well between OSCC and control samples, with an AUC of 0.87, specificity of 0.77, and sensitivity of 0.86 [61]. Furthermore, the ROC analysis performed by He et al. [62] showed that salivary exosomal miR-24-3p has diagnostic accuracy for OSCC, with an AUC of 0.74, while miR-512-3p and miR-412-3p, with AUC values of 0.85 and 0.87, were also reported as perspective diagnostic markers for OSCC [63].
Patients with periodontitis show higher expression of miR-146a/155 in crevicular fluid, showing high diagnostic accuracy for periodontitis, with an AUC >0.9. [52]. In saliva, the AUC, specificity, and sensitivity of salivary miR-155 in diagnosing periodontitis were 0.88, 78%, and 97.14%, respectively, and those of miR-146a in diagnosing PD were 0.75, 58.54%, and 88.57%, respectively [64]. The miR expression profile in saliva may discriminate patients with primary Sjögren’s syndrome from those with Sjögren-like disease. Namely, analysis of the salivary miRs revealed that the AUC for miR-17-5p was 0.87, let-7i-5p was 0.91, and miR-328-3p was 0.84 when used as single biomarkers. Furthermore, the combination of miR-17-5p and let-7i-5p showed an AUC of 0.97 and, similarly, when all three miRNAs were combined while the four miR-17 family members (i.e., miR-17-5p/106a/106b/20b-5p) in combination yielded an AUC of 0.95 [65]. The list of salivary miRs with diagnostic value for oral inflammatory diseases is presented in the Table 2.

Table 2.
The list of salivary miRs with diagnostic value for oral inflammatory diseases.
We performed bioinformatic analysis (miRnet) in an attempt to identify the shared target genes of salivary microRNAs with diagnostic value for oral inflammatory diseases. miRNet is an online network tool that visualizes the interactions between miRs and their targets [66]. According to miRNet, we discovered the shared targets of 17 miR molecules. One gene transcript (PTEN) was the potential target of 12 miRs (degree 12) and 3 gene transcripts (CDKN1A, NFAT5, and KIAA1551) were associated with 10 miRs (degree 10) (Table 3). The three miRs, miR-106a/106b-5p, and miR-20b-5p, interact with all four listed genes. Literature analysis of these four shared genes: CDKN1, PTEN, NFAT5, and KIAA1551 (RESF1), revealed that all genes are significant for immunological responses, mainly via regulation of the T cell responses. Namely, the CDKN1 gene encodes a potent cyclin-dependent kinase inhibitor p21, which has been shown to control autoimmune T cell autoreactivity without affecting normal T cell responses [67]. PTEN gene expression is significantly positively correlated with CD4/CD8A gene expression and T cell infiltration, especially T helper cells, central memory T cells, and effector memory T cells, in multiple tumor types [68] while PTEN loss predicts a poor therapeutic response and worse survival outcomes in patients receiving immunotherapy. NFAT5 plays a role in the development and activation of immune cells, especially T cells and macrophages contributing to autoimmune and inflammatory diseases [69]. KIAA1551 (RESF1, C12orf35) is highly expressed in the thymus, spleen, bone marrow, and liver, organs associated with the immune system and involved in T cell immunology and transplants [70]. Noteworthy, in silico analysis revealed that upregulation of salivary miR-146a/155 is predicted to upregulate ACE2 expression and essential SARS-CoV-2 receptors, and modulate the host antiviral response; thus, it could be related to the susceptibility of these patients to SARS-CoV-2 infection [71]. Evaluation of PTEN immunoexpression in human oral mucosa specimens showed that alteration of PTEN mediates oral submucous fibrosis pathogenesis and oral carcinogenesis [72]. Likewise, a cell culture study revealed that the NFAT5 transcription factor is able to promote oral cancer cell proliferation via changes in the subcellular localization of the epidermal growth factor receptor [73].

Table 3.
Bioinformatic analysis (miRnet) of salivary miRs with diagnostic value for oral inflammatory diseases showing the top four target genes with the highest degree in the miR-mRNA regulatory network.
4.2. ncRNAs in Regenerative Medicine in the Field of Oral Inflammatory Diseases
As important players in the processes of differentiation and proliferation of stem cells, ncRNAs may represent an option for regenerative treatment (Figure 2). There is a growing body of evidence on the role of ncRNAs in human periodontal ligament stem cells (PDLSCs). Periodontal ligament is a highly specialized connective tissue that surrounds the tooth root, which contains mesenchymal stem cells capable of differentiating into osteoblasts, cementoblasts, and adipocytes; thus, it is considered to be a highly promising stem cell population for alveolar bone repair and regeneration in periodontal disease [74]. Substantial evidence has demonstrated that some lncRNAs, including MEG3, H19, and lncRNA-ANCR, may guide osteogenic differentiation of stem cells under physiological and pathological conditions [75]. In the study of Hao et al. [74], microarray analysis identified an miRNA profile of human PDLSCs that induced osteogenic differentiation. A total of 116 miRNAs were found to be differentially regulated, and the expression of 6 of them was validated: miR-654-3p/4288/34c-5p were found to be upregulated while miR-218-5p/663a/874-3p were downregulated during osteogenesis. However, it seems that the miR regulatory role depends on the microenvironment conditions. For instance, miR-17 seems to inhibit osteogenic differentiation of healthy human PDLSCs, but it has a promoting effect when the cells originate from periodontitis patients or are cultured under inflammatory conditions [76,77].
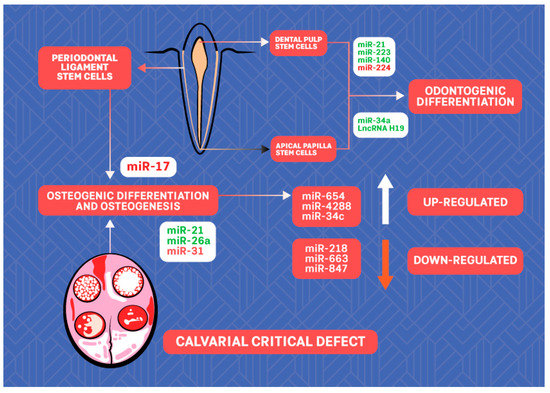
Figure 2.
The scheme of ncRNAs with potential for regenerative dentistry. ncRNAs labeled in green promote while ncRNAs labeled in red inhibit osteogenic/odontogenic differentiation of stem cells.
To illustrate the significance of miRNA in osteogenesis, induction or inhibition of several miRs, including miR-31, miR-26a, and miR-21, for calvarial defect repair was evaluated in vivo [78,79,80,81].
Using miRnet, we investigated the miRs-genes network of selected osteogenic miRs: hsa-mir-654-3p/4288/34c-5p/218-5p/663a/874-3p/21-5p/26a/31-5p (Table 4), and the top three genes with the highest degree (number of miR–mRNA interactions) were found: CDK6, E2F2, and FOXO3, represent perspective targets of bone regenerative medicine since they are involved in bone cell survival, proliferation, differentiation, and angiogenesis [82,83,84]. Noteworthy, FOXO3 transcription factor was found to associate with chronic periapical inflammation in periapical lesion specimens via IL-1β release regulation [85]. Furthermore, an animal study showed that FOXO3a signaling promotion could improve the mandibular bone loss caused by 1,25 dihydroxy vitamin D deficiency [86].

Table 4.
Bioinformatic analysis (miRnet) of miRs involved in the regulation of osteogenic differentiation of human periodontal ligament stem cells and osteogenesis in vivo showing the top four target genes with the highest degree in the miR-mRNA regulatory network.
Based on previous studies, the research on ncRNAs during odontogenic differentiation of dental tissue-derived stem cells has mainly focused on miRs. Xu et al. [87] showed that upregulated expression of miR-21 and expression of signal transducer and activator of transcription 3 (STAT3) are associated with increased odontogenic differentiation of human dental pulp stem cells (DPSCs), promoted by tumor necrosis factor-α. Huang et al. [88] showed that miR-223-3p is expressed at a higher level in inflamed pulp and that overexpression of miR-223-3p in DPSCs significantly increased the levels of markers of odontoblast differentiation: dentine sialophosphoprotein and dentine matrix protein 1. Sun et al. [89] showed that miR-140-5p enhanced the proliferation of human DPSCs but inhibited the differentiation of human DPSCs via regulation of the lipopolysaccharide/toll-like receptor 4 signaling pathway. Downregulation of miR-224-5p may promote DPSCs’ proliferation and migration [90]. miR-34a promotes odontogenic differentiation of human stem cells from the apical papilla (SCAPs), a significant perspective for regenerative endodontics [91]. In this line, LncRNA H19 was reported to lead to enhanced odontogenesis of SCAPs via the miR-141/ SPAG9 signaling pathway [92].
We investigated the shared targets of six selected odontogenic miRNA molecules-hsa-miR-223-3p/21-5p/34a/140-5p-141/224-5p, and the top four genes with the highest number of interactions with the investigated miRs are presented in Table 5. These genes represent widespread regulators of dental pulp stem cell processes, including: quiescence, proliferation, metabolism, differentiation and lineage choice, cell death and survival, self-renewal, and angio-/vasculogenesis [93,94,95,96]. Immunohistochemical data in humans shows that the expression of VEGF is strongly positive in the inflammatory infiltrate in irreversible pulpitis, reflecting the decrease in the microvessel density in irreversible pulpitis [97]. Both VEGF and IGF, by contributing to odontogenic differentiation of DPSCs, represent perspective bioactive molecules in dental pulp tissue engineering [98,99].

Table 5.
Bioinformatic analysis (miRnet) of miRs involved in the regulation of odontogenic differentiation of human dental pulp and apical papilla stem cells showing the top four target genes with the highest degree in the miR-mRNA regulatory network.
4.3. ncRNAs as Biomarkers and Perspective Therapeutics for Neuropathic and Inflammatory Pain
Chronic orofacial pain is usually caused by inflammation and tissue or nerve injury, but it continues even after the initial injury has healed. It is usually characterized by ongoing or intermittent burning pain, an enhanced response to noxious stimuli (hyperalgesia), or pain in response to normally innocuous stimuli (allodynia), accompanied by distress, fatigue, and depression. Current treatment for this disorder has had limited success and new therapeutic strategies, based on precision pain medicine, are warranted. In this regard, engagement of ncRNA in precision oral neuropathic pain medicine represents a potential strategy (Figure 3).
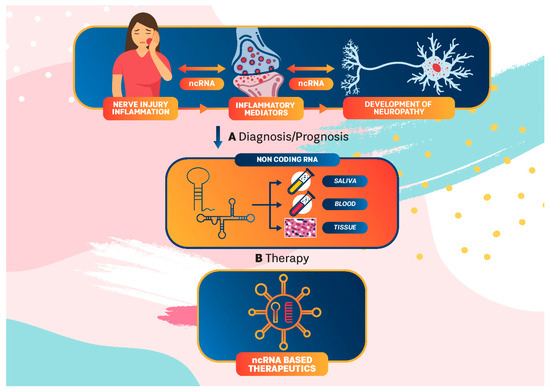
Figure 3.
ncRNA in precision medicine for orofacial neuropathic pain. ncRNA interplay between injury/inflammation and neuropathy development could be used in diagnosis/prediction and in patient-tailored treatments as an integral part of precision medicine for neuropathic pain.
ncRNAs have been identified in pain-related regions in the human nervous system and, following nerve injury in humans, there are highly significant correlations between the abundance of miR-29a and miR-500a in human lingual nerve neuromas and the pain VAS score [100], which suggests a potential contribution of specific miRNAs to the development of chronic neuropathic pain. Lutz et al. [101] proposed a model for the mechanism by which microRNAs contribute to chronic inflammatory and neuropathic pain, and it includes engagement of inflammatory mediators. Namely, the increase in inflammatory mediators, such as interleukin (IL)-1β, after injury induces a change in the expression of miRs in dorsal root ganglion (DRG) neurons, resulting in an alteration of pain-related genes and an increase in DRG neuronal excitability and pain hypersensitivity (hyperalgesia and allodynia). Indeed, miR-146a/199a/558 were shown to be involved in pain-related pathophysiology of osteoarthritis, linked to the expression of cyclooxygenase-2 [102,103,104]. MicroRNA profiles could serve as blood biomarkers of neuropathic pain in humans. For instance, differential expression of 18 miRNAs was reported in blood from patients with complex regional pain syndrome [105]. Human miR-132-3p/146a/miR-21 were upregulated in the white blood cells of patients suffering from neuropathic pain [106,107]. Heyn et al. [108] found that in blood samples from neuropathic pain patients, upregulated miR-124a/155 were associated with reduced expression of Sirtuin 1 mRNA, leading to the development of neuropathic pain. On the other side, Liu et al. [109] found that downregulation of hsa-miR-101 expression in plasma from patients with neuropathic pain led to nuclear factor kappa B activation and consequent development of neuropathic pain.
Trigeminal neuralgia (TN), a common type of orofacial neuropathic pain, is characterized by severe, sudden pain in the trigeminal nerve distribution. In humans with TN, upregulation of circulatory miR-132-3p/146b-5p/155-5p/384 was observed compared to healthy controls while functional analysis indicated that miR-155-5p could directly target and downregulate nuclear factor-E2-related factor 2, which modulates the expression of inflammatory genes [110]. In animal models of TN, Xiong et al. [111] found that knockdown of lncRNA uc.48+ by siRNA could inhibit transduction of TN signals in rats. Li et al. [112] reported that lncRNA MRAK009713 expression was markedly increased in DRG in a rat model of TN, and downregulation of MRAK009713 significantly inhibited the nociceptive transmission and reduced both mechanical and thermal hyperalgesia. On the other side, lncRNAGm14461 expression was upregulated in the trigeminal ganglion in a mice model of TN and is associated with the pain transmission of TN via regulation of proinflammatory cytokines and CGRP expression [113].
Burning mouth syndrome (BMS) is a chronic pain condition characterized by burning sensation or pain felt in the oral mucosa. The etiology of BMS is multifactorial, including oral parafunctional habit, salivary gland dysfunction, or nerve injury, while menopausal disorders and diabetes may contribute to the severity. A recent study by Kim et al. [114] found that salivary exosomal miRNAs (miR-1273h-5p/1273a/1304-3p/4449/1285-3p/6802-5p/1268a/1273d/1273f/423-5p) were upregulated while 18 exosomal miRNAs (miR-27b-3p/16-5p/186-5p/142-3p/141-3p/150-5p/374a-5p/93-5p/29c-3p/29a-3p/148a-3p/22-3p/27a-3p/424-5p/19b-3p/99a-5p/548d-3p/19a-3p) were downregulated in BMS patients compared to controls, suggesting miRs could play an important role in the diagnosis and progression surveillance of BMS.
Patients with temporomandibular disorders (TMDs) frequently report pain deriving from either intra-articular or extra-articular structures. Differences in the perceived TMD pain between individuals make diagnosis and management of the TMD complex, requiring a personalized approach. A research study by Xu et al. [115] found that in synovial fibroblasts from patients suffering from osteoarthritis of the temporomandibular joint, eight miRNAs were upregulated and six miRNAs were downregulated, with miRNA221-3p being the most downregulated. The miRNA221-3p downregulation was attributed to an abundance of IL-1β (inflammation), and associated with induction of matrix metalloproteinases, MMP1 and MMP9, involved in joint injury. In another study, miR-140-5p was found to regulate temporomandibular joint osteoarthritis (TMJOA) via the TGF-β/Smad signaling pathway and might serve as a novel prognostic factor of TMJ degenerative changes [116]. A very recent study showed that the levels of miR-101a-3p were significantly lower in a rat inflammation model with TMJOA and involved in apoptosis of chondrocytes [117]. Using miR21 knockout mice, Zhang et al. [118], reported that miR21, via critical regulation of growth differentiation factor 5 in chondrocytes, regulates cartilage matrix degradation and contributes to the progression of TMJ-OA. A list of ncRNAs with potential as biomarkers of neuropathic orofacial pain is shown in Table 6.

Table 6.
ncRNAs as potential biomarkers of neuropathic orofacial pain.
5. Conclusions
Although still in its infancy, the implementation of precision medicine for oral inflammatory diseases is expected to have a significant impact on patient well-being. At the forefront of precision medicine (in dentistry) is the ability to identify unique characteristics in individual patients with oral inflammatory disease, allowing selection of a tailored treatment. Among the engaged methods, assessment of the in vivo molecular characterization (signature) of the disease and the host (mal)adaptive immune responses, regenerative medicine, and monitoring of drug and patient outcomes could rely on ncRNAs.
Author Contributions
Invitation received, J.R.; Conceptualization, J.R. and N.P.; Data interpretation, J.R. and N.P.; Bioinformatics analysis, J.R. and N.P.; Writing—original draft preparation, review and editing, J.R. and N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Grant [No.451-03-9/2021-14/200129] of the Ministry of Education, Science and Technological Development of the Republic of Serbia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or data interpretation; in the writing of the manuscript, or in the decision to publish the results.
References
- Shankpiece, F.A.; Cantos, A. Oral Inflammation and Infection, and Chronic Medical Diseases: Implications for the Elderly. Periodontology 2000 2016, 72, 153–175. [Google Scholar] [CrossRef]
- ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Angrand, P.-O.; Vennin, C.; Le Bourhis, X.; Adriaenssens, E. The Role of Long Non-Coding RNAs in Genome Formatting and Expression. Front. Genet. 2015, 6, 165. [Google Scholar] [CrossRef] [PubMed]
- Diamantopoulos, M.A.; Tsiakanikas, P.; Scorilas, A. Non-Coding RNAs: The Riddle of the Transcriptome and Their Perspectives in Cancer. Ann. Transl. Med. 2018, 6, 241. [Google Scholar] [CrossRef]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and Abundance Analysis of Human LncRNAs at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, G.; Teng, Z.-Q.; Wingo, T.S. Editorial: Non-Coding RNAs and Human Diseases. Front. Genet. 2020, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Sadik, N.; Cruz, L.; Gurtner, A.; Rodosthenous, R.S.; Dusoswa, S.A.; Ziegler, O.; Van Solinge, T.S.; Wei, Z.; Salvador-Garicano, A.M.; Gyorgy, B.; et al. Extracellular RNAs: A New Awareness of Old Perspectives. Methods Mol. Biol. 2018, 1740, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123. [Google Scholar] [CrossRef]
- Diederichs, S. The Four Dimensions of Noncoding RNA Conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of Long Noncoding RNA Classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Qiu, W.; Wu, B.; Fang, F. Long Non-coding RNAs Are Novel Players in Oral Inflammatory Disorders, Potentially Premalignant Oral Epithelial Lesions and Oral Squamous Cell Carcinoma (Review). Int. J. Mol. Med. 2020, 46, 535–545. [Google Scholar] [CrossRef]
- Hackermüller, J.; Reiche, K.; Otto, C.; Hösler, N.; Blumert, C.; Brocke-Heidrich, K.; Böhlig, L.; Nitsche, A.; Kasack, K.; Ahnert, P.; et al. Cell Cycle, Oncogenic and Tumor Suppressor Pathways Regulate Numerous Long and Macro Non-Protein-Coding RNAs. Genome Biol. 2014, 15, R48. [Google Scholar] [CrossRef]
- Watson, C.N.; Belli, A.; Di Pietro, V. Small Non-Coding RNAs: New Class of Biomarkers and Potential Therapeutic Targets in Neurodegenerative Disease. Front. Genet. 2019, 10, 364. [Google Scholar] [CrossRef] [PubMed]
- Leighton, L.J.; Bredy, T.W. Functional Interplay between Small Non-Coding RNAs and RNA Modification in the Brain. Non-Coding RNA 2018, 4, 15. [Google Scholar] [CrossRef] [PubMed]
- Guglas, K.; Kołodziejczak, I.; Kolenda, T.; Kopczyńska, M.; Teresiak, A.; Sobocińska, J.; Bliźniak, R.; Lamperska, K. YRNAs and YRNA-Derived Fragments as New Players in Cancer Research and Their Potential Role in Diagnostics. Int. J. Mol. Sci. 2020, 21, 5682. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- von Brandenstein, M.; Bernhart, S.H.; Pansky, A.; Richter, C.; Kohl, T.; Deckert, M.; Heidenreich, A.; Stadler, P.F.; Montesinos-Rongen, M.; Fries, J.W.U. Beyond the 3′UTR Binding-MicroRNA-Induced Protein Truncation via DNA Binding. Oncotarget 2018, 9, 32855–32867. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear Functions of Mammalian MicroRNAs in Gene Regulation, Immunity and Cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Choudury, S.G.; Slotkin, R.K. TRNA-Derived Small RNAs Target Transposable Element Transcripts. Nucleic Acids Res. 2017, 45, 5142–5152. [Google Scholar] [CrossRef]
- Pederson, T. Regulatory RNAs Derived from Transfer RNA? RNA 2010, 16, 1865–1869. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, B.; Kim, V.N. Re-Evaluation of the Roles of DROSHA, Exportin 5, and DICER in MicroRNA Biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef]
- Ala, U. Competing Endogenous RNAs, Non-Coding RNAs and Diseases: An Intertwined Story. Cells 2020, 9, 1574. [Google Scholar] [CrossRef]
- Micheel, J.; Safrastyan, A.; Wollny, D. Advances in Non-Coding RNA Sequencing. Non-Coding RNA 2021, 7, 70. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-Coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Menini, M.; Pesce, P.; Baldi, D.; Coronel Vargas, G.; Pera, P.; Izzotti, A. Prediction of Titanium Implant Success by Analysis of microRNA Expression in Peri-Implant Tissue. A 5-Year Follow-Up Study. J. Clin. Med. 2019, 8, 888. [Google Scholar] [CrossRef] [PubMed]
- Bjørndal, L.; Simon, S.; Tomson, P.L.; Duncan, H.F. Management of Deep Caries and the Exposed Pulp. Int. Endod. J. 2019, 52, 949–973. [Google Scholar] [CrossRef]
- Santonocito, S.; Polizzi, A.; Palazzo, G.; Isola, G. The Emerging Role of MicroRNA in Periodontitis: Pathophysiology, Clinical Potential and Future Molecular Perspectives. Int. J. Mol. Sci. 2021, 22, 5456. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Han, S.; Li, Z.; Li, Z. Emerging Role of Exosomes in Craniofacial and Dental Applications. Theranostics 2020, 10, 8648–8664. [Google Scholar] [CrossRef] [PubMed]
- Menini, M.; Pesce, P.; Pera, F.; Baldi, D.; Pulliero, A.; Izzotti, A. MicroRNAs in Peri-Implant Crevicular Fluid Can Predict Peri-Implant Bone Resorption: Clinical Trial with a 5-Year Follow-Up. Int. J. Oral Maxillofac. Implants 2021, 36, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Romão, V.C.; Talarico, R.; Scirè, C.A.; Vieira, A.; Alexander, T.; Baldini, C.; Gottenberg, J.-E.; Gruner, H.; Hachulla, E.; Mouthon, L.; et al. Sjögren’s Syndrome: State of the Art on Clinical Practice Guidelines. RMD Open 2018, 4, e000789. [Google Scholar] [CrossRef]
- Awadallah, M.; Idle, M.; Patel, K.; Kademani, D. Management Update of Potentially Premalignant Oral Epithelial Lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Fang, X.; Chen, J.; Zhang, H.; Tang, Z. Long Non-Coding RNA (LncRNA) in Oral Squamous Cell Carcinoma: Biological Function and Clinical Application. Cancers 2021, 13, 5944. [Google Scholar] [CrossRef] [PubMed]
- Sayad, A.; Mirzajani, S.; Gholami, L.; Razzaghi, P.; Ghafouri-Fard, S.; Taheri, M. Emerging Role of Long Non-Coding RNAs in the Pathogenesis of Periodontitis. Biomed. Pharmacother. 2020, 129, 110362. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Wittfeldt, A.; Sun, J.; Liu, C.; Wang, X.; Gan, L.-M.; Cao, H.; Liang, Z. Long Non-Coding RNA ANRIL Regulates Inflammatory Responses as a Novel Component of NF-ΚB Pathway. RNA Biol. 2016, 13, 98–108. [Google Scholar] [CrossRef]
- Liu, M.; Chen, L.; Wu, J.; Lin, Z.; Huang, S. Long Noncoding RNA MEG3 Expressed in Human Dental Pulp Regulates LPS-Induced Inflammation and Odontogenic Differentiation in Pulpitis. Exp. Cell Res. 2021, 400, 112495. [Google Scholar] [CrossRef]
- Chen, H.; Lan, Z.; Li, Q.; Li, Y. Abnormal Expression of Long Noncoding RNA FGD5-AS1 Affects the Development of Periodontitis through Regulating MiR-142-3p/SOCS6/NF-ΚB Pathway. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2098–2106. [Google Scholar] [CrossRef]
- Li, J.; Wang, M.; Song, L.; Wang, X.; Lai, W.; Jiang, S. LncRNA MALAT1 Regulates Inflammatory Cytokine Production in Lipopolysaccharide-Stimulated Human Gingival Fibroblasts through Sponging MiR-20a and Activating TLR4 Pathway. J. Periodontal Res. 2020, 55, 182–190. [Google Scholar] [CrossRef]
- Wang, J.; Zhai, X.; Guo, J.; Li, Y.; Yang, Y.; Wang, L.; Yang, L.; Liu, F. Long Non-Coding RNA DQ786243 Modulates the Induction and Function of CD4+ Treg Cells through Foxp3-MiR-146a-NF-ΚB Axis: Implications for Alleviating Oral Lichen Planus. Int. Immunopharmacol. 2019, 75, 105761. [Google Scholar] [CrossRef]
- Wang, J.; Peng, H.; Tian, J.; Ma, J.; Tang, X.; Rui, K.; Tian, X.; Wang, Y.; Chen, J.; Lu, L.; et al. Upregulation of Long Noncoding RNA TMEVPG1 Enhances T Helper Type 1 Cell Response in Patients with Sjögren Syndrome. Immunol. Res. 2016, 64, 489–496. [Google Scholar] [CrossRef]
- De Benedittis, G.; Ciccacci, C.; Latini, A.; Novelli, L.; Novelli, G.; Borgiani, P. Emerging Role of MicroRNAs and Long Non-Coding RNAs in Sjögren’s Syndrome. Genes 2021, 12, 903. [Google Scholar] [CrossRef]
- Jia, H.; Wang, X.; Sun, Z. Screening and Validation of Plasma Long Non-coding RNAs as Biomarkers for the Early Diagnosis and Staging of Oral Squamous Cell Carcinoma. Oncol. Lett. 2021, 21, 172. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, D.; Liu, H.; Yang, K. Increased Expression of LncRNA CASC9 Promotes Tumor Progression by Suppressing Autophagy-Mediated Cell Apoptosis via the AKT/MTOR Pathway in Oral Squamous Cell Carcinoma. Cell Death Dis. 2019, 10, 41. [Google Scholar] [CrossRef]
- Tao, D.; Zhang, Z.; Liu, X.; Zhang, Z.; Fu, Y.; Zhang, P.; Yuan, H.; Liu, L.; Cheng, J.; Jiang, H. LncRNA HOTAIR Promotes the Invasion and Metastasis of Oral Squamous Cell Carcinoma through Metastasis-Associated Gene 2. Mol. Carcinog. 2020, 59, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Carrillo, J.L.; Vázquez-Alcaraz, S.J.; Vargas-Barbosa, J.M.; Ramos-Gracia, L.G.; Alvarez-Barreto, I.; Medina-Quiroz, A.; Díaz-Huerta, K.K. The Role of MicroRNAs in Pulp Inflammation. Cells 2021, 10, 2142. [Google Scholar] [CrossRef] [PubMed]
- Nara, K.; Kawashima, N.; Noda, S.; Fujii, M.; Hashimoto, K.; Tazawa, K.; Okiji, T. Anti-inflammatory Roles of MicroRNA 21 in Lipopolysaccharide-stimulated Human Dental Pulp Cells. J. Cell. Physiol. 2019, 234, 21331–21341. [Google Scholar] [CrossRef]
- Ouhara, K.; Savitri, I.J.; Fujita, T.; Kittaka, M.; Kajiya, M.; Iwata, T.; Miyagawa, T.; Yamakawa, M.; Shiba, H.; Kurihara, H. MiR-584 Expressed in Human Gingival Epithelial Cells Is Induced by Porphyromonas Gingivalis Stimulation and Regulates Interleukin-8 Production via Lactoferrin Receptor. J. Periodontol. 2014, 85, e198–e204. [Google Scholar] [CrossRef]
- Olsen, I.; Singhrao, S.K.; Osmundsen, H. Periodontitis, Pathogenesis and Progression: MiRNA-Mediated Cellular Responses to Porphyromonas Gingivalis. J. Oral Microbiol. 2017, 9, 1333396. [Google Scholar] [CrossRef]
- Nik Mohamed Kamal, N.N.S.; Awang, R.A.R.; Mohamad, S.; Shahidan, W.N.S. Plasma- and Saliva Exosome Profile Reveals a Distinct MicroRNA Signature in Chronic Periodontitis. Front. Physiol. 2020, 11, 587381. [Google Scholar] [CrossRef]
- Sipert, C.R.; Morandini, A.C.; Dionísio, T.J.; Trachtenberg, A.J.; Kuo, W.P.; Santos, C.F. MicroRNA-146a and MicroRNA-155 Show Tissue-Dependent Expression in Dental Pulp, Gingival and Periodontal Ligament Fibroblasts in Vitro. J. Oral Sci. 2014, 56, 157–164. [Google Scholar] [CrossRef]
- Radović, N.; Nikolić Jakoba, N.; Petrović, N.; Milosavljević, A.; Brković, B.; Roganović, J. MicroRNA-146a and MicroRNA-155 as Novel Crevicular Fluid Biomarkers for Periodontitis in Non-Diabetic and Type 2 Diabetic Patients. J. Clin. Periodontol. 2018, 45, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-Delivered MicroRNAs Modulate the Inflammatory Response to Endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, R.; Ye, S.; Lin, S.; Yin, G.; Xie, Q. Recent Advances in the Use of Exosomes in Sjögren’s Syndrome. Front. Immunol. 2020, 11, 1509. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Kong, Y.; Hu, X.; Li, Z.; Li, Y.; Zhong, Y.; Wei, X.; Ling, J. MicroRNA-Enriched Small Extracellular Vesicles Possess Odonto-Immunomodulatory Properties for Modulating the Immune Response of Macrophages and Promoting Odontogenesis. Stem Cell Res. Ther. 2020, 11, 517. [Google Scholar] [CrossRef]
- Cortes-Troncoso, J.; Jang, S.-I.; Perez, P.; Hidalgo, J.; Ikeuchi, T.; Greenwell-Wild, T.; Warner, B.M.; Moutsopoulos, N.M.; Alevizos, I. T Cell Exosome-Derived MiR-142-3p Impairs Glandular Cell Function in Sjögren’s Syndrome. JCI Insight 2020, 5, e133497. [Google Scholar] [CrossRef] [PubMed]
- Gassling, V.; Hampe, J.; Açil, Y.; Braesen, J.H.; Wiltfang, J.; Häsler, R. Disease-Associated MiRNA-MRNA Networks in Oral Lichen Planus. PLoS ONE 2013, 8, e63015. [Google Scholar] [CrossRef]
- Scapoli, L.; Palmieri, A.; Muzio, L.L.; Pezzetti, F.; Rubini, C.; Girardi, A.; Farinella, F.; Mazzotta, M.; Carinci, F. MicroRNA Expression Profiling of Oral Carcinoma Identifies New Markers of Tumor Progression. Int. J. Immunopathol. Pharmacol. 2010, 23, 1229–1234. [Google Scholar] [CrossRef]
- Martinez, B.V.; Dhahbi, J.M.; Nunez Lopez, Y.O.; Lamperska, K.; Golusinski, P.; Luczewski, L.; Kolenda, T.; Atamna, H.; Spindler, S.R.; Golusinski, W.; et al. Circulating Small Non Coding RNA Signature in Head and Neck Squamous Cell Carcinoma. Oncotarget 2015, 6, 19246. [Google Scholar] [CrossRef]
- Uma Maheswari, T.N.; Nivedhitha, M.S.; Ramani, P. Expression Profile of Salivary Micro RNA-21 and 31 in Oral Potentially Malignant Disorders. Braz. Oral Res. 2020, 34, e002. [Google Scholar] [CrossRef]
- Scholtz, B.; Horváth, J.; Tar, I.; Kiss, C.; Márton, I.J. Salivary MiR-31-5p, MiR-345-3p, and miR-424-3p Are Reliable Biomarkers in Patients with Oral Squamous Cell Carcinoma. Pathogens 2022, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary Exosomal MiR-24-3p Serves as a Potential Detective Biomarker for Oral Squamous Cell Carcinoma Screening. Biomed. Pharmacother. 2020, 121, 109553. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary Extracellular Vesicle-Associated MiRNAs as Potential Biomarkers in Oral Squamous Cell Carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Feng, J.; Wang, W. Expression of MiR-155 and MiR-146a in the Saliva of Patients with Periodontitis and Its Clinical Value. Am. J. Transl. Res. 2021, 13, 6670–6677. [Google Scholar]
- Sembler-Møller, M.L.; Belstrøm, D.; Locht, H.; Pedersen, A.M.L. Distinct MicroRNA Expression Profiles in Saliva and Salivary Gland Tissue Differentiate Patients with Primary Sjögren’s Syndrome from Non-Sjögren’s Sicca Patients. J. Oral Pathol. Med. 2020, 49, 1044–1052. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. MiRNet 2.0: Network-Based Visual Analytics for MiRNA Functional Analysis and Systems Biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Daszkiewicz, L.; Vázquez-Mateo, C.; Rackov, G.; Ballesteros-Tato, A.; Weber, K.; Madrigal-Avilés, A.; Di Pilato, M.; Fotedar, A.; Fotedar, R.; Flores, J.M.; et al. Distinct P21 Requirements for Regulating Normal and Self-Reactive T Cells through IFN-γ Production. Sci. Rep. 2015, 5, 7691. [Google Scholar] [CrossRef]
- Lin, Z.; Huang, L.; Li, S.L.; Gu, J.; Cui, X.; Zhou, Y. PTEN Loss Correlates with T Cell Exclusion across Human Cancers. BMC Cancer 2021, 21, 429. [Google Scholar] [CrossRef]
- Lee, N.; Kim, D.; Kim, W.-U. Role of NFAT5 in the Immune System and Pathogenesis of Autoimmune Diseases. Front. Immunol. 2019, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Oostvogels, R.; Minnema, M.C.; van Elk, M.; Spaapen, R.M.; te Raa, G.D.; Giovannone, B.; Buijs, A.; van Baarle, D.; Kater, A.P.; Griffioen, M.; et al. Towards Effective and Safe Immunotherapy after Allogeneic Stem Cell Transplantation: Identification of Hematopoietic-Specific Minor Histocompatibility Antigen UTA2-1. Leukemia 2013, 27, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Roganović, J.R. MicroRNA-146a and -155, Upregulated by Periodontitis and Type 2 Diabetes in Oral Fluids, Are Predicted to Regulate SARS-CoV-2 Oral Receptor Genes. J. Periodontol. 2021, 92, e35–e43. [Google Scholar] [CrossRef] [PubMed]
- Angadi, P.V.; Krishnapillai, R. Evaluation of PTEN Immunoexpression in Oral Submucous Fibrosis: Role in Pathogenesis and Malignant Transformation. Head Neck Pathol. 2012, 6, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Morita, H.; Matsuda, M.; Katakura, Y.; Hirata, M.; Hashimoto, S. NFAT5 Promotes Oral Squamous Cell Carcinoma Progression in a Hyperosmotic Environment. Lab. Investig. 2021, 101, 38–50. [Google Scholar] [CrossRef]
- Hao, Y.; Ge, Y.; Li, J.; Hu, Y.; Wu, B.; Fang, F. Identification of MicroRNAs by Microarray Analysis and Prediction of Target Genes Involved in Osteogenic Differentiation of Human Periodontal Ligament Stem Cells. J. Periodontol. 2017, 88, 1105–1113. [Google Scholar] [CrossRef]
- Zhang, J.; Hao, X.; Yin, M.; Xu, T.; Guo, F. Long Non-Coding RNA in Osteogenesis: A New World to Be Explored. Bone Jt. Res. 2019, 8, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, W.; Hu, C.; Xue, Z.; Wang, G.; Ding, B.; Luo, H.; Tang, L.; Kong, X.; Chen, X.; et al. MiR-17 Modulates Osteogenic Differentiation Through a Coherent Feed-Forward Loop in Mesenchymal Stem Cells Isolated from Periodontal Ligaments of Patients with Periodontitis. Stem Cells 2011, 29, 1804–1816. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Guo, T.; Hu, C.; Luo, H.; Zhang, L.; Shi, S.; Cai, T.; Ding, Y.; Jin, Y. TCF3, a Novel Positive Regulator of Osteogenesis, Plays a Crucial Role in MiR-17 Modulating the Diverse Effect of Canonical Wnt Signaling in Different Microenvironments. Cell Death Dis. 2013, 4, e539. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhou, H.; Zou, D.; Xie, Q.; Bi, X.; Gu, P.; Fan, X. The Role of MiR-31-Modified Adipose Tissue-Derived Stem Cells in Repairing Rat Critical-Sized Calvarial Defects. Biomaterials 2013, 34, 6717–6728. [Google Scholar] [CrossRef]
- Liu, Z.; Chang, H.; Hou, Y.; Wang, Y.; Zhou, Z.; Wang, M.; Huang, Z.; Yu, B. Lentivirus-mediated MicroRNA-26a Overexpression in Bone Mesenchymal Stem Cells Facilitates Bone Regeneration in Bone Defects of Calvaria in Mice. Mol. Med. Rep. 2018, 18, 5317–5326. [Google Scholar] [CrossRef]
- Yang, C.; Liu, X.; Zhao, K.; Zhu, Y.; Hu, B.; Zhou, Y.; Wang, M.; Wu, Y.; Zhang, C.; Xu, J.; et al. MiRNA-21 Promotes Osteogenesis via the PTEN/PI3K/Akt/HIF-1α Pathway and Enhances Bone Regeneration in Critical Size Defects. Stem Cell Res. Ther. 2019, 10, 65. [Google Scholar] [CrossRef]
- Baćević, M.; Brković, B.; Lambert, F.; Djukić, L.; Petrović, N.; Roganović, J. Leukocyte- and Platelet-Rich Fibrin as Graft Material Improves MicroRNA-21 Expression and Decreases Oxidative Stress in the Calvarial Defects of Diabetic Rabbits. Arch. Oral Biol. 2019, 102, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, T.; Mori, Y.; Abe, M.; Suenaga, H.; Kawase-Koga, Y.; Saijo, H.; Takato, T. Role of Cyclin-Dependent Kinase (Cdk)6 in Osteoblast, Osteoclast, and Chondrocyte Differentiation and Its Potential as a Target of Bone Regenerative Medicine. Oral Sci. Int. 2011, 8, 2–6. [Google Scholar] [CrossRef]
- Ma, X.; Su, P.; Yin, C.; Lin, X.; Wang, X.; Gao, Y.; Patil, S.; War, A.R.; Qadir, A.; Tian, Y.; et al. The Roles of FoxO Transcription Factors in Regulation of Bone Cells Function. Int. J. Mol. Sci. 2020, 21, 692. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Zhang, Y.; Pan, J. The E2F Transcription Factor 2: What Do We Know? Biosci. Trends 2021, 15, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Tamura, T.; Hatori, K.; Himi, K.; Nakamura, T.; Toyama, Y.; Miyata, T.; Takeichi, O. Elevated Foxo3a and Fas-ligand Expression in Human Periapical Granulomas as a Potential Treatment Target. Oral Dis. 2021, odi.14052. [Google Scholar] [CrossRef]
- Chen, H.; Hu, X.; Yang, R.; Wu, G.; Tan, Q.; Goltzman, D.; Miao, D. SIRT1/FOXO3a Axis Plays an Important Role in the Prevention of Mandibular Bone Loss Induced by 1,25 (OH) 2D Deficiency. Int. J. Biol. Sci. 2020, 16, 2712–2726. [Google Scholar] [CrossRef]
- Xu, K.; Xiao, J.; Zheng, K.; Feng, X.; Zhang, J.; Song, D.; Wang, C.; Shen, X.; Zhao, X.; Wei, C.; et al. MiR-21/STAT3 Signal Is Involved in Odontoblast Differentiation of Human Dental Pulp Stem Cells Mediated by TNF-α. Cell. Reprogramming 2018, 20, 107–116. [Google Scholar] [CrossRef]
- Huang, X.; Liu, F.; Hou, J.; Chen, K. Inflammation-induced Overexpression of MicroRNA-223-3p Regulates Odontoblastic Differentiation of Human Dental Pulp Stem Cells by Targeting SMAD3. Int Endod. J. 2019, 52, 491–503. [Google Scholar] [CrossRef]
- Sun, D.; Xin, B.; Wu, D.; Zhou, L.; Wu, H.; Gong, W.; Lv, J. MiR-140-5p-Mediated Regulation of the Proliferation and Differentiation of Human Dental Pulp Stem Cells Occurs through the Lipopolysaccharide/Toll-like Receptor 4 Signaling Pathway. Eur. J. Oral Sci. 2017, 125, 419–425. [Google Scholar] [CrossRef]
- Ke, Z.; Qiu, Z.; Xiao, T.; Zeng, J.; Zou, L.; Lin, X.; Hu, X.; Lin, S.; Lv, H. Downregulation of MiR-224-5p Promotes Migration and Proliferation in Human Dental Pulp Stem Cells. BioMed Res. Int. 2019, 2019, 4759060. [Google Scholar] [CrossRef]
- Sun, F.; Wan, M.; Xu, X.; Gao, B.; Zhou, Y.; Sun, J.; Cheng, L.; Klein, O.D.; Zhou, X.; Zheng, L. Crosstalk between MiR-34a and Notch Signaling Promotes Differentiation in Apical Papilla Stem Cells (SCAPs). J. Dent. Res. 2014, 93, 589–595. [Google Scholar] [CrossRef]
- Li, Z.; Yan, M.; Yu, Y.; Wang, Y.; Lei, G.; Pan, Y.; Li, N.; Gobin, R.; Yu, J. LncRNA H19 Promotes the Committed Differentiation of Stem Cells from Apical Papilla via MiR-141/SPAG9 Pathway. Cell Death Dis. 2019, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Julian, L.M.; Blais, A. Transcriptional Control of Stem Cell Fate by E2Fs and Pocket Proteins. Front. Genet. 2015, 6, 161. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-C.; Kuo, Y.-C.; Chuong, C.-M.; Huang, Y.-H. Niche Modulation of IGF-1R Signaling: Its Role in Stem Cell Pluripotency, Cancer Reprogramming, and Therapeutic Applications. Front. Cell Dev. Biol. 2021, 8, 625943. [Google Scholar] [CrossRef] [PubMed]
- Dissanayaka, W.L.; Han, Y.; Zhang, L.; Zou, T.; Zhang, C. Bcl-2 Overexpression and Hypoxia Synergistically Enhance Angiogenic Properties of Dental Pulp Stem Cells. Int. J. Mol. Sci. 2020, 21, 6159. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Xu, W.; Li, J.; Chen, Y.; Pan, Y.; Wu, B. Effects of Vascular Endothelial Growth Factor and Insulin Growth Factor-1 on Proliferation, Migration, Osteogenesis and Vascularization of Human Carious Dental Pulp Stem Cells. Mol. Med. Rep. 2019, 20, 3924–3932. [Google Scholar] [CrossRef]
- Artese, L.; Rubini, C.; Ferrero, G.; Fioroni, M.; Santinelli, A.; Piattelli, A. Vascular Endothelial Growth Factor (VEGF) Expression in Healthy and Inflamed Human Dental Pulps. J. Endod. 2002, 28, 20–23. [Google Scholar] [CrossRef]
- Xia, K.; Chen, Z.; Chen, J.; Xu, H.; Xu, Y.; Yang, T.; Zhang, Q. RGD- and VEGF-Mimetic Peptide Epitope-Functionalized Self-Assembling Peptide Hydrogels Promote Dentin-Pulp Complex Regeneration. Int. J. Nanomed. 2020, 15, 6631–6647. [Google Scholar] [CrossRef]
- Lovschall, H.; Fejerskov, O.; Flyvbjerg, A. Pulp-Capping with Recombinant Human Insulin- like Growth Factor I (RhIGF-I) in Rat Molars. Adv. Dent. Res. 2001, 15, 108–112. [Google Scholar] [CrossRef]
- Tavares-Ferreira, D.; Lawless, N.; Bird, E.V.; Atkins, S.; Collier, D.; Sher, E.; Malki, K.; Lambert, D.W.; Boissonade, F.M. Correlation of MiRNA Expression with Intensity of Neuropathic Pain in Man. Mol. Pain 2019, 15, 174480691986032. [Google Scholar] [CrossRef]
- Lutz, B.M.; Bekker, A.; Tao, Y.-X. Noncoding RNAs. Anesthesiology 2014, 121, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Haqqi, T.M. MicroRNA-199a* Regulates the Expression of Cyclooxygenase-2 in Human Chondrocytes. Ann. Rheum. Dis. 2012, 71, 1073–1080. [Google Scholar] [CrossRef]
- Li, X.; Gibson, G.; Kim, J.-S.; Kroin, J.; Xu, S.; van Wijnen, A.J.; Im, H.-J. MicroRNA-146a Is Linked to Pain-Related Pathophysiology of Osteoarthritis. Gene 2011, 480, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Cheon, E.J.; Kim, H.A. MicroRNA-558 Regulates the Expression of Cyclooxygenase-2 and IL-1β-Induced Catabolic Effects in Human Articular Chondrocytes. Osteoarthr. Cartil. 2013, 21, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Orlova, I.A.; Alexander, G.M.; Qureshi, R.A.; Sacan, A.; Graziano, A.; Barrett, J.E.; Schwartzman, R.J.; Ajit, S.K. MicroRNA Modulation in Complex Regional Pain Syndrome. J. Transl. Med. 2011, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Leinders, M.; Üçeyler, N.; Pritchard, R.A.; Sommer, C.; Sorkin, L.S. Increased MiR-132-3p Expression Is Associated with Chronic Neuropathic Pain. Exp. Neurol. 2016, 283, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Leinders, M.; Üçeyler, N.; Thomann, A.; Sommer, C. Aberrant MicroRNA Expression in Patients with Painful Peripheral Neuropathies. J. Neurol. Sci. 2017, 380, 242–249. [Google Scholar] [CrossRef]
- Heyn, J.; Luchting, B.; Hinske, L.C.; Hübner, M.; Azad, S.C.; Kreth, S. MiR-124a and MiR-155 Enhance Differentiation of Regulatory T Cells in Patients with Neuropathic Pain. J. Neuroinflamm. 2016, 13, 248. [Google Scholar] [CrossRef]
- Liu, J.; Xue, D.; Wang, X.; Ai, D.; Qin, P. MiR-101 Relates to Chronic Peripheral Neuropathic Pain through Targeting KPNB1 and Regulating NF-κB Signaling. Kaohsiung J. Med. Sci. 2019, 35, 139–145. [Google Scholar] [CrossRef]
- Li, X.; Wang, D.; Zhou, J.; Yan, Y.; Chen, L. Evaluation of circulating microRNA expression in patients with trigeminal neuralgia: An observational study. Medicine 2020, 99, e22972. [Google Scholar] [CrossRef]
- Xiong, W.; Tan, M.; Tong, Z.; Yin, C.; He, L.; Liu, L.; Shen, Y.; Guan, S.; Ge, H.; Li, G.; et al. Effects of Long Non-Coding RNA Uc.48+ on Pain Transmission in Trigeminal Neuralgia. Brain Res. Bull. 2019, 147, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jiang, H.; Zheng, C.; Zhu, G.; Xu, Y.; Sheng, X.; Wu, B.; Guo, J.; Zhu, S.; Zhan, Y.; et al. Long Noncoding RNA MRAK009713 Is a Novel Regulator of Neuropathic Pain in Rats. Pain 2017, 158, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yan, Y.; Zhu, M.; Wang, Z.; Zhang, X.; Zhang, D. Effects of Long Non-Coding RNA Gm14461 on Pain Transmission in Trigeminal Neuralgia. J. Inflamm. 2020, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Byun, J.; Jung, J.; Choi, J. Profiling of Salivary Exosomal Micro RNAs in Burning Mouth Syndrome Patients. J. Oral Med. Pain 2019, 44, 25–30. [Google Scholar]
- Xu, J.; Liu, Y.; Deng, M.; Li, J.; Cai, H.; Meng, Q.; Fang, W.; Long, X.; Ke, J. MicroRNA221-3p Modulates Ets-1 Expression in Synovial Fibroblasts from Patients with Osteoarthritis of Temporomandibular Joint. Osteoarthr. Cartil. 2016, 24, 2003–2011. [Google Scholar] [CrossRef]
- Li, W.; Zhao, S.; Yang, H.; Zhang, C.; Kang, Q.; Deng, J.; Xu, Y.; Ding, Y.; Li, S. Potential Novel Prediction of TMJ-OA: MiR-140-5p Regulates Inflammation Through Smad/TGF-β Signaling. Front. Pharmacol. 2019, 10, 15. [Google Scholar] [CrossRef]
- Mao, D.; Wu, M.; Wei, J.; Zhou, X.; Yang, L.; Chen, F. MicroRNA-101a-3p Could Be Involved in the Pathogenesis of Temporomandibular Joint Osteoarthritis by Mediating UBE2D1 and FZD4. J. Oral Pathol. Med. 2021, 50, 236–243. [Google Scholar] [CrossRef]
- Zhang, A.; Ma, S.; Yuan, L.; Wu, S.; Liu, S.; Wei, X.; Chen, L.; Ma, C.; Zhao, H. Knockout of MiR-21-5p Alleviates Cartilage Matrix Degradation by Targeting Gdf5 in Temporomandibular Joint Osteoarthritis. Bone Jt. Res. 2020, 9, 689–700. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).