Functional and Transcriptome Analysis of Streptococcus pyogenes Virulence on Loss of Its Secreted Esterase
Abstract
1. Introduction
2. Results
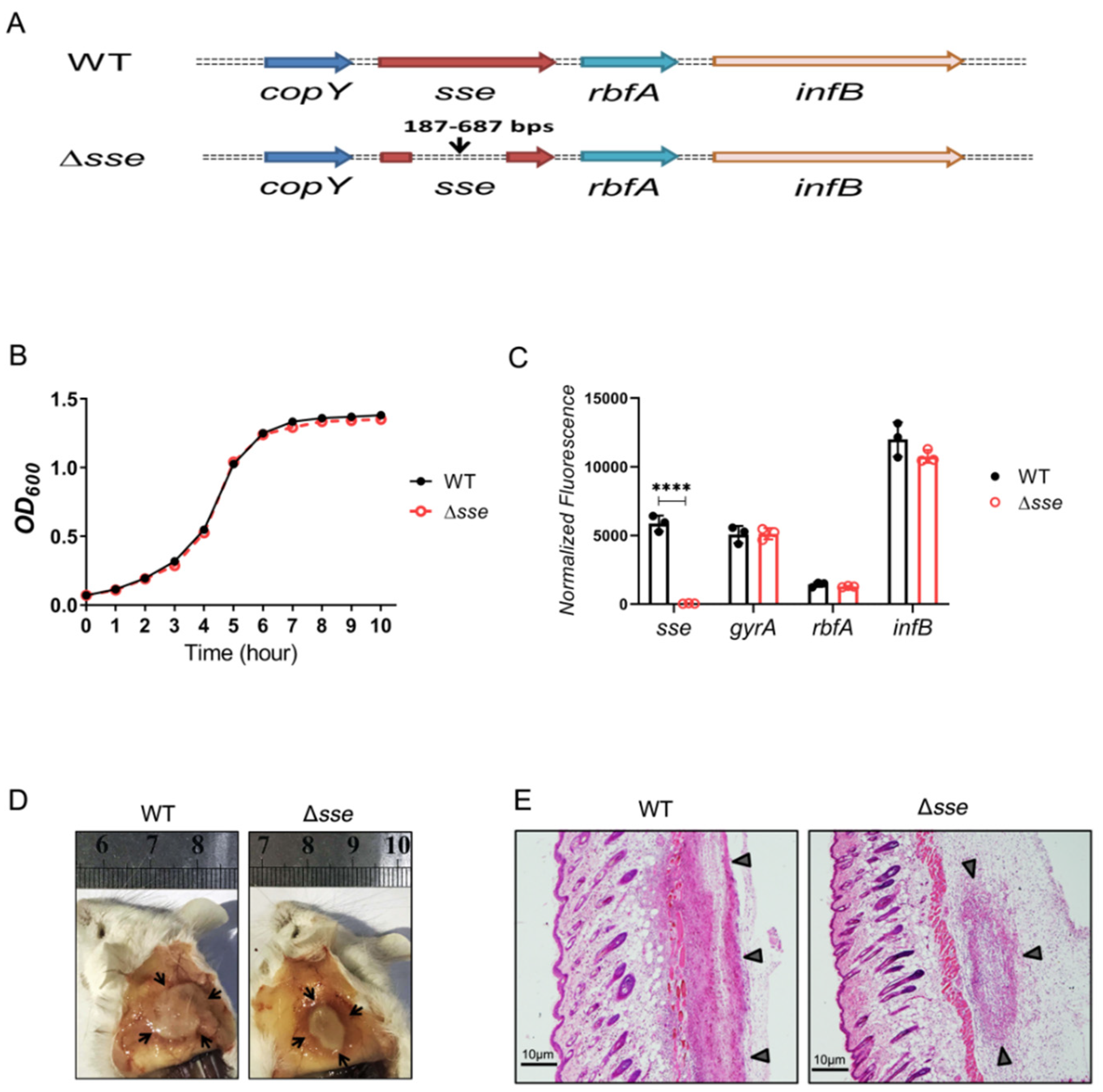
2.1. Sse Enhances GAS Skin Invasion in a Murine Infection Model
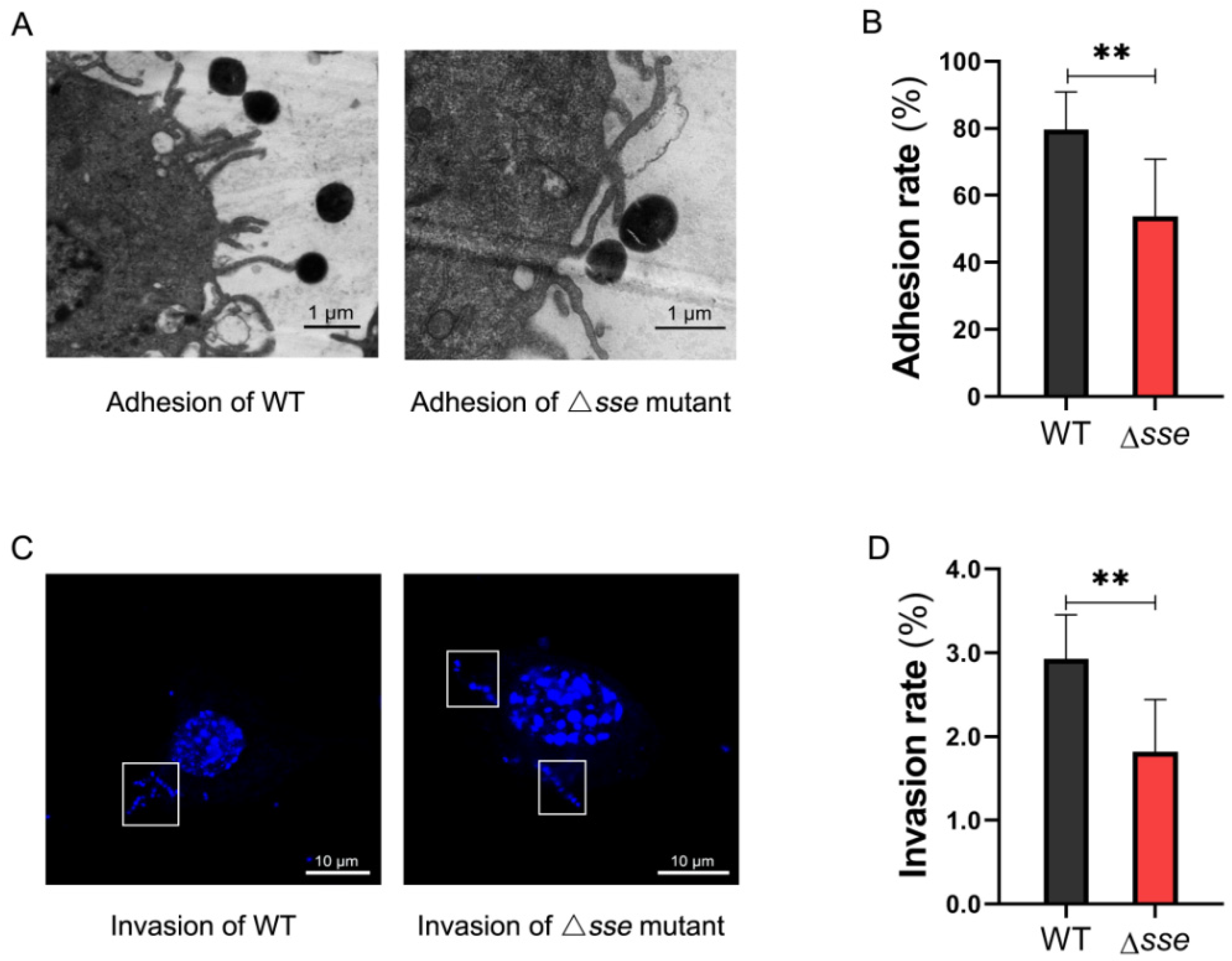
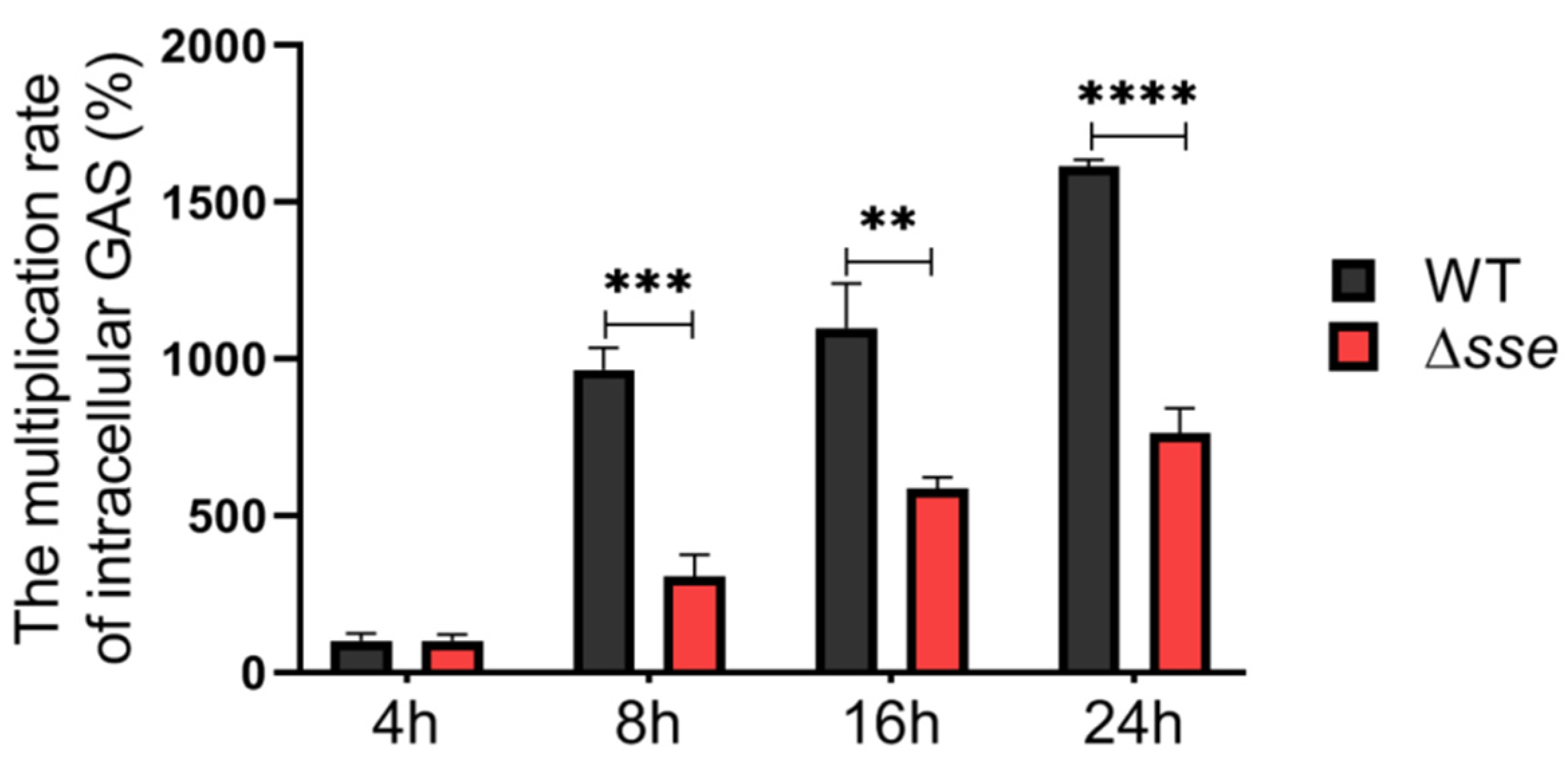
2.2. Sse Significantly Promotes GAS Invasion and Intracellular Survival in the Epithelial Cells
2.3. Sse Probably Inhibits Epithelial Phagocytic Function to Promote Intracellular GAS Proliferation
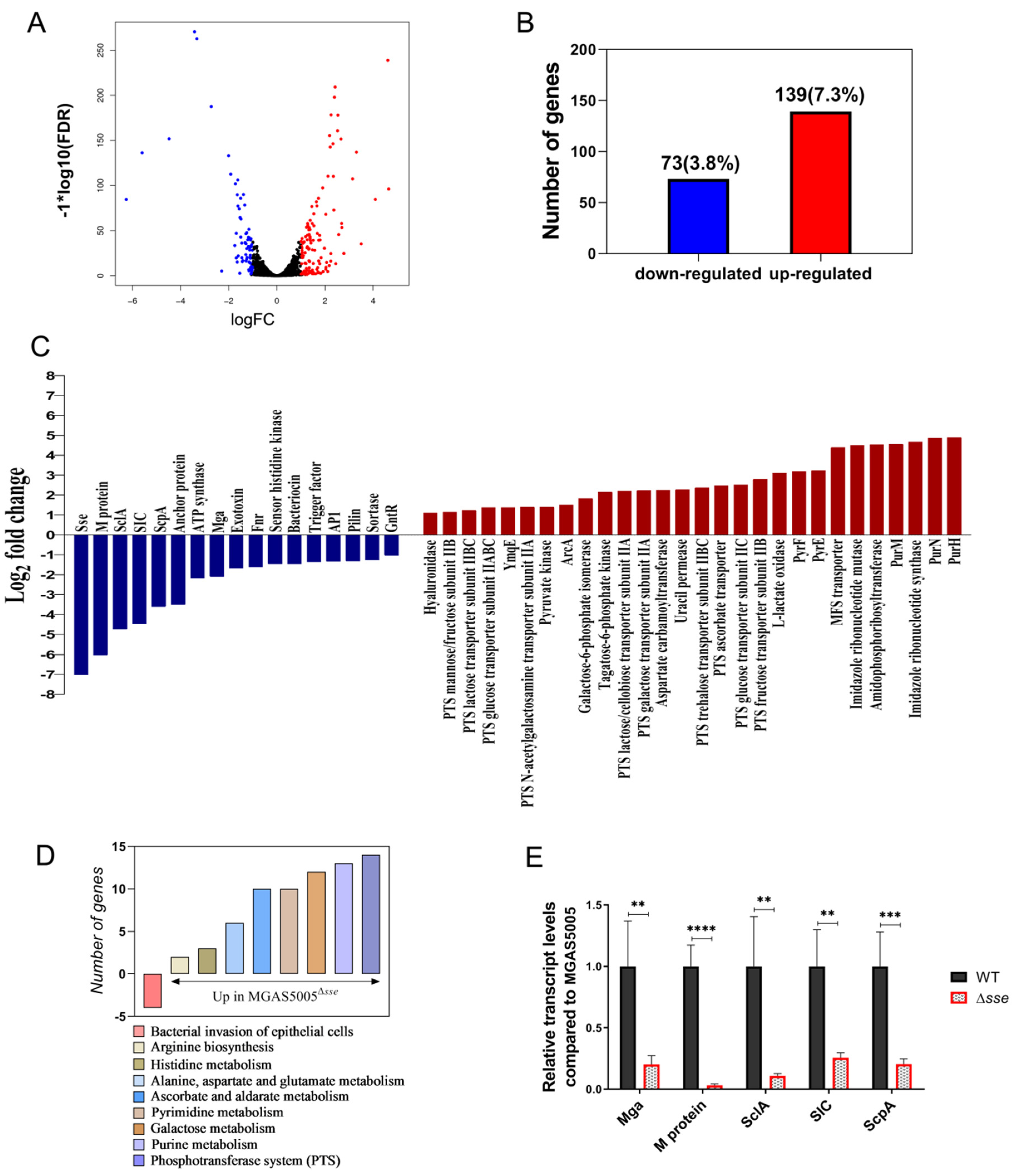
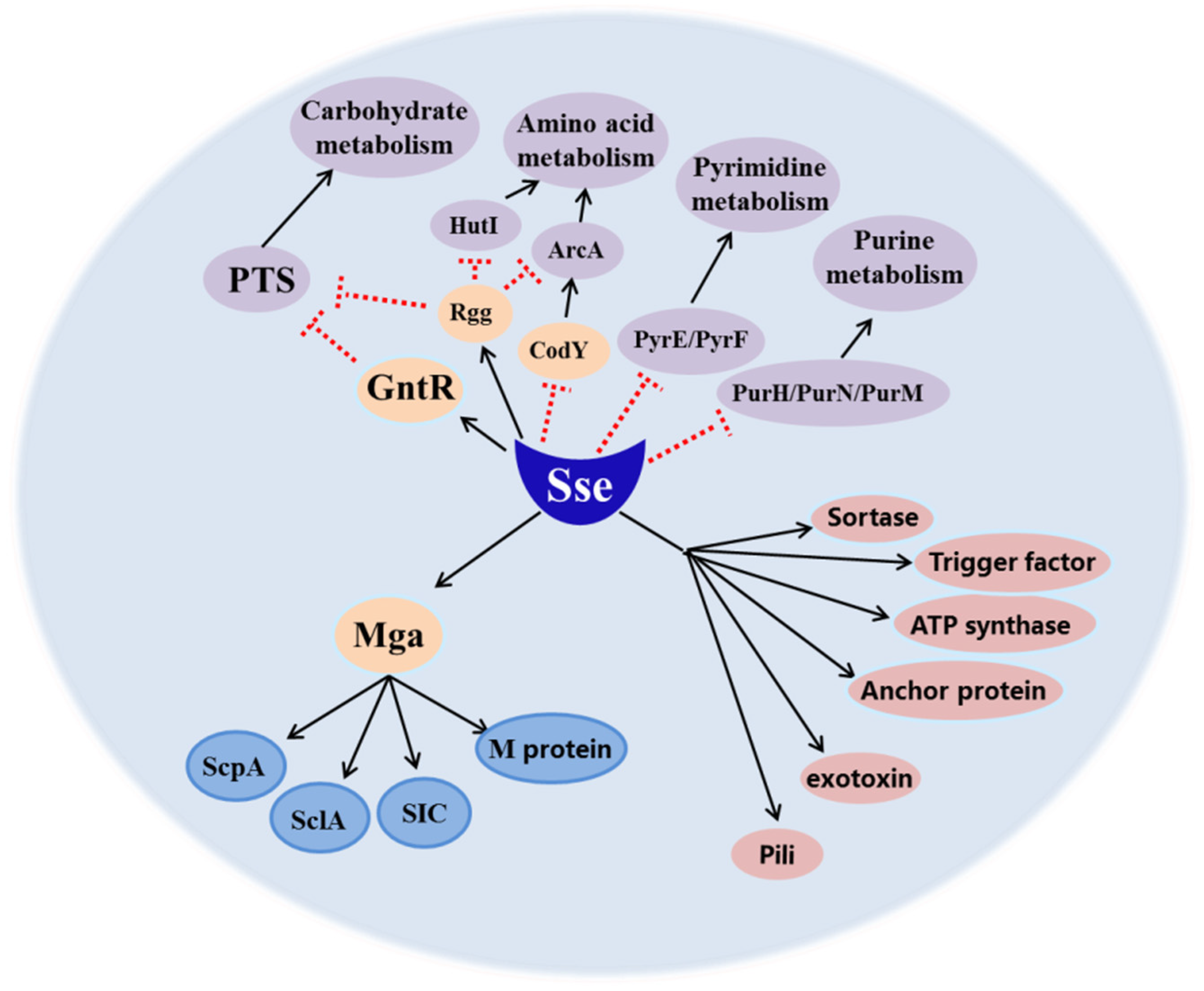
2.4. Sse Increases the Transcription of Invasive Virulence Factors and Affects Multiple Metabolic Pathways
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Mouse Subcutaneous Infection
4.3. Adhesion and Invasion Assays
4.4. RNA Extraction and Transcriptome Sequencing
4.5. Transmission Electron Microscopy
4.6. RT−PCR
4.7. Immunofluorescence Staining
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hua, C.-Z.; Yu, H.; Xu, H.-M.; Yang, L.-H.; Lin, A.-W.; Lyu, Q.; Lu, H.-P.; Xu, Z.-W.; Gao, W.; Chen, X.-J.; et al. A multi-center clinical investigation on invasive Streptococcus pyogenes infection in China, 2010–2017. BMC Pediatr. 2019, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Dooley, L.M.; Ahmad, T.B.; Pandey, M.; Good, M.F.; Kotiw, M. Rheumatic heart disease: A review of the current status of global research activity. Autoimmun. Rev. 2021, 20, 102740. [Google Scholar] [CrossRef]
- Hurst, J.R.; Brouwer, S.; Walker, M.J.; McCormick, J.K. Streptococcal superantigens and the return of scarlet fever. PloS Pathog. 2021, 17, e1010097. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.J.; Barnett, T.C.; McArthur, J.D.; Cole, J.N.; Gillen, C.M.; Henningham, A.; Sriprakash, K.S.; Sanderson-Smith, M.L.; Nizet, V. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin. Microbiol. Rev. 2014, 27, 264–301. [Google Scholar] [CrossRef]
- Liu, M.; Zhu, H.; Li, J.; Garcia, C.C.; Feng, W.; Kirpotina, L.N.; Hilmer, J.; Tavares, L.P.; Layton, A.W.; Quinn, M.T.; et al. Group A Streptococcus secreted esterase hydrolyzes platelet-activating factor to impede neutrophil recruitment and facilitate innate immune evasion. PloS Pathog. 2012, 8, e1002624. [Google Scholar] [CrossRef]
- Liu, G.; Liu, M.; Xie, G.; Lei, B. Characterization of streptococcal platelet-activating factor acetylhydrolase variants that are involved in innate immune evasion. Infect. Immun. 2013, 81, 3128–3138. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Wei, D.; Zhao, Y.; Zhong, Z.; Wang, Y.; Song, Y.; Cai, M.; Zhang, W.; Zhao, J.; Lv, C.; et al. Immunization with a secreted esterase protects mice against multiple serotypes (M1, M3, and M28) of group A Streptococcus. Front. Microbiol. 2020, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Wang, Y.; Cai, M.; Song, Y.; Zhu, H. Attenuation of virulence in multiple serotypes (M1, M3, and M28) of group A Streptococcus after the loss of secreted esterase. J. Microbiol. Immunol. Infect. 2021, S1684-1182(21)00196-1. [Google Scholar] [CrossRef]
- Marouni, M.J.; Sela, S. Fate of Streptococcus pyogenes and epithelial cells following internalization. J. Med. Microbiol. 2004, 53, 1–7. [Google Scholar] [CrossRef]
- Rox, K.; Rohde, M.; Chhatwal, G.S.; Müller, R. Disorazoles block group A streptococcal invasion into epithelial cells via interference with the host factor ezrin. Cell Chem. Biol. 2017, 24, 159–170. [Google Scholar] [CrossRef]
- Sanchez, L.; Chang, Y.Y.; Mellouk, N.; Enninga, J. Time-resolved fluorescence microscopy screens on host protein subversion during bacterial cell invasion. Methods Mol. Biol. 2022, 2523, 113–131. [Google Scholar] [PubMed]
- Iuchi, H.; Ohori, J.; Matsuzaki, H.; Tokushige, T.; Toge, S.; Yamashita, M. Impact of phosphorylcholine expression on the adherence and invasion of Streptococcus pyogenes to epithelial cells. Microorganisms 2022, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Pallon, J.; Sundqvist, M.; Hedin, K. A 2-year follow-up study of patients with pharyngotonsillitis. BMC Infect. Dis. 2018, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cleary, P.P. Intracellular invasion by Streptococcus pyogenes: Invasins, host receptors, and relevance to human disease. Microbiol. Spectr. 2019, 7, 7-4. [Google Scholar] [CrossRef] [PubMed]
- Grijmans, B.J.M.; van der Kooij, S.B.; Varela, M.; Meijer, A.H. LAPped in proof: LC3-associated phagocytosis and the arms race against bacterial pathogens. Front. Cell. Infect. Microbiol. 2022, 11, 809121. [Google Scholar] [CrossRef]
- Herb, M.; Gluschko, A.; Schramm, M. LC3-associated phagocytosis—The highway to hell for phagocytosed microbes. Semin. Cell Dev. Biol. 2020, 101, 68–76. [Google Scholar] [CrossRef]
- Hause, L.L.; McIver, K.S. Nucleotides critical for the interaction of the Streptococcus pyogenes Mga virulence regulator with Mga-regulated promoter sequences. J. Bacteriol. 2012, 194, 4904–4919. [Google Scholar] [CrossRef]
- Laabei, M.; Ermert, D. Catch me if you can: Streptococcus pyogenes complement evasion strategies. J. Innate Immun. 2019, 11, 3–12. [Google Scholar] [CrossRef]
- Lynskey, N.N.; Reglinski, M.; Calay, D.; Siggins, M.K.; Mason, J.C.; Botto, M.; Sriskandan, S. Multi-functional mechanisms of immune evasion by the streptococcal complement inhibitor C5a peptidase. PloS Pathog. 2017, 13, e1006493. [Google Scholar] [CrossRef]
- Humtsoe, J.O.; Kim, J.K.; Xu, Y.; Keene, D.R.; Höök, M.; Lukomski, S.; Wary, K.K. A streptococcal collagen-like protein interacts with the alpha2beta1 integrin and induces intracellular signaling. J. Biol. Chem. 2005, 280, 13848–13857. [Google Scholar] [CrossRef]
- Nakata, M.; Kawabata, S. Detection of fibronectin-binding proteins of Streptococcus pyogenes using ligand blot analysis. Methods Mol. Biol. 2020, 2136, 181–190. [Google Scholar] [PubMed]
- Ayinuola, Y.A.; Tjia-Fleck, S.; Readnour, B.M.; Liang, Z.; Ayinuola, O.; Paul, L.N.; Lee, S.W.; Fischetti, V.A.; Ploplis, V.A.; Castellino, F.J. Relationships between plasminogen-binding M-protein and surface enolase for human plasminogen acquisition and activation in Streptococcus pyogenes. Front. Microbiol. 2022, 13, 905670. [Google Scholar] [CrossRef] [PubMed]
- Khakzad, H.; Happonen, L.; Karami, Y.; Chowdhury, S.; Bergdahl, G.E.; Nilges, M.; van Nhieu, G.T.; Malmström, J.; Malmström, L. Structural determination of Streptococcus pyogenes M1 protein interactions with human immunoglobulin G using integrative structural biology. PLoS Comput. Biol. 2021, 17, e1008169. [Google Scholar] [CrossRef]
- Voyich, J.M.; Musser, J.M.; DeLeo, F.R. Streptococcus pyogenes and human neutrophils: A paradigm for evasion of innate host defense by bacterial pathogens. Microbes Infect. 2004, 6, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, Y.; Wang, G.; Feng, S.; Guo, Z.; Gu, G. Group A Streptococcus Cell Wall Oligosaccharide-Streptococcal C5a Peptidase Conjugates as Effective Antibacterial Vaccines. ACS Infect. Dis. 2020, 6, 281–290. [Google Scholar] [CrossRef]

| Target | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| Mga | TACAAGGGACACTCTGCCGTCTAC | GAGGTGAAACAAGCGTGAAAGGTC |
| M protein | AGGTAAAGGTCAAGCACCACAAGC | GGCTGCCGCTGTGAAGAATGG |
| SclA | CGCAATGGCAATATGGCTAAG | CTACTGCTGCTGCTGTAAAGA |
| SIC | GGAGCATTAGGTACAGGGTATG | CTGAGGAGGTTCAGGAATATGAG |
| ScpA | ATGCTGCGATCTCTCCAAATGGG | CTTGCTCGGTTACCTCACTTGTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, Y.; Zhu, H.; Zhong, Z. Functional and Transcriptome Analysis of Streptococcus pyogenes Virulence on Loss of Its Secreted Esterase. Int. J. Mol. Sci. 2022, 23, 7954. https://doi.org/10.3390/ijms23147954
Zhang X, Wang Y, Zhu H, Zhong Z. Functional and Transcriptome Analysis of Streptococcus pyogenes Virulence on Loss of Its Secreted Esterase. International Journal of Molecular Sciences. 2022; 23(14):7954. https://doi.org/10.3390/ijms23147954
Chicago/Turabian StyleZhang, Xiaolan, Yue Wang, Hui Zhu, and Zhaohua Zhong. 2022. "Functional and Transcriptome Analysis of Streptococcus pyogenes Virulence on Loss of Its Secreted Esterase" International Journal of Molecular Sciences 23, no. 14: 7954. https://doi.org/10.3390/ijms23147954
APA StyleZhang, X., Wang, Y., Zhu, H., & Zhong, Z. (2022). Functional and Transcriptome Analysis of Streptococcus pyogenes Virulence on Loss of Its Secreted Esterase. International Journal of Molecular Sciences, 23(14), 7954. https://doi.org/10.3390/ijms23147954







