The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Results
2.1. Regulation of HBV Replication from the Genome in HepAD38 Cells
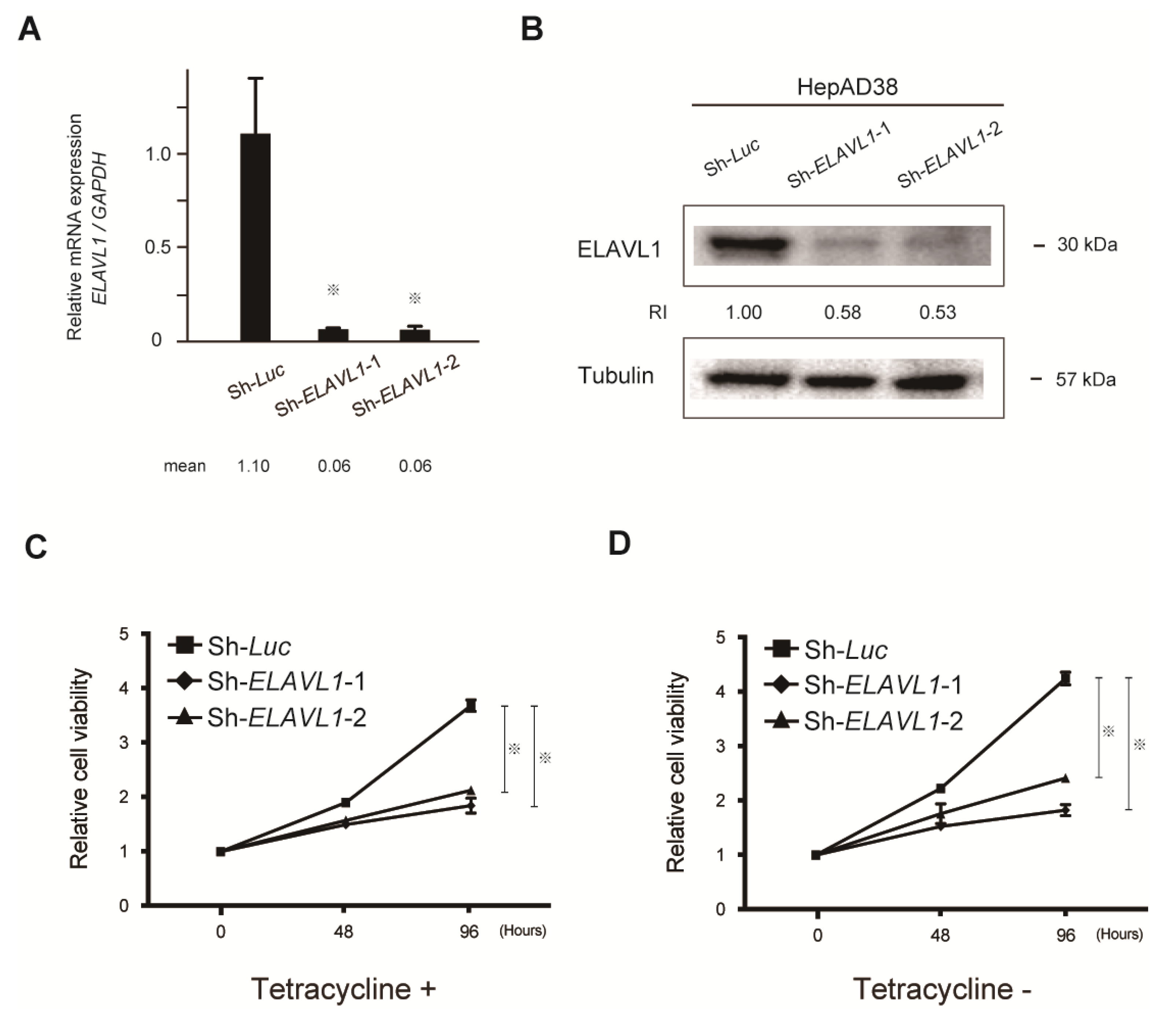
2.2. Stable Knockdown of ELAVL1 in HCC Cells
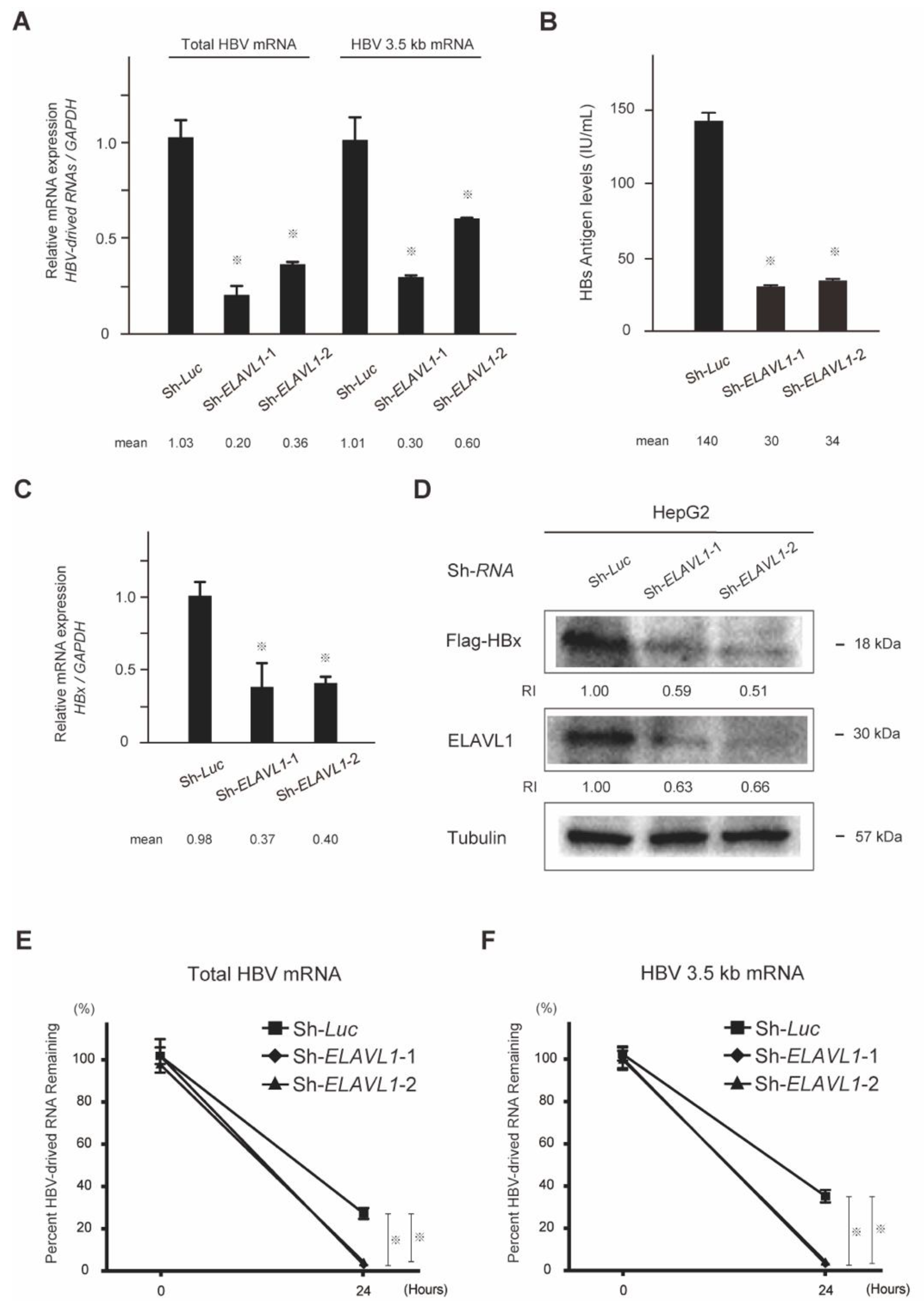
2.3. The Effect of ELAVL1 on HBV-Derived RNAs and Proteins
2.4. The Role of ELAVL1 in Cell Proliferation Ability
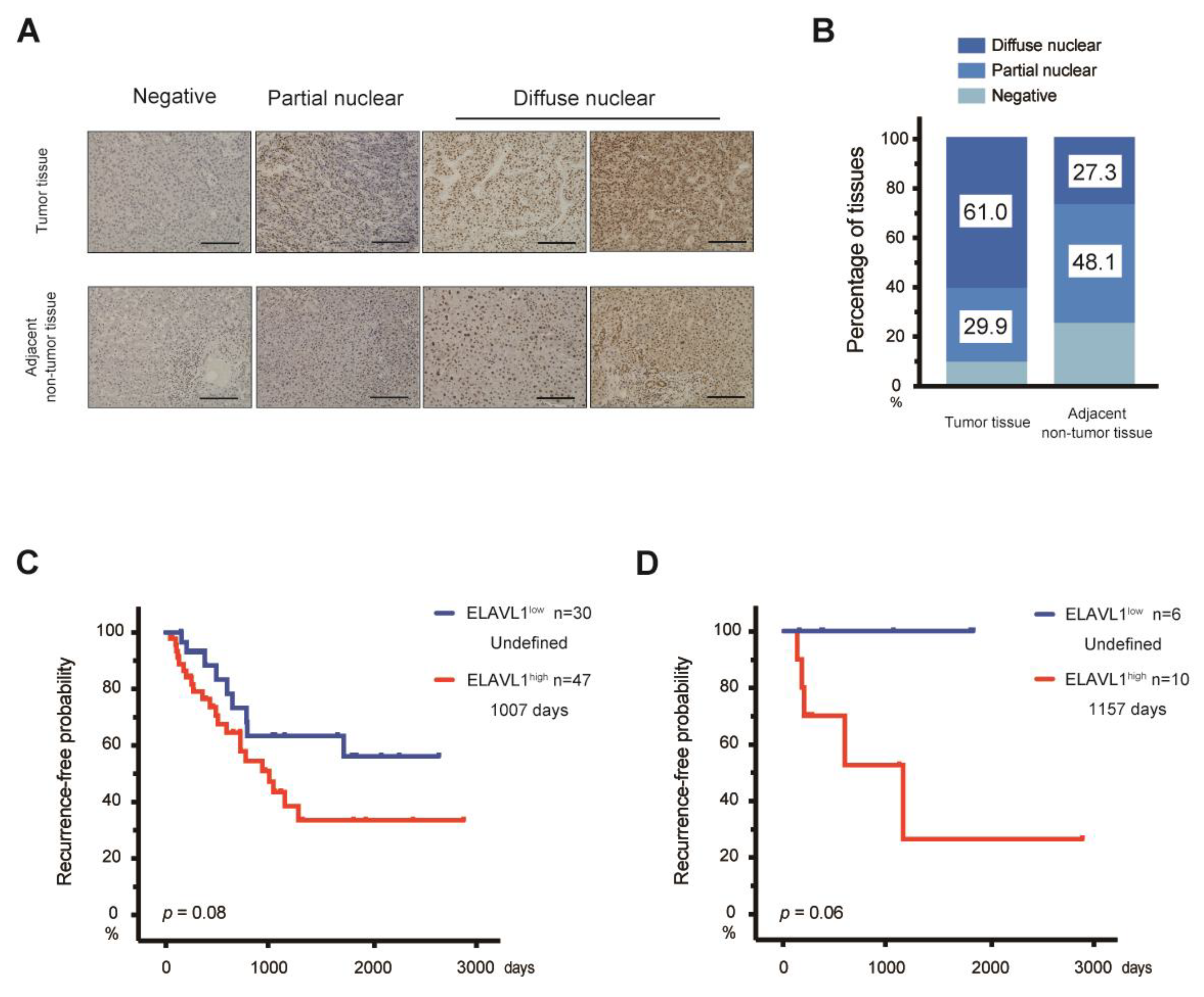
2.5. Expression of ELAVL1 in Primary HCC Tissues
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Quantitative Real-Time PCR
4.3. Western Blotting
4.4. Measurement of HBs Antigen Levels
4.5. Lentiviral Production and Transduction
4.6. Cell Proliferation Assay
4.7. Patients and Surgical Specimens
4.8. Data Collection and Analysis from the Cancer Genome Atlas (TCGA)-Liver Hepatocellular Carcinoma (LIHC)
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, L.S.Y.; Covert, E.; Wilson, E.; Kottilil, S. Chronic hepatitis B infection: A Review. JAMA 2018, 319, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, L.; Charlson, F.; Stanaway, J.; Larney, S.; Alexander, L.T.; Hickman, M.; Cowie, B.; Hall, W.D.; Strang, J.; Whiteford, H.; et al. Estimating the burden of disease attributable to injecting drug use as a risk factor for HIV, hepatitis C, and hepatitis B: Findings from the Global Burden of Disease Study 2013. Lancet Infect Dis. 2016, 16, 1385–1398. [Google Scholar] [CrossRef]
- Levrero, M.; Pollicino, T.; Petersen, J.; Belloni, L.; Raimondo, G.; Dandri, M. Control of cccDNA function in hepatitis B virus infection. J. Hepatol. 2019, 51, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.T.; Schwinn, S.; Locarnini, S.; Fyfe, J.; Manns, M.P.; Trautwein, C.; Zentgraf, H. Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol. 2001, 307, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Mason, W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000, 64, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Takata, A.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yoshikawa, T.; Koike, K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci. Rep. 2016, 6, 23237. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.W.; Wang, N.; Wang, Y.; Wang, F.; Fu, Z.; Yan, X.; Zhu, H.; Diao, W.; Ding, Y.; Chen, X.; et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016, 64, 278–291. [Google Scholar] [CrossRef]
- Duriez, M.; Mandouri, Y.; Lekbaby, B.; Wang, H.; Schnuriger, A.; Redelsperger, F.; Guerrera, C.I.; Lefevre, M.; Fauveau, V.; Ahodantin, J.; et al. Alternative splicing of hepatitis B virus: A novel virus/host interaction altering liver immunity. J. Hepatol. 2017, 67, 687–699. [Google Scholar] [CrossRef]
- Marcellin, P.; Heathcote, E.J.; Buti, M.; Gane, E.; de Man, R.A.; Krastev, Z.; Germanidis, G.; Lee, S.S.; Flisiak, R.; Kaita, K.; et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N. Engl. J. Med. 2008, 359, 2442–2455. [Google Scholar] [CrossRef]
- Hadziyannis, S.J.; Tassopoulos, N.C.; Heathcote, E.J.; Chang, T.T.; Kitis, G.; Rizzetto, M.; Marcellin, P.; Lim, S.G.; Goodman, Z.; Wulfsohn, M.S.; et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-negative chronic hepatitis B. N. Engl. J. Med. 2003, 348, 800–807. [Google Scholar] [CrossRef]
- Sekiba, K.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yamagami, M.; Suzuki, T.; Ishibashi, R.; Seimiya, T.; Tanaka, E.; Koike, K. DHX9 regulates production of hepatitis B virus-derived circular RNA and viral protein levels. Oncotarget 2018, 9, 20953–20964. [Google Scholar] [CrossRef]
- Meisner, N.C.; Hackermüller, J.; Uhl, V.; Aszódi, A.; Jaritz, M.; Auer, M. mRNA openers and closers: Modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. ChemBioChem 2004, 5, 1432–1447. [Google Scholar] [CrossRef] [PubMed]
- Liaw, Y.F.; Sung, J.J.Y.; Chow, W.C.; Farrell, G.; Lee, C.Z.; Yuen, H.; Tanwandee, T.; Tao, Q.M.; Shue, K.; Keene, O.N.; et al. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Eng. J. Med. 2004, 351, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Suzuki, F.; Kobayashi, M.; Seko, Y.; Kawamura, Y.; Sezaki, H.; Akuta, N.; Suzuki, Y.; Saitoh, S.; Arase, Y.; et al. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology 2013, 58, 98–107. [Google Scholar] [CrossRef]
- Xia, Y.; Guo, H. Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential. Antivir. Res. 2020, 180, 104824. [Google Scholar] [CrossRef]
- Tada, T.; Kumada, T.; Toyoda, H.; Kiriyama, S.; Tanikawa, M.; Hisanaga, Y.; Kanamori, A.; Kitabatake, S.; Yama, T.; Tanaka, J. HBcrAg predicts hepatocellular carcinoma development: An analysis using time-dependent receiver operating characteristics. J. Hepatol. 2016, 65, 48–56. [Google Scholar] [CrossRef]
- Martinez, M.G.; Villeret, F.; Testoni, B.; Zoulim, F. Can we cure hepatitis B virus with novel direct-acting antivirals? Liver Int. 2020, 40 (Suppl. 1), 27–34. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.G.; Boyd, A.; Combe, E.; Testoni, B.; Zoulim, F. Covalently closed circular DNA: The ultimate therapeutic target for curing HBV infections. J. Hepatol. 2021, 75, 706–717. [Google Scholar] [CrossRef]
- Ross, J. mRNA stability in mammalian cells. Microbiol. Rev. 1995, 59, 423–450. [Google Scholar] [CrossRef]
- Zubiaga, A.M.; Greenberg, M.E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 1995, 15, 2219–2230. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Chu, H.; Guan, Y.; Bi, J.; Wang, B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int. J. Mol. Sci. 2013, 14, 10015–10041. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Govoni, S. The complex world of post-transcriptional mechanisms: Is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell Mol. Life Sci. 2012, 69, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Caldwell, M.C.; Lin, S.; Furneaux, H.; Gorospe, M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000, 19, 2340–2350. [Google Scholar] [CrossRef]
- Wang, W.; Yang, X.; Cristofalo, V.J.; Holbrook, N.J.; Gorospe, M. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol. Cell. Biol. 2001, 21, 5889–5898. [Google Scholar] [CrossRef][Green Version]
- de Silanes, I.L.; Lal, A.; Gorospe, M. HuR: Post-transcriptional paths to malignancy. RNA Biol. 2005, 2, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Giaginis, C.; Theocharis, S. Clinical significance of HuR expression in human malignancy. Med. Oncol. 2014, 31, 161. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Zhou, Y.; Zheng, W.; Chambers, S.K. HuR expression in the nucleus correlates with high histological grade and poor disease-free survival in ovarian cancer. Aust. N. Z. J. Obstet. Gynaecol. 2009, 49, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Yoo, P.S.; Sullivan, C.A.W.; Kiang, S.; Gao, W.; Uchio, E.M.; Chung, G.G.; Cha, C.H. Tissue microarray analysis of 560 patients with colorectal adenocarcinoma. high expression of HuR predicts poor survival. Ann. Surg. Oncol. 2009, 16, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Wang, Y.; Ye, L.H. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol. Med. 2014, 11, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Slagle, B.L.; Bouchard, M.J. Role of HBx in hepatitis B virus persistence and its therapeutic implications. Curr. Opin. Virol. 2018, 30, 32–38. [Google Scholar] [CrossRef]
- Heinonen, M.; Bono, P.; Narko, K.; Chang, S.H.; Lundin, J.; Joensuu, H.; Furneaux, H.; Hla, T.; Haglund, C.; Ristimäki, A. Cytoplasmic HuR Expression Is a Prognostic Factor in Invasive Ductal Breast Carcinoma. Cancer Res. 2005, 65, 2157–2161. [Google Scholar] [CrossRef]
- Miyata, Y.; Watanabe, S.; Sagara, Y.; Nitsunari, K.; Natsuo, T.; Ohba, K.; Sakai, H. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS ONE 2013, 8, e59095. [Google Scholar] [CrossRef] [PubMed]
- Papatheofani, V.; Levidou, G.; Sarantis, P.; Koustas, E.; Karamouzis, M.V.; Pergaris, A.; Kouraklis, G.; Theocharis, S. HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment. Biomedicines 2021, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Kakizaki, M.; Yamamoto, Y.; Yabuta, S.; Kurosaki, N.; Kagawa, T.; Kotani, A. The immunological function of extracellular vesicles in hepatitis B virus-infected hepatocytes. PLoS ONE 2018, 13, e205886. [Google Scholar] [CrossRef] [PubMed]
- Ladner, S.K.; Otto, M.J.; Barker, C.S.; Zaifert, K.; Wang, G.H.; Guo, J.T.; Seeger, C.; King, R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997, 41, 1715–1720. [Google Scholar] [CrossRef] [PubMed]
- Sekiba, K.; Otsuka, M.; Ohno, M.; Yamagami, M.; Kishikawa, T.; Suzuki, T.; Ishibashi, R.; Seimiya, T.; Tanaka, E.; Koike, K. Inhibition of HBV Transcription From cccDNA With Nitazoxanide by Targeting the HBx–DDB1 Interaction. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 297–312. [Google Scholar] [CrossRef]
- Imai, Y.; Chiba, T.; Kondo, T.; Kanzaki, H.; Kanayama, K.; Ao, J.; Kojima, R.; Kusakabe, Y.; Nakamura, M.; Tomoko, S.; et al. Interferon-γ induced PD-L1 expression and soluble PD-L1 production in gastric cancer. Oncol. Lett. 2020, 20, 2161–2168. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Miyagi, S.; Saraya, A.; Aoki, R.; Seki, A.; Morita, Y.; Yonemitsu, Y.; Yokosuka, O.; Taniguchi, H.; Nakauchi, H.; et al. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008, 68, 7742–7749. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | ELAVL1low | ELAVL1high | p-Value |
|---|---|---|---|
| (n = 30) | (n = 47) | ||
| Age (years) (median (IQR)) | 67 (11) | 70 (13) | 0.257 |
| Gender: male/female | 23/7 | 37/10 | 0.832 |
| Etiology: HBV/HCV/others | 6/12/12 | 10/16/21 | 0.866 |
| Fibrosis stage: CH/LC | 25/5 | 34/13 | 0.266 |
| AFP (ng/mL) (median (IQR)) | 12.9 (206.7) | 9.2 (116.9) | 0.703 |
| BCLC stage: A/B | 27/3 | 38/9 | 0.280 |
| Edmondson–Steiner grade: I/II/III/IV | 2/8/16/4 | 1/11/26/9 | 0.699 |
| Characteristics | ELAVL1low | ELAVL1high | p-Value |
|---|---|---|---|
| (n = 6) | (n = 10) | ||
| Age (years ) (median (IQR)) | 66 (12) | 62 (15) | 0.713 |
| Gender: male/female | 4/2 | 10/0 | 0.051 |
| Fibrosis stage: CH/LC | 5/1 | 8/2 | 0.869 |
| AFP (ng/mL) (median (IQR)) | 662.7 (1651.7) | 3.9 (268.7) | 0.263 |
| BCLC stage: A/B | 5/1 | 8/2 | 0.869 |
| Edmondson–Steiner grade: I/II/III/IV | 0/0/3/3 | 0/2/6/2 | 0.309 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanzaki, H.; Chiba, T.; Kaneko, T.; Ao, J.; Kan, M.; Muroyama, R.; Nakamoto, S.; Kanda, T.; Maruyama, H.; Kato, J.; et al. The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2022, 23, 7878. https://doi.org/10.3390/ijms23147878
Kanzaki H, Chiba T, Kaneko T, Ao J, Kan M, Muroyama R, Nakamoto S, Kanda T, Maruyama H, Kato J, et al. The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences. 2022; 23(14):7878. https://doi.org/10.3390/ijms23147878
Chicago/Turabian StyleKanzaki, Hiroaki, Tetsuhiro Chiba, Tatsuya Kaneko, Junjie Ao, Motoyasu Kan, Ryosuke Muroyama, Shingo Nakamoto, Tatsuo Kanda, Hitoshi Maruyama, Jun Kato, and et al. 2022. "The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells" International Journal of Molecular Sciences 23, no. 14: 7878. https://doi.org/10.3390/ijms23147878
APA StyleKanzaki, H., Chiba, T., Kaneko, T., Ao, J., Kan, M., Muroyama, R., Nakamoto, S., Kanda, T., Maruyama, H., Kato, J., Zen, Y., Kotani, A., Sekiba, K., Otsuka, M., Ohtsuka, M., & Kato, N. (2022). The RNA-Binding Protein ELAVL1 Regulates Hepatitis B Virus Replication and Growth of Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences, 23(14), 7878. https://doi.org/10.3390/ijms23147878







