CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis
Abstract
:1. Introduction
2. Results
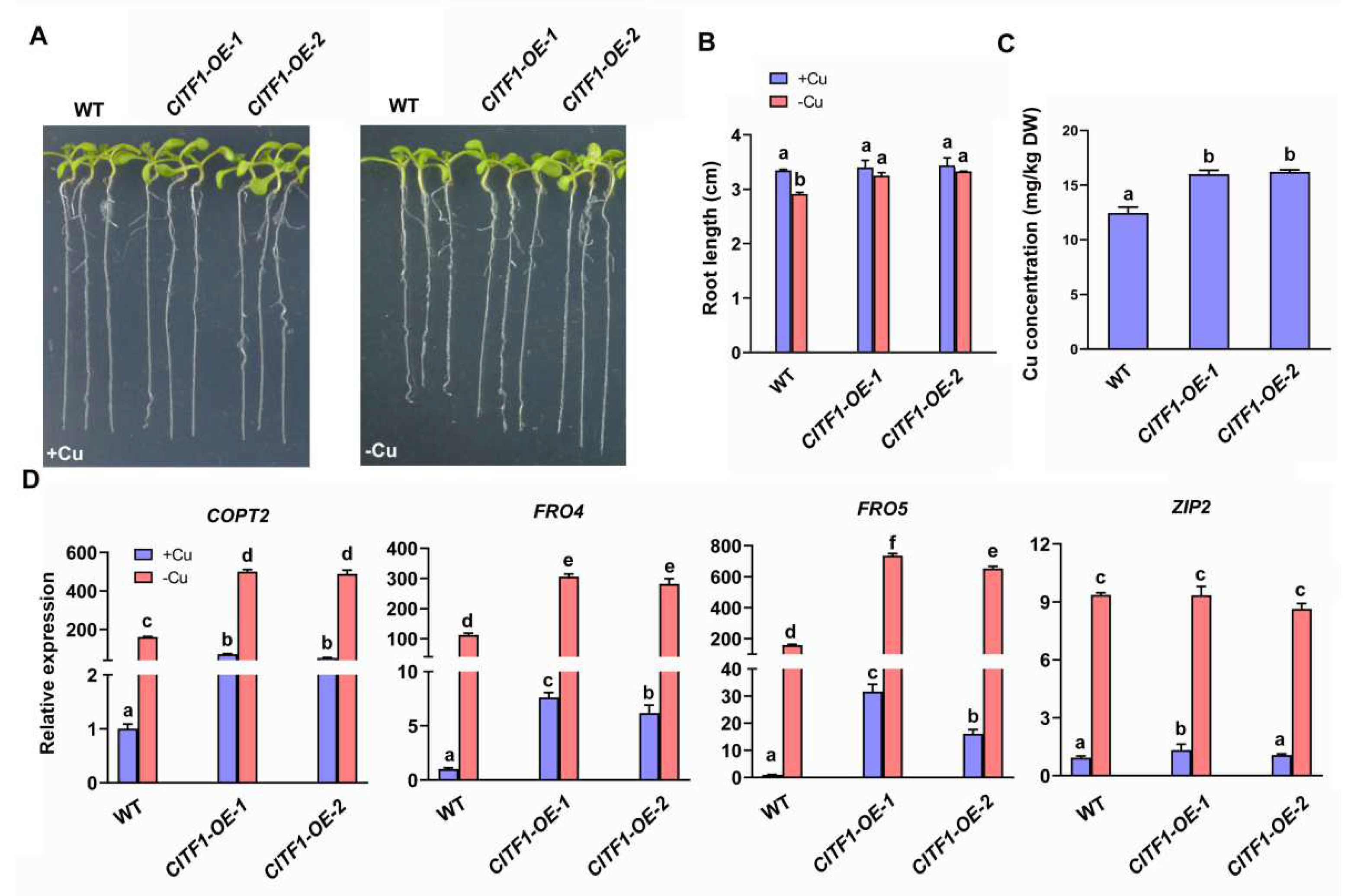
2.1. CITF1 Specifically Affects the Expression of Cu Uptake Genes COPT2, FRO4 and FRO5
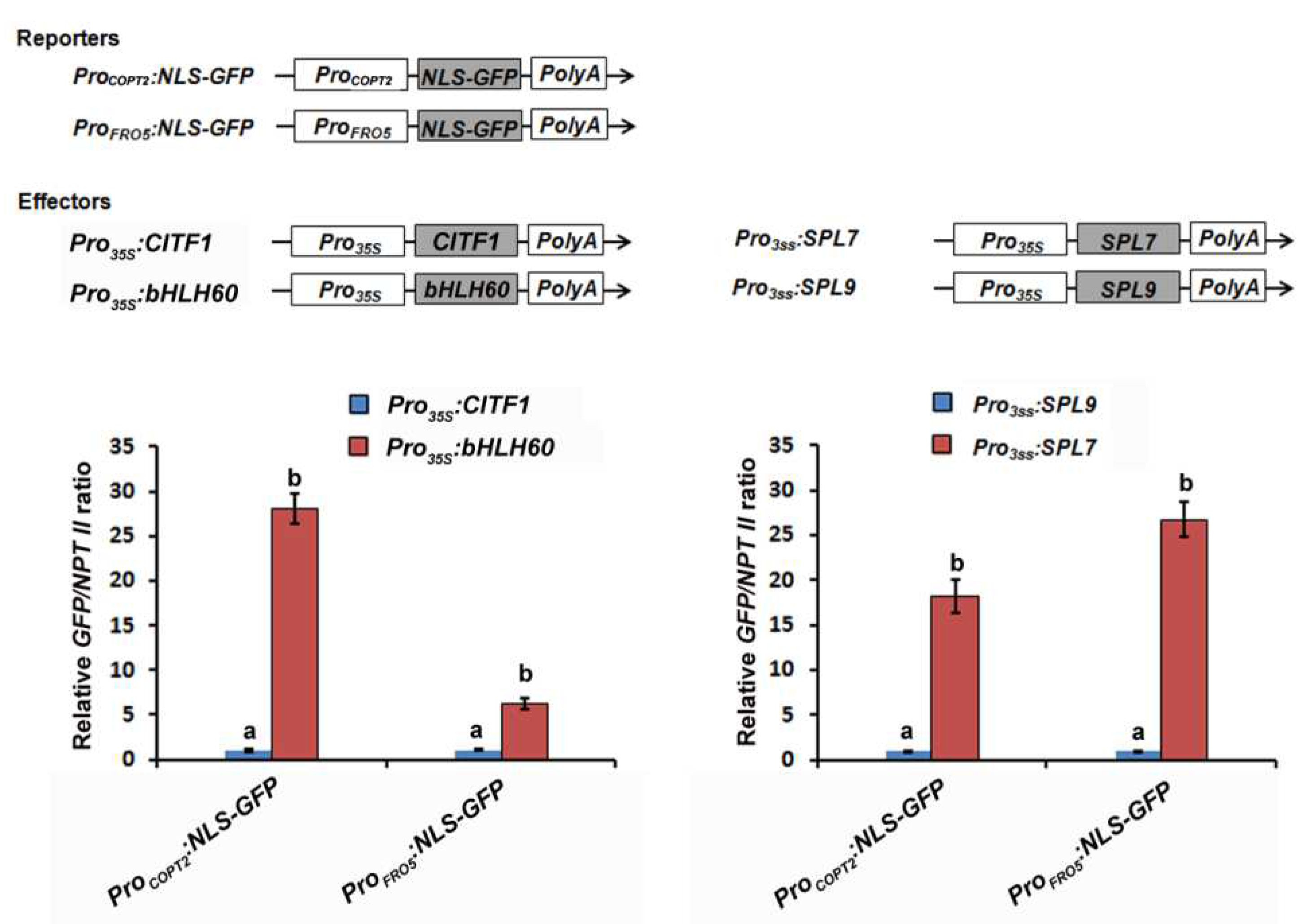
2.2. CITF1 Is a Direct Target of SPL7
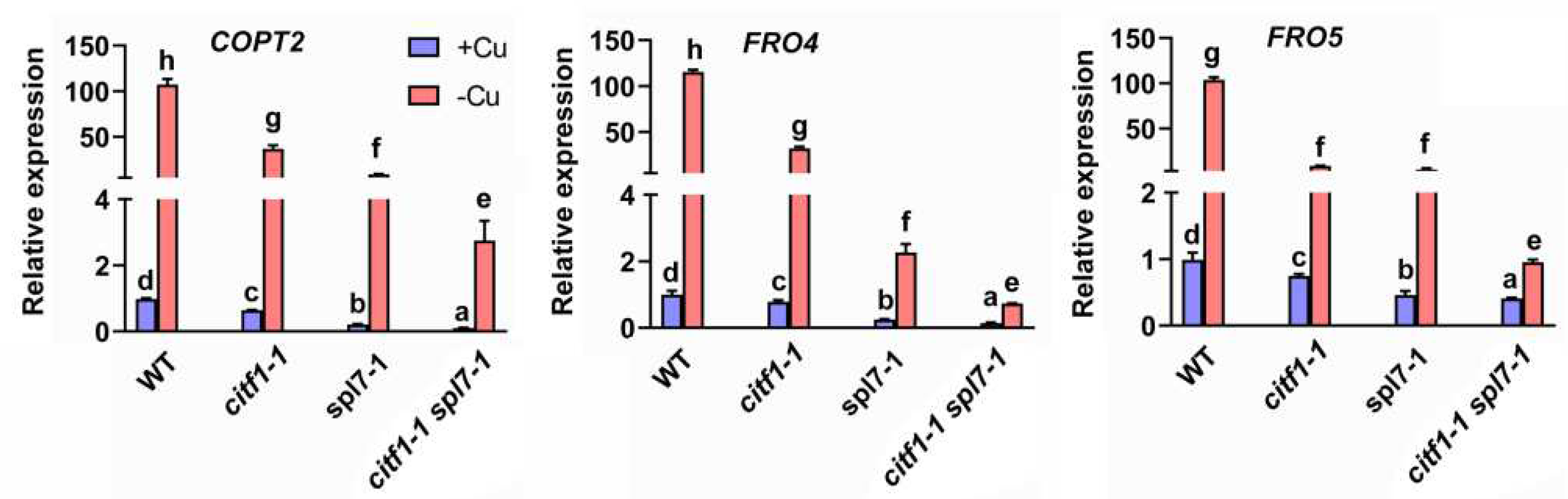
2.3. Genetic Interaction of CITF1 and SPL7
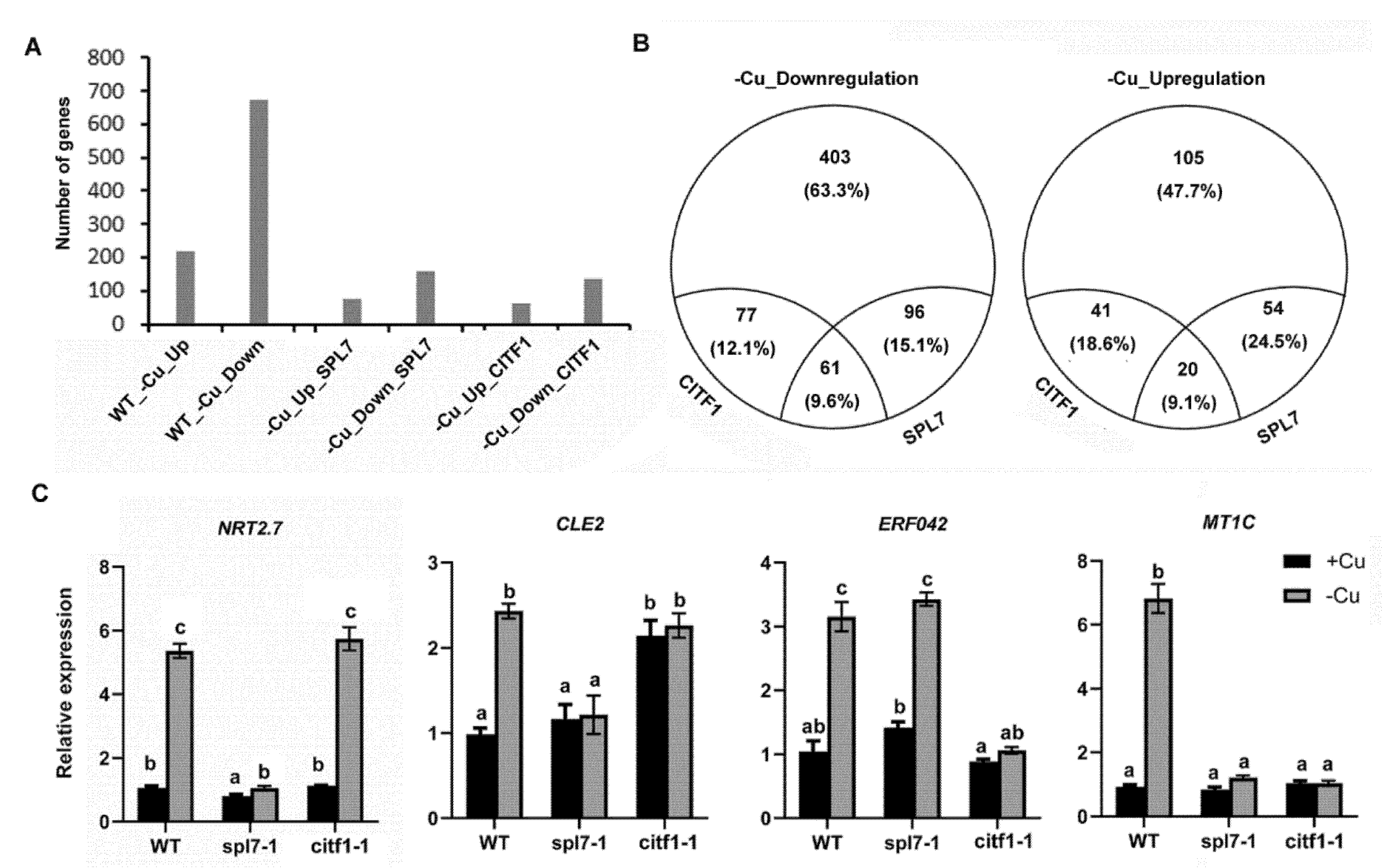
2.4. CITF1 and SPL7 Coregulate Cu-Deficiency Response
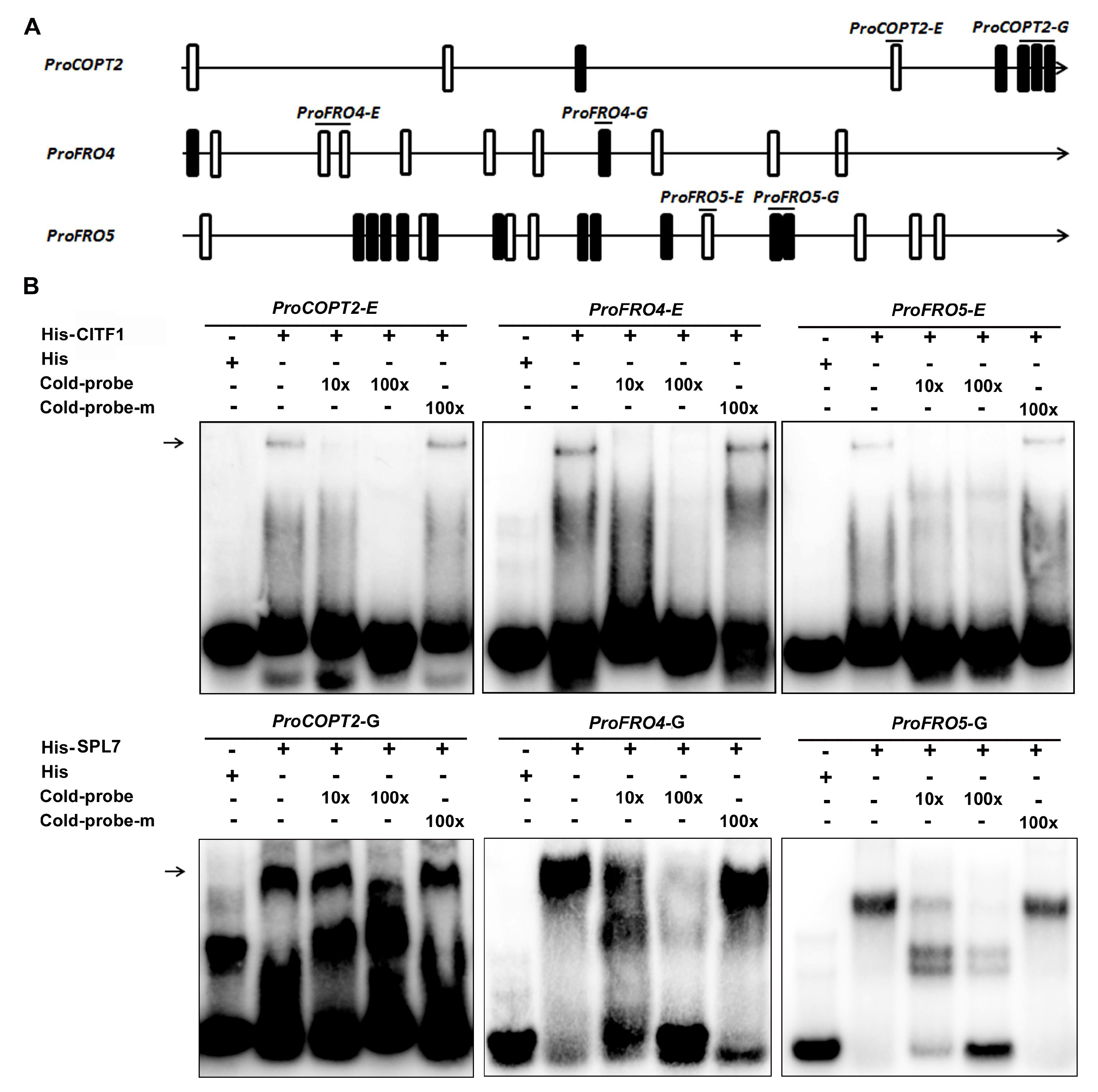
2.5. CITF1 and SPL7 Directly Bind to the Promoters of Cu Uptake Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Plant Transformation
4.3. Transient Expression Assays
4.4. EMSA
4.5. Cu Measurement
4.6. Gene Expression Analysis
4.7. High-Throughput Sequencing of mRNA, and Differential Gene Expression Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 2010, 22, 3164–3176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravet, K.; Pilon, M. Copper and iron homeostasis in plants: The challenges of oxidative stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef] [Green Version]
- Yruela, I. Copper in plants: Acquisition, transport and interactions. Funct. Plant Biol. 2009, 36, 409–430. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Ghany, S.E.; Müller-Moulé, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef] [Green Version]
- Andrés-Colás, N.; Perea-García, A.; Mayo de Andrés, S.; Garcia-Molina, A.; Dorcey, E.; Rodríguez-Navarro, S.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Comparison of global responses to mild deficiency and excess copper levels in Arabidopsis seedlings. Metallomics 2013, 5, 1234–1246. [Google Scholar] [CrossRef]
- Hänsch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef]
- Puig, S.; Peñarrubia, L. Placing metal micronutrients in context: Transport and distribution in plants. Curr. Opin. Plant Biol. 2009, 12, 299–306. [Google Scholar] [CrossRef]
- Palmer, C.M.; Guerinot, M.L. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 2009, 5, 333–340. [Google Scholar] [CrossRef] [Green Version]
- Mira, H.; Martínez, N.; Peñarrubia, L. Expression of a vegetative-storage-protein gene from Arabidopsis is regulated by copper, senescence and ozone. Planta 2002, 214, 939–946. [Google Scholar] [CrossRef]
- Sancenón, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 577–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, M.; Casero, D.; Singh, V.; Wilson, G.T.; Grande, A.; Yang, H.; Dodani, S.C.; Pellegrini, M.; Huijser, P.; Connolly, E.L.; et al. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 2012, 24, 738–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.I.; Gayomba, S.R.; Rutzke, M.A.; Craft, E.; Kochian, L.V.; Vatamaniuk, O.K. COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J. Biol. Chem. 2012, 287, 33252–33267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig, S. Function and Regulation of the Plant COPT Family of High-Affinity Copper Transport Proteins. Adv. Bot. 2014, 2014, 476917. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Molina, A.; Xing, S.; Huijser, P. A conserved KIN17 curved DNA-binding domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability. Plant Physiol. 2014, 164, 828–840. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Chia, J.C.; Sheng, H.; Jung, H.I.; Zavodna, T.O.; Zhang, L.; Huang, R.; Jiao, C.; Craft, E.J.; Fei, Z.; et al. Arabidopsis Pollen Fertility Requires the Transcription Factors CITF1 and SPL7 That Regulate Copper Delivery to Anthers and Jasmonic Acid Synthesis. Plant Cell 2017, 29, 3012–3029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depège, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef] [Green Version]
- Waters, B.M.; McInturf, S.A.; Stein, R.J. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5903–5918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, F.I.; Esch, J.J.; Hall, A.E.; Binder, B.M.; Schaller, G.E.; Bleecker, A.B. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 1999, 283, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Yoo, M.J.; Davenport, R.; Boatwright, J.L.; Koh, J.; Chen, S.; Barbazuk, W.B. Jasmonate induced alternative splicing responses in Arabidopsis. Plant Direct 2020, 4, e00245. [Google Scholar] [CrossRef] [PubMed]
- Mermod, M.; Takusagawa, M.; Kurata, T.; Kamiya, T.; Fujiwara, T.; Shikanai, T. SQUAMOSA promoter-binding protein-like 7 mediates copper deficiency response in the presence of high nitrogen in Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 835–846. [Google Scholar] [CrossRef]
- Chopin, F.; Orsel, M.; Dorbe, M.F.; Chardon, F.; Truong, H.N.; Miller, A.J.; Krapp, A.; Daniel-Vedele, F. The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 2007, 19, 1590–1602. [Google Scholar] [CrossRef] [Green Version]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [Green Version]
- Fisher, F.; Goding, C.R. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992, 11, 4103–4109. [Google Scholar] [CrossRef]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, E.P.; Guerinot, M.L. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 2004, 16, 3400–3412. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, B.; Bauer, P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cui, Y.; Liu, Y.; Fan, H.; Du, J.; Huang, Z.; Yuan, Y.; Wu, H.; Ling, H.Q. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 2013, 6, 503–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, B.; Li, M.; Feng, D.; Jin, H.; Wang, P.; Liu, J.; Xiong, F.; Wang, J.; Wang, H.B. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 2015, 27, 787–805. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, Y.; Yao, X.; Liang, G.; Yu, D. Positive Regulator of Iron Homeostasis1, OsPRI1, Facilitates Iron Homeostasis. Plant Physiol. 2017, 175, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, Y.; Pu, M.; Xu, P.; Liang, G.; Yu, D. Oryza sativa Positive Regulator of Iron Deficiency Response 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant Cell Environ. 2020, 43, 261–274. [Google Scholar] [CrossRef]
- Kim, S.A.; LaCroix, I.S.; Gerber, S.A.; Guerinot, M.L. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. USA 2019, 116, 24933–24942. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Robe, K.; Bettembourg, M.; Navarro, N.; Rofidal, V.; Santoni, V.; Gaymard, F.; Vignols, F.; Roschzttardtz, H.; Izquierdo, E.; et al. The Transcription Factor bHLH121 Interacts with bHLH105 (ILR3) and Its Closest Homologs to Regulate Iron Homeostasis in Arabidopsis. Plant Cell 2020, 32, 508–524. [Google Scholar] [CrossRef]
- Lei, R.; Li, Y.; Cai, Y.; Li, C.; Pu, M.; Lu, C.; Yang, Y.; Liang, G. bHLH121 Functions as a Direct Link that Facilitates the Activation of FIT by bHLH IVc Transcription Factors for Maintaining Fe Homeostasis in Arabidopsis. Mol. Plant 2020, 13, 634–649. [Google Scholar] [CrossRef]
- Liang, G.; Zhang, H.; Li, Y.; Pu, M.; Yang, Y.; Li, C.; Lu, C.; Xu, P.; Yu, D. Oryza sativa FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (OsFIT/OsbHLH156) interacts with OsIRO2 to regulate iron homeostasis. J. Integr. Plant Biol. 2020, 62, 668–689. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y.; Liang, G. FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis. Plant Cell Environ. 2021, 44, 1679–1691. [Google Scholar] [CrossRef]
- Perea-García, A.; Garcia-Molina, A.; Andrés-Colás, N.; Vera-Sirera, F.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol. 2013, 162, 180–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastoori Ramamurthy, R.; Xiang, Q.; Hsieh, E.J.; Liu, K.; Zhang, C.; Waters, B.M. New aspects of iron-copper crosstalk uncovered by transcriptomic characterization of Col-0 and the copper uptake mutant spl7 in Arabidopsis thaliana. Metallomics 2018, 10, 1824–1840. [Google Scholar] [CrossRef] [PubMed]
- Hangarter, R.P.; Stasinopoulos, T.C. Effect of fe-catalyzed photooxidation of EDTA on root growth in plant culture media. Plant Physiol. 1991, 96, 843–847. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Liang, G. CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 7239. https://doi.org/10.3390/ijms23137239
Cai Y, Liang G. CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(13):7239. https://doi.org/10.3390/ijms23137239
Chicago/Turabian StyleCai, Yuerong, and Gang Liang. 2022. "CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis" International Journal of Molecular Sciences 23, no. 13: 7239. https://doi.org/10.3390/ijms23137239
APA StyleCai, Y., & Liang, G. (2022). CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis. International Journal of Molecular Sciences, 23(13), 7239. https://doi.org/10.3390/ijms23137239





