Abstract
Aluminum (Al) toxicity causes severe reduction in crop yields in acidic soil. The natural resistance-associated macrophage proteins (NRAMPs) play an important role in the transport of mineral elements in plants. Recently, OsNrat1 and SbNrat1 were reported specifically to transport trivalent Al ions. In this study, we functionally characterized ZmNRAMP4, a gene previously identified from RNA-Seq data from Al-treated maize roots, in response to Al exposure in maize. ZmNRAMP4 was predominantly expressed in root tips and was specifically induced by Al stress. Yeast cells expressing ZmNRAMP4 were hypersensitive to Al, which was associated with Al accumulation in yeast. Furthermore, overexpression of ZmNRAMP4 in Arabidopsis conferred transgenic plants with a significant increase in Al tolerance. However, expression of ZmNRAMP4, either in yeast or in Arabidopsis, had no effect on the response to cadmium stress. Taken together, these results underlined an internal tolerance mechanism involving ZmNRAMP4 to enhance Al tolerance via cytoplasmic sequestration of Al in maize.
1. Introduction
Aluminum (Al) is the most abundant metal element in the Earth’s crust [1]. About 50% of the Earth’s arable land is acidic. In acidic soil (pH < 5.0), Al exists in the form of trivalent Al (Al3+), which is toxic to plants, especially at high concentrations [2]. Therefore, Al toxicity is an important limiting factor for crop yield in acidic soil [3].
To cope with the high availability of soil Al, plants have evolved two mechanisms involving external exclusion and internal tolerance [4,5]. External exclusion of Al via exudation of organic acid anions from roots is an important Al resistance mechanism in many plants. External exclusion of Al via exudation of organic acid anions from the roots is an important Al tolerance mechanism in many plant species. Malate and citrate transporters have been functionally characterized in many species, including AtMATE and AtMLT1 in Arabidopsis, OsFRDL2 and OsFRDL4 in rice, SbMATE in sorghum, and TaALMT1 in wheat [6]. With regard to an internal detoxification mechanism, some transporters associated with Al tolerance can be so classified. For example, STAR1 and STAR2 form a transporter protein complex that is responsible specifically for efflux of UDP-glucose, which is likely to be used to modify the cell wall [7]. In Arabidopsis, AtSTAR1 and ALS3 are involved in Al tolerance [8,9]. Overexpression of OsPIN2, which encodes an auxin transporter, can decrease the binding of Al to the cell wall [10]. Certain transcription factors (AtSTOP1 [11], OsART1 [12], VuNAR1 [13], WRKY47 [14], HvHOX9 [15], GsMAS1 [16] and abscisic acid (ABA)-stress and ripening (ASR5) [17]) and ROS-related genes (ZmAT6) [18] are also involved in plant Al stress response.
NRAMP proteins comprise a group of highly conserved integral membrane proteins widespread among bacteria, animals, and plants [19]. The genomes of sorghum, rice, maize, and Arabidopsis contain 7, 7, 8, and 6 NRAMP members, respectively [20]. NRAMP proteins play a major role in the transport of mineral elements in plants, including the metal ions Fe2+, Mn2+, Cd2+, Zn2+, Co2+, and Cu2+ [21]. For example, OsNRAMP1 transports Cd, As, and Fe, but not Mn [22]; OsNRAMP5 is the main transporter for Cd2+ influx in rice [23,24]; AtNRAMP3 and AtNRAMP4 transport Fe2+ and Mn2+ in Arabidopsis [25]; BcNRAMP1 is a plasma membrane-localized protein and transports Mn and Cd [26]; and FeNRAMP5 is a transporter for the uptake of Mn and Cd [27]. Interestingly, two NRAMP members, OsNrat1 and SbNrat1, transport Al3+ specifically but not divalent metal ions, such as Fe2+, Mn2+, and Cd2+, and are required for a prior step of final Al detoxification through vacuolar sequestration of Al [3,28,29]. The aim of the present study was to functionally characterize the role of ZmNRAMP4, which was previously identified from RNA-Seq data for Al-stressed maize roots, in Al transport in maize. The results showed that ZmNRAMP4 was upregulated by Al treatment. Combined heterologous expression in yeast and Arabidopsis indicated that ZmNRAMP4 is an Al transporter and its expression is stimulated by Al. Overexpression of ZmNRAMP4 enhanced the Al tolerance of transgenic Arabidopsis plants via Al influx in the roots.
2. Results
2.1. Cloning and Phylogenetic Analysis of ZmNRAMP4
ZmNRAMP4 was cloned from the Al-tolerant maize inbred line 178 (Figure S1). The gene encoded a putative protein of 556 amino acids, which belongs to the NRAMP family of maize. The multiple sequence alignment showed that ZmNRAMP4 included all conserved elements of NRAMP members, such as Motif A and Motif B, and had the highest identity with SbNrat1 of 96% (Figure S2). A phylogenetic tree for NRAMP members was composed of five main clades. Notably, ZmNRAMP4 was placed in the same clade as OsNrat1 and SbNrat1, two previously reported transporters of trivalent Al ions, but was separated from divalent ion transporters (Figure 1).
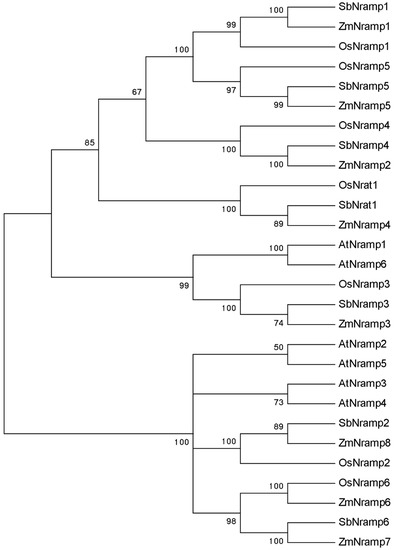
Figure 1.
Phylogenic tree of ZmNRAMP4 with NRAMPs from Arabidopsis, rice, sorghum, and maize. The information of accession number and species for NRAMPs is as follows, including AtNRAMP1 to AtNRAMP6 (AT1G80830, AT1G47240, AT2G23150, AT5G67330, AT4G18790, AT1G15960), OsNRAMP1 to OsNRAMP6 (Os07g15460, Os03g11010, Os06g46310, Os07g15370, Os12g39180, Os01g0503400), OsNrat1 (Os02g0131800), SbNRAMP1 to SbNRAMP6 (XP_002459640, XP_002465667, XP_002438846, XP_021317241, XP_002461772, XP_002464246), SbNrat1 (XP_002451480), ZmNRAMP1 to ZmNRAMP8 (Zm00001d005479, Zm00001d007397, Zm00001d014391, Zm00001d015133, Zm00001d019327, Zm00001d030846, Zm00001d033367, Zm00001d048129).
2.2. Expression Patterns of ZmNRAMP4
To explore if the eight ZmNRAMP members were stimulated by Al stress, the expression patterns of all eight ZmNRAMP genes were investigated. The results revealed that ZmNRAMP4 was the only ZmNRAMP gene that was induced by Al stress (Figure 2A). Therefore, the expression pattern of ZmNRAMP4 in response to Al stress was studied further. The Al-induced expression of ZmNRAMP4 was predominantly detected in the root and was highest in the root tip (Figure 2B). A time-course experiment showed that ZmNRAMP4 expression increased after Al exposure and peaked at 24 h, and thereafter decreased with prolonged treatment (Figure 2C). The expression of ZmNRAMP4 was significantly affected by the AlCl3 concentration; a broad range of AlCl3 concentrations (40–100 µM) stimulated ZmNRAMP4 expression effectively and 80 μM AlCl3 induced the optimum stimulation (Figure 2D). In addition, ZmNRAMP4 expression was specifically affected by Al rather than other metal ions, such as Zn, Cu, La, or Cd, as well as low pH (Figure 2E).
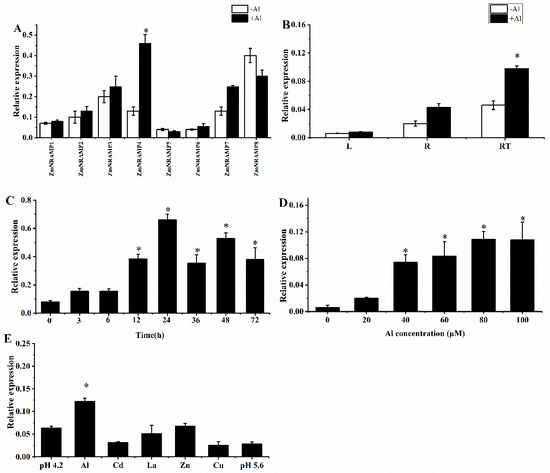
Figure 2.
Expression pattern of the ZmNRAMP4 gene. (A), Relative expression of ZmNramp gene family in the presence of Al. (B), Relative expression of ZmNRAMP4 in root tips (0–1 cm), root regions (behind removed root tips, 1–10 cm), and leaves. (C), Time-dependent expression of ZmNRAMP4 in the root tips. The seedlings were cultured in the nutrient solution added with 60 µM AlCl3 for 0, 3, 6, 12, 24, 36, 48, and 72 h. (D), Expression analysis of ZmNRAMP4 under different concentration of Al. Seedlings were treated for 6 h by 0, 10 μM, 20 μM, 40 μM, 60 μM, 80 μM, and 100 μM Al. (E), Expression of ZmNRAMP4 in response to other metals. Seedlings were exposed to a solution containing 60 µM AlCl3, 10 µM LaCl3, 100 µM ZnSO4, 2 µM CuCl2, and 30 µM CdCl2 or pH 4.2 for 12 h. Asterisk indicated significant difference from control at p < 0.05 by Tukey’s test with three independently biological replicates.
2.3. Subcellular Localization of ZmNRAMP4
To visualize the subcellular localization of ZmNRAMP4, ZmNRAMP4-GFP was transiently expressed in Nicotiana benthamiana leaves. The green fluorescent signal of GFP of the control vector was ubiquitous throughout the cell (Figure 3A). In contrast, the ZmNRAMP4-GFP signal was only detected at the plasma membrane (Figure 3B), consistent with the prediction of ZmNRAMP4 as a putative metal ion transporter.
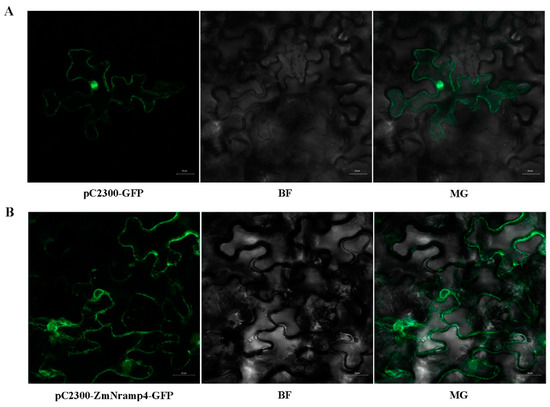
Figure 3.
Subcellular localization of ZmNRAMP4 in tobacco leaves. The ZmNRAMP4-GFP fusion protein was transiently expressed in the leaves of N. benthamiana (Scale bar: 50 μm).
2.4. Functional Expression of ZmNRAMP4 in Yeast
To investigate the Al transport ability of ZmNRAMP4, ZmNRAMP4 was expressed in the yeast strain BY4741. OsNrat1, an Al transporter in rice, its expression vector, and the empty vector were used as a positive control and negative control, respectively. In the absence of Al, all yeast cells, either carrying ZmNRAMP4, OsNrat1, or the empty vector, showed similar growth rates (Figure 4A). However, compared with the negative control, the growth of yeast cells expressing ZmNRAMP4 or OsNrat1 were slightly inhibited in the presence of Al (Figure 4A), suggesting that Al influx into the yeast cells occurred. Moreover, the yeast cells expressing ZmNRAMP4 or OsNrat1 accumulated a higher concentration of Al (Figure 4C), accompanied by stronger inhibition of growth (Figure 4B) when compared with the negative control. We also investigated the Cd transport activity of ZmNRAMP4 in yeast because some NRAMP proteins are involved in the transport of other metal ions. The pYES2-ZmNRAMP4 construct or the empty vector pYES2 were expressed both in a Cd-sensitive mutant strain Δycf1 and the wild-type strain BY4741. No growth difference was observed among the transformed yeasts in the absence of Cd stress (Figure S3). In response to treatment with 50 µM CdCl2, BY4741 yeast cells carrying ZmNRAMP4 grew similarly to those carrying the empty vector. In contrast, Cd severely inhibited the growth of Δycf1 yeast cells transformed with the empty vector but not cells transformed with pYES2-ZmNRAMP4 (Figure S3). These results revealed that ZmNRAMP4 has transport activity for Al but not Cd in yeast.
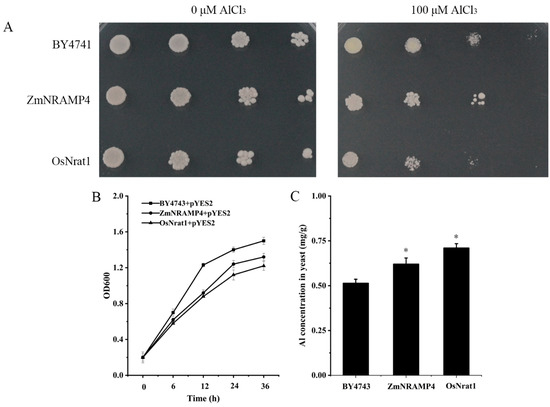
Figure 4.
Al transport activity of ZmNRAMP4 in yeast. (A), the growth of yeast cells carrying ZmNRAMP4 and OsNrat1 on LPM medium plate with 0 or 100 μM AlCl3. (B), Growth curves of yeast cells transformed with empty vector pYES2, pYES2-OsNrat1, or pYES2-ZmNRAMP4 in a galactose containing LPM liquid medium with or without 60 μM AlCl3. (C), Al concentration in yeast. Asterisk indicated significant difference from control at p < 0.05 by Tukey’s test with three independently biological replicates.
2.5. Overexpression of ZmNRAMP4 Enhanced Al Tolerance of Arabidopsis Plants
To determine whether ZmNRAMP4 affected Al tolerance in other plants, ZmNRAMP4 was overexpressed in Arabidopsis under the control of the 35S promoter. The Al-tolerant phenotype conferred by ZmNRAMP4 was investigated in two transgenic lines that highly expressed ZmNRAMP4 (Figure S4). Under normal condition, root growth of transgenic plants was comparable with that of the wild type, whereas the roots were longer than those of the wild type after Al exposure (Figure 5A). Similarly, the relative root growth of the transgenic lines was significantly higher than that of the wild type under Al stress (Figure 5B). In addition, the transgenic lines accumulated greater concentrations of Al in the root than the wild type (Figure 5C). However, overexpression of ZmNRAMP4 did not affect the root growth of the transgenic Arabidopsis plants under Cd stress (Figure 6). These results demonstrated that overexpression of ZmNRAMP4 in Arabidopsis conferred a notable increase in Al tolerance, but had no effect on tolerance of Cd stress.
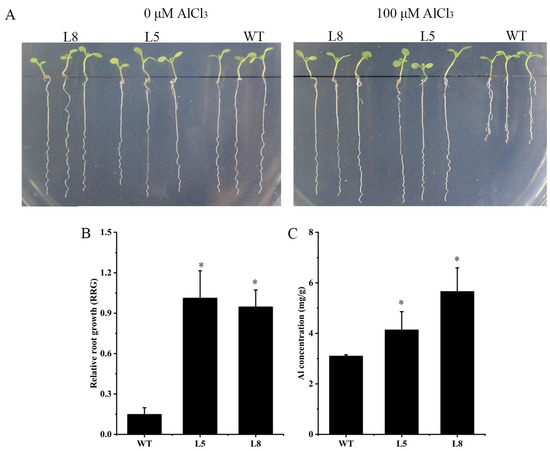
Figure 5.
Phenotype of ZmNRAMP4-overexpressed transgenic Arabidopsis plants under Al stress. (A) Al sensitivity, (B) relative root growth (RRG), and (C) Al concentration in wild-type and ZmNRAMP4 transgenic plants. The 14-day-old seedlings were exposed to one-fifth Hoagland solution (pH 4.7) containing 100 μM Al for 12 h. Asterisk indicated a significant difference from control at p < 0.05 by Tukey’s test with three independently biological replicates.
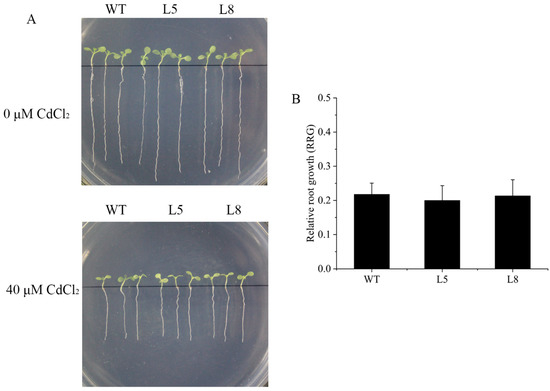
Figure 6.
Phenotype of ZmNRAMP4-overexpressed transgenic Arabidopsis plants under Cd stress. (A) Cd sensitivity and (B) relative root growth (RRG) in wild-type and ZmNRAMP4 transgenic plants. Relative root growth (RRG) of wild-type and ZmNRAMP4 transgenic plants under 0 or 50 µM CdCl2.
3. Discussion
Most NRAMP proteins are capable of transporting Cd and essential metals, such as Mn and Fe [30]. Recent studies have shown that OsNrat1 and SbNrat1 have transport activity for Al [3]. ZmNRAMP4 is a candidate gene for Al tolerance QTL co-localization and upregulated after 6 h of Al exposure in the root tip [31]. However, the functional characterization of ZmNRAMP genes in response to Al stress remains unclear. To verify the function of ZmNRAMP4, we investigated the effects of ZmNRAMP4 expression on metal tolerance and accumulation in yeast and Arabidopsis.
In rice, OsNrat1 expression is stimulated rapidly by Al, particularly in the root, and expression is maintained at a low level in the shoot. Furthermore, the stimulation by Al is specific because other metals do not induce OsNrat1 expression. The expression pattern of ZmNRAMP4 was consistent with that of OsNrat1 but differed notably from that of SbNrat1, which was not responsive to Al regardless of the Al concentration and exposure duration [3]. Given that ZmNRAMP4 was placed in the same subclade as OsNrat1 and SbNrat1 in the present phylogenic analysis (Figure 1), the difference in expression is probably attributable to differences among cereal species in the fine-tuning of environmental adaptation during evolution. This suggestion is consistent with the evolution of diverse functions among NRAMP members in the transport of metal ions, such as Fe2+, Mn2+, Cd2+, Zn2+, Co2+, and Cu2+.
Different from other Nramp members that could transport divalent metals, OsNRAT1 specifically transports trivalent aluminum [32]. In the present study, NRAMP transporters were resolved into five clades in a phylogenetic analysis, of which ZmNRAMP4 was placed in the same clade as the previously reported Al3+ transporters OsNrat1 and SbNrat1 (Figure 1). The metal binding site of NRAMP proteins is characterized by two conserved sequence motifs, i.e., the Asp–Pro–Ser–Asn motif (motif A) and the Ala–Ile–Ile–Thr motif (motif B) [33]. The Nrat1-type motif B is both effective and essential for Al transport by OsNrat1, as one crucial determinant of Al selectivity [34]. Missense mutations of amino acids in motif B of Al-tolerant rice lines reduce Al uptake in yeast cells, and the full function of OsNrat1 in Al influx is dependent on the most closely matched sequence of motif B [32]. Therefore, the exactly matched sequence of motif B in ZmNRAMP4 may be the predominant determinant of Al selectivity (Figure S2). In the present study, ZmNRAMP4-expressing yeast cells exhibited transport activity for Al but not Cd, which was consistent with accumulation of a higher concentration of Al. This was also the case for the Al influx transporters OsNrat1 and SbNrat1 when expressed in yeast [3]. In addition, the plasma membrane localization of ZmNRAMP4 was compatible with its function as a metal ion transporter.
NRAMP genes have been reported to be widely involved in the transport of metal ions. Recent studies have shown that overexpression of SbNrat1 recovers the Al tolerance of the rice mutant osnrat1 [3]. Similarly, overexpression of OsNRAT1 improves the Al tolerance of transgenic Arabidopsis and results in a significant increase in Al content [32]. In the present study, overexpression of ZmNRAMP4 in Arabidopsis conferred significantly elevated Al tolerance compared with the control, but had no effect on Cd tolerance. Consistent with the Al influx transport function of ZmNRAMP4, the roots of transgenic Arabidopsis plants accumulated higher concentrations of Al. The expression of endogenous AtALS1 is elevated (Figure S5), consistent with significantly enhanced expression of NRAT1 and Al accumulation in transgenic Arabidopsis roots [32]. Given that AtALS1 has the capacity to sequester the cytosolic Al in the vacuole, it is speculated that orthologs of AtALS1 coordinate with ZmNRAMP4 to enhance the Al tolerance of maize via Al sequestration.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The Al-tolerant maize inbred line 178 was used in this experiment [18]. Seeds were sterilized in 10% NaClO for 15 min, washed three times with deionized water, then cultured in an incubator at 28 °C. After germination, the seedlings were transplanted to pots filled with Hoagland’s nutrient solution and grown in a greenhouse maintained at 28/20 °C under a 16 h/8 h (day/night) photoperiod. The nutrient solution was replaced every 3 days.
4.2. Phylogenetic Analysis
The sequences of NRAMP family members were retrieved from the ARAMEMNON plant membrane protein database (http://aramemnon.botanik.uni-koeln.de/index.ep; accessed on 12 December 2018). A phylogenetic tree was constructed using the neighbor-joining algorithm with MEGA 5.0.
4.3. Relative Expression Analysis of ZmNRAMP4
For metal stress treatment, seedlings at the three-leaf stage were treated individually with 60 µM AlCl3, 10 µM LaCl3, 100 µM ZnSO4, 2 µM CuCl2, 30 µM CdCl2, or pH 4.2 for 12 h. To examine the expression pattern of ZmNRAMP4, maize seedlings were exposed to Al (0, 10, 20, 40, 60, 80, or 100 µM) for 12 h or 60 µM AlCl3 for different durations (0, 3, 6, 12, 24, 36, 48, and 72 h). Root and leaf samples were individually collected, rapidly frozen in liquid nitrogen, and stored at −80 °C for RNA extraction. The primers used for RT-PCR of ZmNRAMP4 are listed in Supplementary Table S2.
4.4. Subcellular Localization of ZmNRAMP4
The cDNA of ZmNRAMP4 was subcloned into the expression vector pC2300-GFP under the control of the 35S promoter using the primers listed in Supplementary Table S2. The pC2300-GFP construct was introduced into Agrobacterium tumefaciens strain EHA105. Agrobacterium-mediated transient expression assays were conducted in Nicotiana benthamiana leaves [35].
4.5. Mental Ion Transport Activity of ZmNRAMP4 in Yeast
The coding sequence of ZmNRAMP4 was cloned into the pYES2 vector (negative control) to generate the construct pYES2-ZmNRAMP4. The construct pYES2-OsNrat1 was generated to serve as a positive control. The yeast strains used in this study were BY4741 (MATa his2Δ0 met15Δ0 ura3Δ0) and the Cd-sensitive strain Δycf1 (MATa his2Δ0 met15Δ0 ura3Δ0 YOL122c::KanMX4). Aluminum and Cd sensitivity was tested on agar, whereas Al uptake was measured in liquid culture. For Al-sensitivity evaluation, the constructs pYES2-ZmNRAMP4 and pYES2-OsNrat1 and the empty vector pYES2 were transformed into yeast strain BY4741 and cultured on solid medium (LPM with galactose as the carbon source) supplemented with 0 or 100 µM Al buffered with 5 mM succinic acid. For Cd-sensitivity evaluation, pYES2-ZmNRAMP4 and pYES2 were transformed into yeast strains BY4741 and Δycf1, and cultured in liquid SD-U medium supplemented with 0 or 50 µM Cd. For estimation of Al uptake in liquid culture, transformants were selected on uracil-deficient medium and grown in synthetic complete yeast solution using glucose as the carbon source. Cells at the mid-exponential phase were harvested, transferred to LPM medium using galactose as the carbon source, and were cultured for 2 h. Next, AlCl3 was added to the medium at the final concentration of 60 µM Al. After incubation for 4 h with shaking, the cells were harvested by centrifugation, washed three times with deionized water, and then digested with 2 M HCl. The concentration of Al in the digest solution was determined by inductively coupled plasma–mass spectrometry.
4.6. Overexpression of ZmNRAMP4 in Arabidopsis thaliana
ZmNRAMP4 was inserted into the pC2300 vector under the control of the 35S promoter. Transgenic plants were screened with kanamycin (40 mg/L) and confirmed by PCR analysis. Two dependent T3 transgenic lines with elevated expression levels of ZmNRAMP4 were used for assessment of Al tolerance (Figure S4).
4.7. Evaluation of Al Tolerance
To quantify the effect of Al toxicity, the wild type and two transgenic lines were surface sterilized and germinated for 4 days on solid Murashige and Skoog (MS) medium. Seedlings of uniform growth were transferred to half-strength MS solid medium supplemented with AlCl3 (0 or 100 μM) or CdCl2 (0 or 25 μM) and cultured for 7 days. The root length of the seedlings was measured individually with a ruler before and after treatment.
4.8. Statistical Analysis and Reproducibility
All treatments were repeated at least three times. Statistical analysis including student’s t test was performed using the SPSS software. The figures were drawn using the Origin 8.0 software (OriginLab Corporation, Northampton, MA, USA).
5. Conclusions
We investigated a ZmNRAMP4 gene encoding a plasma membrane-localized Al influx transporter from the Al-tolerant maize inbred line 178. Its expression is significantly upregulated in response to Al exposure, but not by other metals, in maize roots. ZmNRAMP4 confers Al influx activity in yeast cells. Ectopic overexpression of ZmNRAMP4 in Arabidopsis enhances the Al tolerance of transgenic plants. The results suggested that ZmNRAMP4 may increase resistance to Al toxicity via cytoplasmic sequestration of Al in transgenic Arabidopsis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23158162/s1.
Author Contributions
Conceptualization, H.L.; Formal analysis, N.W.; Funding acquisition, Y.Y.; Investigation, H.L.; Project administration, C.L. and S.Z.; Resources, X.J. and C.D.; Software, H.L.; Supervision, S.Z.; Validation, W.Y. and W.H. (Wenzhu He); Visualization, W.H. (Wanpeng Hu); Writing—original draft, H.L.; Writing—review and editing, S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China, grant number 32171951, the Applied Basic Research Programs of Science and Technology Department of Sichuan, grant number 2021YJ0300.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are available within the article or upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H.; Delhaize, E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochian, V.L. Cellular Mechanisms of Aluminum Toxicity and Resistance in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Lu, M.; Wang, Z.; Fu, S.; Yang, G.; Shi, M.; Lu, Y.; Wang, X.; Xia, J. Functional characterization of the SbNrat1 gene in sorghum. Plant Sci. Int. J. Exp. Plant Biol. 2017, 262, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol.-A Surv. Cell Biol. 2007, 264, 225–252. [Google Scholar]
- Furukawa, J.; Yamaji, N.; Wang, H.; Mitani, N.; Murata, Y.; Sato, K.; Katsuhara, M.; Takeda, K.; Ma, J.F. An Aluminum-Activated Citrate Transporter in Barley. Plant Cell Physiol. 2007, 8, 1081–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Feng, X.; Cao, F.; Wu, D.; Wu, F. An ATP Binding Cassette Transporter HvABCB25 Confers Aluminum Detoxification in Wild Barley. J. Hazard. Mater. 2020, 401, 123371. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A Bacterial-Type ABC Transporter Is Involved in Aluminum Tolerance in Rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef]
- Wu, D.; Shen, H.; Yokawa, K.; Baluška, F. Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 2014, 65, 5305–5315. [Google Scholar] [CrossRef] [Green Version]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A Zinc Finger Transcription Factor ART1 Regulates Multiple Genes Implicated in Aluminum Tolerance in Rice. Plant Cell 2009, 10, 3339–3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Zheng, S.J. A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 2020, 43, 463–478. [Google Scholar] [CrossRef]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2020, 8, 1176–1192. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Wenxing, L.; Huaxin, D.; Yue, Q.; Guoping, Z.; Zhong-Hua, C.; Feibo, W. HvHOX9, a Novel Homeobox Leucine Zipper Transcription Factor Revealed by Root miRNA and RNA Sequencing in Tibetan Wild Barley, Positively Regulates Al Tolerance. J. Exp. Bot. 2020, 71, 6057–6073. [Google Scholar]
- Zhang, X.; Li, L.; Yang, C.; Cheng, Y.; Ma, Q. GsMAS1 Encoding a MADS-box Transcription Factor Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arenhart, R.A.; Bai, Y.; de Oliveira, L.F.; Neto, L.B.; Schunemann, M.; Maraschin Fdos, S.; Mariath, J.; Silverio, A.; Sachetto-Martins, G.; Margis, R.; et al. New insights into aluminum tolerance in rice: The ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol. Plant 2014, 7, 709–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Huang, Y.; Qu, M.; Li, Y.; Zhang, S. A Maize ZmAT6 Gene Confers Aluminum Tolerance via Reactive Oxygen Species Scavenging. Front. Plant Sci. 2020, 9, 1016. [Google Scholar] [CrossRef]
- Courville, P.; Chaloupka, R.; Cellier, M.F.M. Recent progress in structure-function analyses of Nramp proton-dependent metal-ion transporters. Biochem. Cell Biol.-Biochim. Biol. Cell. 2006, 84, 960–978. [Google Scholar] [CrossRef]
- Sui, F.Q.; Chang, J.D.; Tang, Z.; Liu, W.J.; Huang, X.Y.; Zhao, F.J. Nramp5 expression and functionality likely explain higher cadmium uptake in rice than in wheat and maize. Plant Soil 2018, 433, 377–389. [Google Scholar] [CrossRef]
- Gunshin, H.; Mackenzie, B.; Berger, U.V.; Gunshin, Y.; Romero, M.F.; Boron, W.F.; Nussberger, S.; Gollan, J.L.; Hediger, M.A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 1997, 388, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Curie, C.; Alonso, J.M.; JEAN, M.L.; Ecker, J.R.; Briat, J.F. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem. J. 2000, 347, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Nishizawa, N.K. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef]
- Ishikawa, S.; Ishimaru, Y.; Igura, M.; Kuramata, M.; Abe, T.; Senoura, T.; Hase, Y.; Arao, T.; Nishizawa, N.K.; Nakanishi, H. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proc. Natl. Acad. Sci. USA 2012, 109, 19166–19171. [Google Scholar] [CrossRef] [Green Version]
- Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium. Plant J. 2015, 83, 625–637. [CrossRef] [PubMed] [Green Version]
- Yue, X.; Song, J.; Fang, B.; Wang, L.; Cui, J. BcNRAMP1 promotes the absorption of cadmium and manganese in Arabidopsis. Chemosphere 2021, 283, 131113. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Ma, J.F. Buckwheat FeNramp5 mediates high Mn uptake in roots. Plant Cell Physiol. 2020, 62, 600–609. [Google Scholar] [CrossRef]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice [Agricultural_Sciences]. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jixing, X.; Naoki, Y.; Jing, C.; Fang, S.R.; Feng, M.J. Differential expression of Nrat1 is responsible for Al-tolerance QTL on chromosome 2 in rice. J. Exp. Bot. 2014, 65, 4297–4304. [Google Scholar]
- Peng, F.; Wang, C.; Cheng, Y.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zeng, J.; Zhou, Y.; Wang, Y. Cloning and Characterization of TpNRAMP3, a Metal Transporter From Polish Wheat (Triticum polonicum L.). Front. Plant Sci. 2018, 29, 1354. [Google Scholar] [CrossRef]
- Matonyei, T.K.; Barros, B.A.; Guimaraes, R.G.N.; Ouma, E.O.; Cheprot, R.K.; Apolinário, L.C.; Ligeyo, D.O.; Costa, M.B.R.; Were, B.A.; Kisinyo, P.O. Aluminum tolerance mechanisms in Kenyan maize germplasm are independent from the citrate transporter ZmMATE1. Sci. Rep. 2020, 10, 7320. [Google Scholar] [CrossRef]
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; Mccouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrnstorfer, I.A.; Manatschal, C.; Arnold, F.M.; Laederach, J.; Dutzler, R. Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat. Commun. 2017, 8, 14033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Yang, G.; Li, P.; Wang, Z.; Fu, S.; Zhang, X.; Chen, X.; Shi, M.; Ming, Z.; Xia, J. Bioinformatic and Functional Analysis of a Key Determinant Underlying the Substrate Selectivity of the Al Transporter, Nrat1. Front. Plant Sci. 2018, 7, 606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xian, P.; Cai, Z.; Cheng, Y.; Lin, R.; Hai, N. Wild Soybean Oxalyl-CoA Synthetase Degrades Oxalate and Affects the Tolerance to Cadmium and Aluminum Stresses. Int. J. Mol. Sci. 2020, 21, 8869. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).