scAPAmod: Profiling Alternative Polyadenylation Modalities in Single Cells from Single-Cell RNA-Seq Data
Abstract
1. Introduction
2. Results
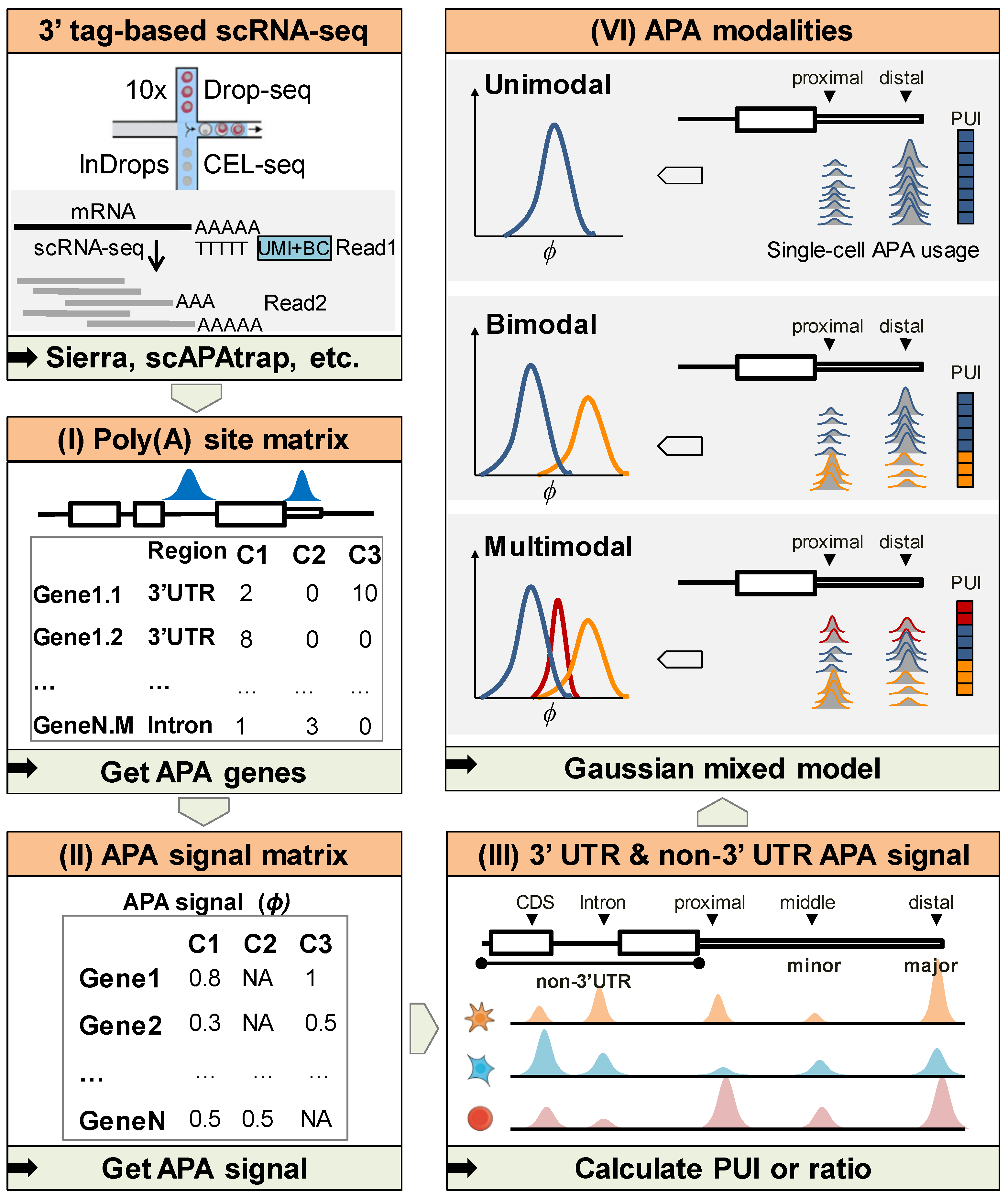
2.1. Overview of scAPAmod
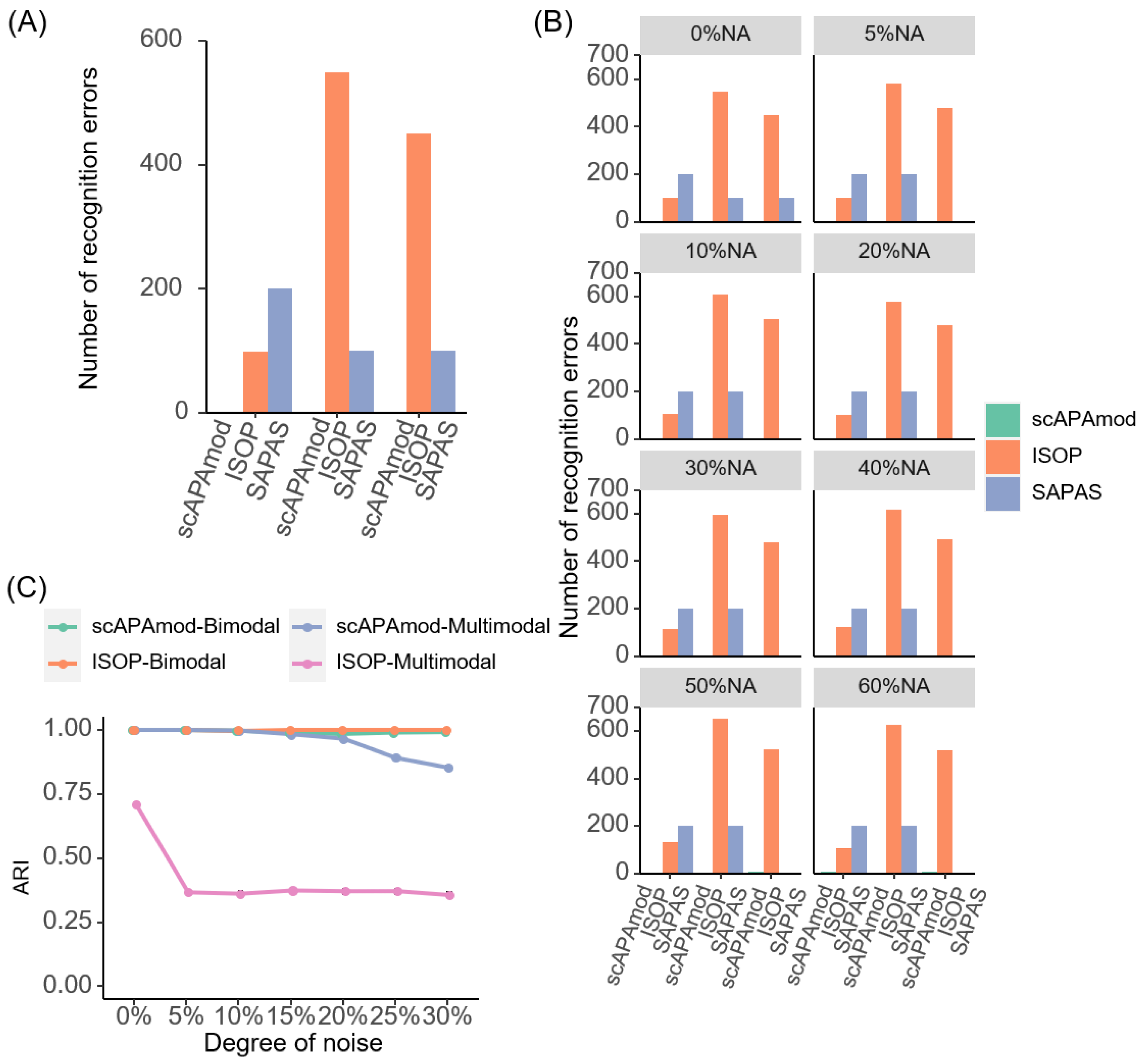
2.2. Evaluation of scAPAmod for APA Modality Identification
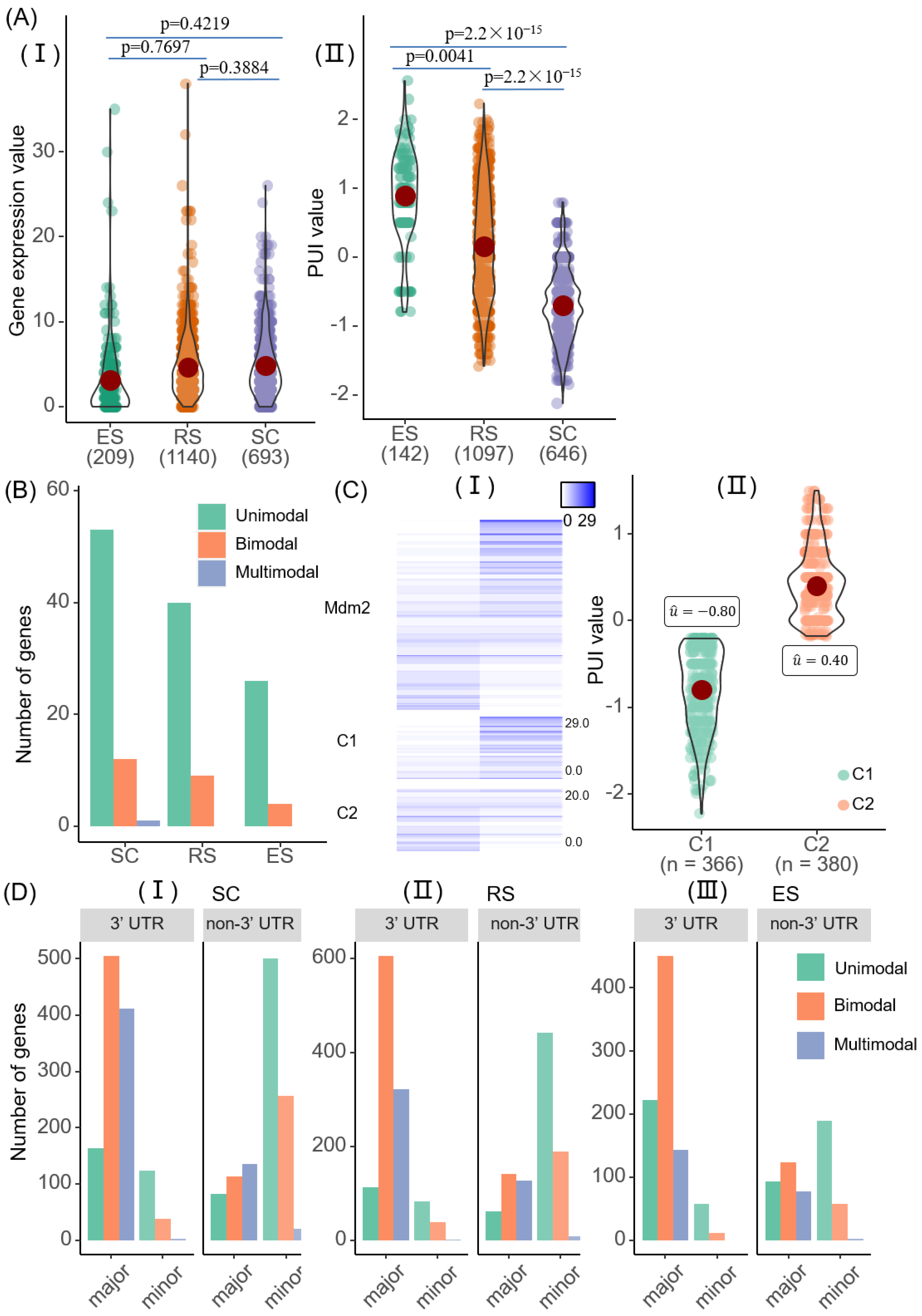
2.3. Modalities of 3′ UTR APA Sites in Mouse Sperm Data
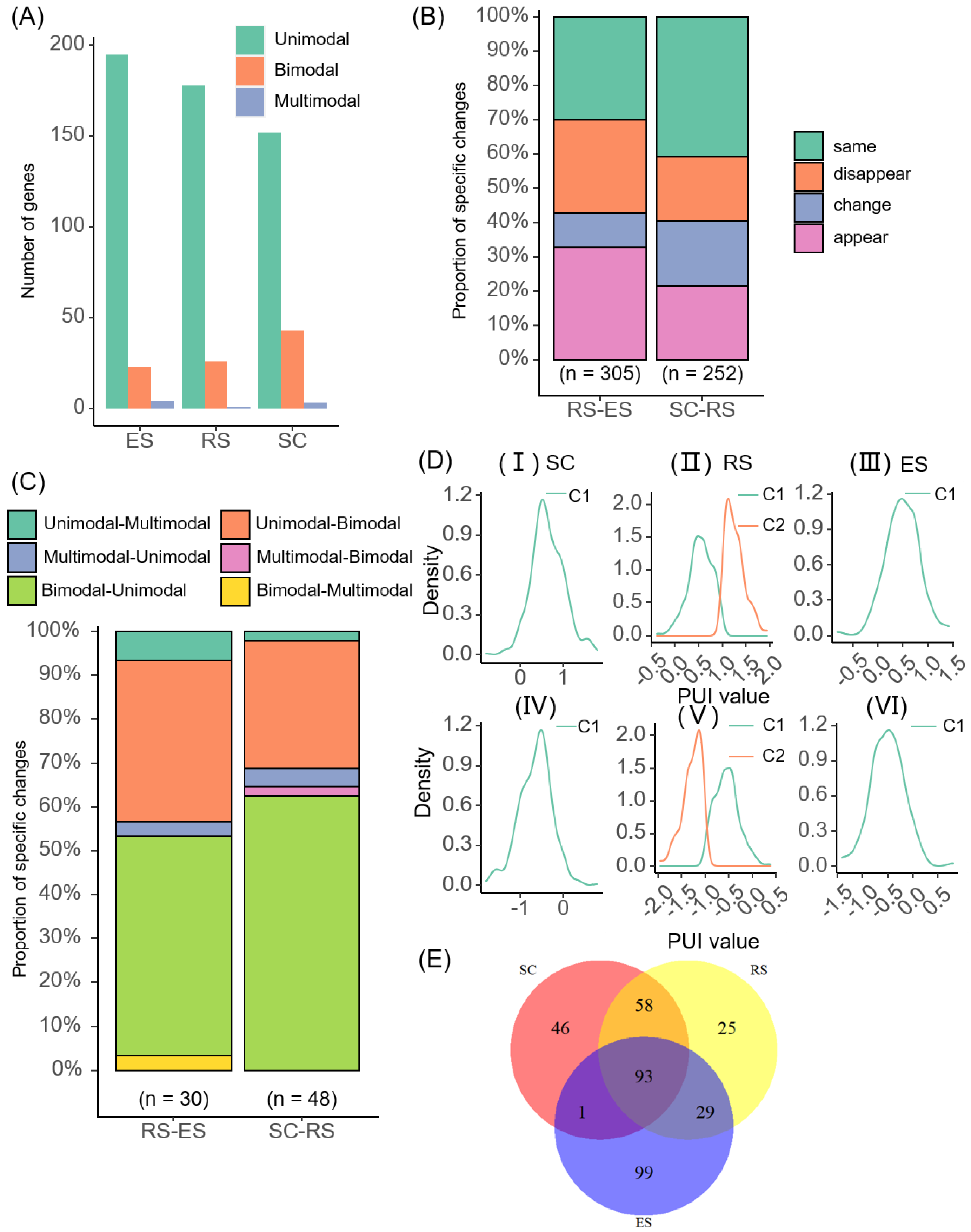
2.4. Cell-Type-Specific APA Modalities
2.5. Non-3′ UTR-APA Modality
3. Discussion
4. Materials and Methods
4.1. Data
4.2. Calculation of APA Index
4.3. Identification of Modalities of APA Usage
| Algorithm 1 The pseudocode of scAPAmod |
| Input: PUI value of a gene or a poly(A) site, or the ratio of a poly(A) site η |
| Output: number of components M Note: BIC(K) represents the BIC value corresponding to the Gaussian mixture model obeying K Gaussian distributions |
4.4. Identification of 3′ UTR-APA Modality
4.5. Identification of Non-3′ UTR-APA Modality
4.6. Performance Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, M.; Ji, G.; Fu, H.; Lin, Q.; Ye, C.; Ye, W.; Su, Y.; Wu, X. A survey on identification and quantification of alternative polyadenylation sites from RNA-seq data. Brief Bioinform. 2020, 21, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Guan, J.; Zeng, Y.; Li, Q.Q.; Wu, X. Genome-wide identification and predictive modeling of polyadenylation sites in eukaryotes. Brief. Bioinform. 2015, 16, 304–313. [Google Scholar] [CrossRef]
- Yeh, H.S.; Zhang, W.; Yong, J. Analyses of alternative polyadenylation: From old school biochemistry to high-throughput technologies. BMB Rep. 2017, 50, 201–207. [Google Scholar] [CrossRef]
- Velten, L.; Anders, S.; Pekowska, A.; Jarvelin, A.I.; Huber, W.; Pelechano, V.; Steinmetz, L.M. Single-cell polyadenylation site mapping reveals 3′ isoform choice variability. Mol. Syst. Biol. 2015, 11, 812. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jia, Q.; Song, Y.; Fu, H.; Wei, G.; Ni, T. Alternative Polyadenylation: Methods, Findings, and Impacts. Genom. Proteom. Bioinform. 2017, 15, 287–300. [Google Scholar] [CrossRef]
- Hwang, H.W.; Saito, Y.; Park, C.Y.; Blachere, N.E.; Tajima, Y.; Fak, J.J.; Zucker-Scharff, I.; Darnell, R.B. cTag-PAPERCLIP Reveals Alternative Polyadenylation Promotes Cell-Type Specific Protein Diversity and Shifts Araf Isoforms with Microglia Activation. Neuron 2017, 95, 1334–1349.e5. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef]
- Hashimshony, T.; Wagner, F.; Sher, N.; Yanai, I. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Rep. 2012, 2, 666–673. [Google Scholar] [CrossRef]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Ye, C.; Zhou, Q.; Hong, Y.; Li, Q.Q. Role of alternative polyadenylation dynamics in acute myeloid leukaemia at single-cell resolution. RNA Biol. 2019, 16, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhou, Q.; Wu, X.; Yu, C.; Ji, G.; Saban, D.R.; Li, Q.Q. scDAPA: Detection and visualization of dynamic alternative polyadenylation from single cell RNA-seq data. Bioinformatics 2019, 36, 1262–1264. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Chung, W.; Eum, H.H.; Lee, H.O.; Park, W.Y. Alternative polyadenylation of single cells delineates cell types and serves as a prognostic marker in early stage breast cancer. PLoS ONE 2019, 14, e0217196. [Google Scholar] [CrossRef]
- Levin, M.; Zalts, H.; Mostov, N.; Hashimshony, T.; Yanai, I. Gene expression dynamics are a proxy for selective pressures on alternatively polyadenylated isoforms. Nucleic Acids Res. 2020, 48, 5926–5938. [Google Scholar] [CrossRef]
- Shulman, E.D.; Elkon, R. Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Res. 2019, 47, 10027–10039. [Google Scholar] [CrossRef] [PubMed]
- Patrick, R.; Humphreys, D.T.; Janbandhu, V.; Oshlack, A.; Ho, J.W.K.; Harvey, R.; Lo, K.K. Sierra: Discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 2020, 21, 167. [Google Scholar] [CrossRef]
- Wu, X.; Liu, T.; Ye, C.; Ye, W.; Ji, G. scAPAtrap: Identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data. Brief Bioinform. 2021, 22, bbaa273. [Google Scholar] [CrossRef]
- Wen, W.X.; Mead, A.J.; Thongjuea, S. Technological advances and computational approaches for alternative splicing analysis in single cells. Comput. Struct. Biotechnol. J. 2020, 18, 332–343. [Google Scholar] [CrossRef]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef]
- Ramsköld, D.; Luo, S.; Wang, Y.-C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012, 30, 777. [Google Scholar] [CrossRef]
- Song, Y.; Botvinnik, O.B.; Lovci, M.T.; Kakaradov, B.; Liu, P.; Xu, J.L.; Yeo, G.W. Single-Cell Alternative Splicing Analysis with Expedition Reveals Splicing Dynamics during Neuron Differentiation. Mol. Cell 2017, 67, 148–161.e5. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sanguinetti, G. BRIE: Transcriptome-wide splicing quantification in single cells. Genome Biol. 2017, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.D.; Hu, Y.; Prins, J.F. Robust detection of alternative splicing in a population of single cells. Nucleic Acids Res. 2016, 44, e73. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Paul, A.; Bach, T.N.; Huang, Z.J.; Zhang, M.Q. Single-cell alternative polyadenylation analysis delineates GABAergic neuron types. BMC Biol. 2021, 19, 144. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Li, W.; Park, J.Y.; Zheng, D.; Hoque, M.; Yehia, G.; Tian, B. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016, 14, 1–17. [Google Scholar] [CrossRef]
- Liu, D.; Brockman, J.M.; Dass, B.; Hutchins, L.N.; Singh, P.; McCarrey, J.R.; Macdonald, C.C.; Graber, J.H. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007, 35, 234–246. [Google Scholar] [CrossRef]
- Lukassen, S.; Bosch, E.; Ekici, A.B.; Winterpacht, A. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Sci. Rep. 2018, 8, 6521. [Google Scholar] [CrossRef]
- Papoutsopoulou, S.; Nikolakaki, E.; Chalepakis, G.; Kruft, V.; Chevaillier, P.; Giannakouros, T. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 1999, 27, 2972–2980. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Kline, D.; Lu, J.; Orth, J.; Pilder, S.; Vijayaraghavan, S. Analysis of Ppp1cc-null mice suggests a role for PP1gamma2 in sperm morphogenesis. Biol. Reprod. 2007, 76, 992–1001. [Google Scholar] [CrossRef]
- Lin, C.L.; Jennen, D.G.J.; Ponsuksili, S.; Tholen, E.; Tesfaye, D.; Schellander, K.; Wimmers, K. Haplotype analysis of β-actin gene for its association with sperm quality and boar fertility. J. Anim. Breed. Genet. 2006, 123, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, N.S.; Schlotter, F.; Pévet, P.; Sassone-Corsi, P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature 1993, 362, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Berkovits, B.D.; Wang, L.; Guarnieri, P.; Wolgemuth, D.J. The testis-specific double bromodomain-containing protein BRDT forms a complex with multiple spliceosome components and is required for mRNA splicing and 3’-UTR truncation in round spermatids. Nucleic Acids Res. 2012, 40, 7162–7175. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.E.; Glantz, J.N.; Orth, J.D.; Poynter, G.M.; Salisbury, J.L. Testis-Specific Murine Centrin, Cetn1: Genomic Characterization and Evidence for Retroposition of a Gene Encoding a Centrosome Protein. Genomics 1999, 60, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Salces-Ortiz, J.; Ramon, M.; Gonzalez, C.; Perez-Guzman, M.D.; Garde, J.J.; Garcia-Alvarez, O.; Maroto-Morales, A.; Calvo, J.H.; Serrano, M.M. Differences in the ovine HSP90AA1 gene expression rates caused by two linked polymorphisms at its promoter affect rams sperm DNA fragmentation under environmental heat stress conditions. PLoS ONE 2015, 10, e0116360. [Google Scholar] [CrossRef]
- Ozturk, S.; Guzeloglu-Kayisli, O.; Demir, N.; Sozen, B.; Ilbay, O.; Lalioti, M.D.; Seli, E. Epab and Pabpc1 are differentially expressed during male germ cell development. Reprod. Sci. 2012, 19, 911–922. [Google Scholar] [CrossRef]
- Castaneda, J.M.; Miyata, H.; Archambeault, D.R.; Satouh, Y.; Yu, Z.; Ikawa, M.; Matzuk, M.M. Mouse t-complex protein 11 is important for progressive motility in spermdagger. Biol. Reprod. 2020, 102, 852–862. [Google Scholar] [CrossRef]
- Sinnar, S.A.; Small, C.L.; Evanoff, R.M.; Reinholdt, L.G.; Griswold, M.D.; Kopito, R.R.; Ryu, K.Y. Altered testicular gene expression patterns in mice lacking the polyubiquitin gene Ubb. Mol. Reprod. Dev. 2011, 78, 415–425. [Google Scholar] [CrossRef][Green Version]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.B.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Wang, H.; Wang, G.; Dai, Y.; Li, Z.; Zhu, Y.; Sun, F. Functional role of GKAP1 in the regulation of male germ cell spontaneous apoptosis and sperm number. Mol. Reprod. Dev. 2019, 86, 1199–1209. [Google Scholar] [CrossRef]
- Yant, L.J.; Ran, Q.; Rao, L.; van Remmen, H.; Shibatani, T.; Belter, J.G.; Motta, L.; Richardson, A.; Prolla, T.A. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free. Radic. Biol. Med. 2003, 34, 496–502. [Google Scholar] [CrossRef]
- Gao, Y.; Li, L.; Amos, C.I.; Li, W. Analysis of alternative polyadenylation from single-cell RNA-seq using scDaPars reveals cell subpopulations invisible to gene expression. Genome Res. 2021, 31, 1856–1866. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5488362/pdf/13059_2017_Article_1248.pdf (accessed on 15 June 2022).
- Mouselimis, L. ClusterR: Gaussian Mixture Models, K-Means, Mini-Batch-Kmeans, K-Medoids and Affinity Propagation Clustering. 2022. Available online: https://CRAN.R-project.org/package=ClusterR (accessed on 15 June 2022).
- Vu, T.N.; Wills, Q.F.; Kalari, K.R.; Niu, N.; Wang, L.; Pawitan, Y.; Rantalainen, M. Isoform-level gene expression patterns in single-cell RNA-sequencing data. Bioinformatics 2018, 34, 2392–2400. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, L.; Fu, H.; Mou, Y.; Lin, W.; Ye, L.; Ji, G. scAPAmod: Profiling Alternative Polyadenylation Modalities in Single Cells from Single-Cell RNA-Seq Data. Int. J. Mol. Sci. 2022, 23, 8123. https://doi.org/10.3390/ijms23158123
Qian L, Fu H, Mou Y, Lin W, Ye L, Ji G. scAPAmod: Profiling Alternative Polyadenylation Modalities in Single Cells from Single-Cell RNA-Seq Data. International Journal of Molecular Sciences. 2022; 23(15):8123. https://doi.org/10.3390/ijms23158123
Chicago/Turabian StyleQian, Lingwu, Hongjuan Fu, Yunwen Mou, Weixu Lin, Lishan Ye, and Guoli Ji. 2022. "scAPAmod: Profiling Alternative Polyadenylation Modalities in Single Cells from Single-Cell RNA-Seq Data" International Journal of Molecular Sciences 23, no. 15: 8123. https://doi.org/10.3390/ijms23158123
APA StyleQian, L., Fu, H., Mou, Y., Lin, W., Ye, L., & Ji, G. (2022). scAPAmod: Profiling Alternative Polyadenylation Modalities in Single Cells from Single-Cell RNA-Seq Data. International Journal of Molecular Sciences, 23(15), 8123. https://doi.org/10.3390/ijms23158123




