Abstract
Although 99mTc is not an ideal Auger electron (AE) emitter for Targeted Radionuclide Therapy (TRT) due to its relatively low Auger electron yield, it can be considered a readily available “model” radionuclide useful to validate the design of new classes of AE-emitting radioconjugates. With this in mind, we performed a detailed study of the radiobiological effects and mechanisms of cell death induced by the dual-targeted radioconjugates 99mTc-TPP-BBN and 99mTc-AO-BBN (TPP = triphenylphosphonium; AO = acridine orange; BBN = bombesin derivative) in human prostate cancer PC3 cells. 99mTc-TPP-BBN and 99mTc-AO-BBN caused a remarkably high reduction of the survival of PC3 cells when compared with the single-targeted congener 99mTc-BBN, leading to an augmented formation of γH2AX foci and micronuclei. 99mTc-TPP-BBN also caused a reduction of the mtDNA copy number, although it enhanced the ATP production by PC3 cells. These differences can be attributed to the augmented uptake of 99mTc-TPP-BBN in the mitochondria and enhanced uptake of 99mTc-AO-BBN in the nucleus, allowing the irradiation of these radiosensitive organelles with the short path-length AEs emitted by 99mTc. In particular, the results obtained for 99mTc-TPP-BBN reinforce the relevance of targeting the mitochondria to promote stronger radiobiological effects by AE-emitting radioconjugates.
1. Introduction
In the past few years, Targeted Radionuclide Therapy (TRT) with β− emitters has shown significant progress, which has led to the recent approval of the somatostatin analog 177Lu-DOTATATE (LUTATHERA®) for clinical use in the treatment of neuroendocrine neoplasms. Additionally, the prostate-specific membrane antigen (PSMA) inhibitor 177Lu-PSMA-617 (PLUVICTOTM) has been recently approved by the FDA to treat metastatic castration-resistant prostate cancer (PCa) [1,2]. However, the use of β− emitters in the TRT of cancer has several limitations, such as the nephrotoxicity and beta radiation resistance encountered in a non-negligible number of patients. Targeted alpha therapy (TAT) can be an alternative, and promising preclinical and clinical data were reported in a few instances [3]. Unfortunately, most alpha emitters (e.g., 211At, 225Ac and 231Bi) have a low availability that limits their clinical use. Auger electron (AE) emitters can be a more feasible alternative, because this class of radionuclides has an easier availability than alpha emitters, as many of them are already commonly used in nuclear medicine imaging (e.g., 123I, 125I, 67Ga, 99mTc or 111In). Most relevantly, there is an increasing availability of new and more suitable AE emitters obtained through innovative production methods (e.g., 161Tb, 195mPt, 197Hg, 103mRh and 165Er) [4].
TRT using AE emitters presents the additional advantage of having potential in the treatment of small size cancers and metastases because of their high level of cytotoxicity, high linear energy transfer (LET) and short-range biological effectiveness. From a dosimetric point of view, the highest relative biological effectiveness (RBE) of the AE emitters results when these radionuclides are internalized in highly radiosensitive organelles, such as the cell nucleus and the mitochondria, or even when linked to the cell membrane [5,6,7]. A great challenge is to avoid the undue irradiation of nontarget tissues by γ-photons often emitted by AE-emitting radionuclides. The specific delivery of AE emitters to these radiosensitive organelles (nucleus or mitochondria) in cancer cells might enhance the radiotherapeutic effects at lower doses, thus minimizing any undesired side effects (e.g., hematological toxicity and kidney damage). Towards this goal, a variety of AE-emitting radionuclides have been studied in the past few years by several research groups, using a variety of chemical forms spanning from simple radiochemical precursors (e.g., 99mTcO4− or 201TlCl) to target-specific compounds obtained based on small molecules, monoclonal antibodies or peptides recognizing antigens, such as, receptors or enzymes overexpressed in cancer cells [4,8,9].
Historically, 125I has played a pioneer role in the study of the therapeutic potential of AE emitters, mainly due to its easy incorporation into nucleotides (e.g., 5-[(125)I]iodo-2¢- deoxyuridine (125I-UdR)), which allowed to show that there is an inverse relationship between the 125I-distance to the DNA and DNA damage and cell killing [10]. Hence, the cell’s nucleus has been considered the most important subcellular target to obtain efficient AE therapy outcomes. Interestingly, it has been reported that the radioiodinated PSMA inhibitor [125I]I-DCIBzL has a high potential for clinical translation to treat prostate cancer, which was attributed to the perinuclear localization of this radioconjugate in PSMA-positive PCa cells [11,12,13].
The influence of the proximity to DNA and accumulation in the cell nucleus on DNA damage and other radiobiological effects was thoroughly investigated for several other AE emitters, namely SPECT radiometals in clinical use such as 67Ga, 111In or 99mTc or less common but more efficient AE emitters such as 195mPt and 191Pt [14,15]. For instance, Terry’s group demonstrated how the DNA damage induced by 67Ga and 111In depends on their proximity to the DNA using a cell-free plasmid DNA assay [16,17]. Several classical DNA intercalators or groove binders have been studied as nuclear-targeting vectors for AE-emitting radionuclides, such as radiolabeled derivatives of acridine orange (AO) [18], pyrene [19], DAPI [20] and doxorubicin [21]. Our group radiolabeled several AO derivatives with the AE emitters 99mTc and 125I and demonstrated that some of the compounds homed to the nucleus, resulting in an increased number of double-stranded breaks (DSBs) [18]. In a similar way, Alberto’s group explored the effect of 99mTc-labeled pyrene conjugates on cell survival and found that these compounds exhibited a DNA-damaging effect, leading to mitotic catastrophe [22]. The need for close association with DNA molecules for AE-generated DNA lesion formation was also demonstrated by Reissig [19] using related 99mTc-labeled pyrene constructs carrying alkyne groups of variable lengths.
Other strategies to carry AE emitters to the cell nucleus rely on the use of nuclear localization sequences (NLS) and cell-penetrating peptides. Among those studies, the anti-HER2 antibody trastuzumab was functionalized with two cationic peptides (NLS5-10 or TAT1-3) to improve the nuclear uptake in HER2-positive breast cancer, but this effect was limited and nonspecific [23]. Another approach for the in vivo targeting of DNA with AE emitters takes advantage of the nuclear trafficking properties of some cell surface receptors. Of particular interest are members of the human epidermal growth factor receptor (EGFR) family that contain NLS sequences in the transmembrane region. The Reilly group extensively explored this concept for nuclear targeting with AE emitters [4]. One of the studied radioconjugates ([111In]-In-DTPA-EGF) has come the closest to clinical translation [24]. However, its evaluation in patients did not confirm the promising results obtained in the animal models [25].
Although the nucleus and DNA have been considered the primary cellular targets of radiation damage, it has been shown that internalization into cancer cells and delivery to the cell nucleus is not required for cell killing with AE-emitting radionuclides. An important breakthrough was reported by Pouget and collaborators, who showed that targeting the cell membrane can be an effective strategy for killing cancer cells with AEs [7].
The mitochondrion is another interesting extranuclear target for ionizing radiation (IR)-based cancer therapies but remains relatively understudied when compared with the cell nucleus. However, circular mitochondrial DNA, like genomic DNA, is sensitive to IR-induced damage, being less prone to undergoing repair processes. Moreover, IR can alter the mitochondrial function, induce mitochondrial oxidative stress and cause mitochondrial-induced apoptosis, either by direct or indirect effects. Most studies have focused on mitochondria-targeted radiosensitizers for external beam radiation therapy (EBRT), using triphenylphosphonium (TPP) derivatives or small peptides [26]. In contrast, studies with mitotropic therapeutic radioconjugates, namely those carrying AE emitters, are scarce and almost limited to the work that we recently reported for the dual-targeted radioconjugate 99mTc-TPP-BBN (TPP = triphenylphosphonium; BBN = bombesin derivative) [27].
Although 99mTc is not an ideal AE for TRT due to its relatively low Auger electron yield, it can be considered a readily available “model” radionuclide useful in validating the design of new classes of AE-emitting radioconjugates. Thus, with this in mind, we designed the dual-targeted 99mTc-TPP-BBN as an AE-emitting mitotropic therapeutic radioconjugate carrying: (i) a BBN sequence to target the gastrin-releasing peptide receptor (GRPR) and promote a selective uptake in GRPR (+) human prostate cancer PC3 cells and (ii) a TPP pharmacophore to provide an increased accumulation in the mitochondria of PC3 cells (Figure 1). Our initial studies showed that 99mTc-TPP-BBN caused a remarkably high reduction in the survival of human prostate cancer PC3 cells when compared with the single-targeted congener 99mTc-BBN [27]. This difference was attributed to the enhanced uptake of 99mTc-TPP-BBN in the mitochondria and the consequent irradiation of this radiosensitive organelle with the AEs emitted by 99mTc, which present a high-energy deposition and ultra-short trajectories. Previously, other researchers have also argued that the differences in the radiobiological effects induced in a rat thyroid cell line by 99mTcO4− and by the radiopharmaceuticals [99mTc]Tc-hexamethyl-propylene-aminoxime (99mTc-HMPAO) and [99mTc]Tc-hexakis-2-methoxyisobutylisonitrile (99mTc-MIBI) were due to their different subcellular distribution, namely in terms of mitochondrial accumulation [28].
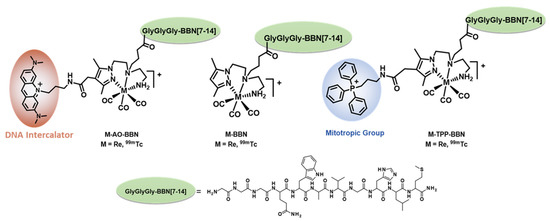
Figure 1.
99mTc-labeled BBN derivatives carrying or not organelle-specific pharmacophores evaluated in this work.
These encouraging results obtained for 99mTc-TPP-BBN prompted us to further validate the relevance of mitochondrial targeting in AE cancer therapy. Towards this goal, we performed a more detailed study of the radiobiological effects induced by 99mTc-TPP-BBN in PC3 cells in comparison with a related dual-targeted 99mTc-AO-BBN (AO = acridine orange) (Figure 1). Instead of the TPP moiety, the latter contains a DNA intercalator to promote a preferential accumulation in the cell nucleus [5], the canonical target of radiotoxic effects by AE-emitting radionuclides. In this manuscript, we describe the subcellular localization of complexes 99mTc-TPP-BBN and 99mTc-AO-BBN to assess their targeting ability towards the nucleus or the mitochondria and report on the detailed study of the radiobiological effects exerted by each dual-targeted radioconjugate in PC3 cells. This study included the evaluation of the influence of 99mTc-TPP-BBN and 99mTc-AO-BBN in the cellular proliferation rate, nuclear DNA damage and mitochondrial function in comparison with 99mTc-BBN, which does not contain any organelle-specific pharmacophore.
2. Results and Discussion
2.1. Chemistry and Radiochemistry
Previously, we showed that the length of the methylenic linker (n = 3, 5 or 8) between the AO group and the fourth position of the pyrazolyl ring in the 99mTc(I)-tricarbonyl complexes, anchored by tridentate pyrazolyldiamine chelators, has a strong influence on the DNA damage exerted by these AO-containing 99mTc complexes [18,29]. This trend was found either in vitro in plasmid DNA or in vivo in PC3 cells and was attributed to the placement of 99mTc at the shortest distance to the DNA for the complex with the shortest methylenic linker (propylenic; n = 3), upon AO intercalation into the double helix. Such a short distance enhanced the formation of DSBs by the short path-length AEs emitted by 99mTc.
Thus, we considered the same propylenic linker to attach the AO to the dual-targeted chelator AO-Pz-BBN used to obtain the M-AO-BBN (M = Re and 99mTc) complexes evaluated in this work (Figure 1). As for the BBN peptide, it contains the G3-BBN [7-14] sequence with a triglycine (G3) linker cleavable by cathepsin B, as we recently demonstrated for the Re-BBN complex [27]. The presence of the G3 linker is crucial to obtain an augmented accumulation of 99mTc or AO chromophore in the nucleus of PC3 cells for dual-targeted M(I) (M = Re and 99mTc) tricarbonyl complexes similar to M-AO-BBN. Most probably, the G3 linker is cleaved by cathepsin B inside the cells, leading to a small-sized AO-containing Re(I)/99mTc(I) fragment with a better ability to reach the cell nucleus.
The dual-targeted bifunctional chelator AO-Pz-BBN was synthesized, as we have previously reported for similar compounds [30]. As shown in Scheme 1, the synthesis of AO-Pz-BBN was done in solution by the in situ activation of the pendant carboxylic acid function of a pyrazolyl diamine prochelator (compound 4), followed by an amidation reaction with the terminal amino group from the G3-BBN [7-14] peptide. The synthesis of the respective pyrazolyldiamine Re(I) complex, Re-AO-BBN, was also performed in a solution by reacting the Re-AO-COOH complex with the G3-BBN [7-14] sequence (Scheme 1). The compounds AO-Pz-BBN and Re-AO-BBN were recovered by precipitation from the respective reaction mixtures with diethyl ether, followed by semipreparative HPLC purification of the crude solids. Their characterization was done by ESI-MS and analytical HPLC (Figure 2, Figures S1 and S2). The obtained ESI(+) mass spectra confirmed the formation of the desired conjugates being observed the expected molecular ion peaks with isotope distributions in agreement with the proposed formulations.
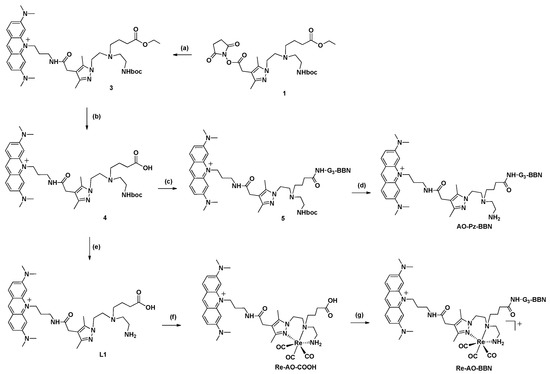
Scheme 1.
Synthesis of the BBN derivatives AO-Pz-BBN and Re-AO-BBN. (a) 3,6-bis(dimethylamino)-10-(3-(1,3-dioxoisoindolin-2-yl)-propyl)acridinium iodide (2), DIPEA, dry DMF, 3 days; (b) NaOH (aq.), 75 °C, overnight; (c) G3-BBN [7-14], DMF, DIPEA, HBTU, 4 h; (d) CH2Cl2, TFA, 1,2-Ethanedithiol (2.5 %), r.t., 2 h; (e) CH2Cl2, TFA, R.T., 5 h; (f) H2O, [Re(CO)3(H2O)3]Br, reflux, 12 h; (g) G3-BBN [7-14], DMF, DIPEA, HBTU, 3 h.
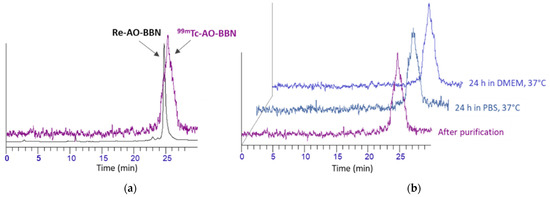
Figure 2.
RP-HPLC radiochromatograms (γ-detection) of 99mTc-AO-BBN: (a) immediately after purification and in comparison with the congener Re-AO-BBN (UV detection); (b) after incubation in PBS pH 7.4 and DMEM cell culture at 37 °C for 24 h.
The complex 99mTc-AO-BBN was synthesized by the reaction of the fac-[99mTc(CO)3(H2O)3]+ precursor with the dual-targeted chelator AO-Pz-BBN in an aqueous medium (Scheme 2), as we described previously for 99mTc-BBN and 99mTc-TPP-BBN [27]. All these 99mTc complexes were purified by HPLC prior to their evaluation in the several biological studies reported in this work to separate the respective cold chelators. The chemical identification of the new complex 99mTc-AO-BBN was done by HPLC comparison with the Re congener (Figure 2). The characterization of 99mTc-AO-BBN also involved the assessment of its lipophilicity/hydrophilicity by determination of the n-octanol/PBS partition coefficient (Po/w) using the shake flask method (Table 1) [31]. The radiopeptide 99mTc-AO-BBN has a hydrophilic nature, showing a similar log Po/w value (−0.84) with that exhibited by 99mTc-BBN (−0.75), in contrast with 99mTc-TPP-BBN, which is moderately lipophilic (log Po/w = 0.45). This trend can be accounted for by a higher hydrophilic character for the AO moiety when compared with TPP, due to the presence of several tertiary amine groups in the aromatic scaffold of the former.
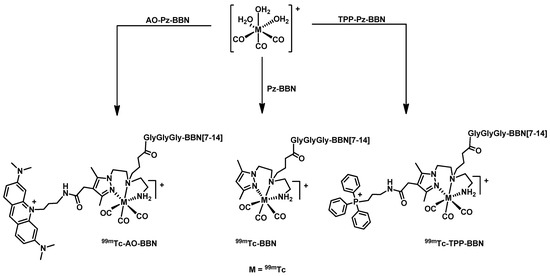
Scheme 2.
Synthesis of 99mTc complexes: 99mTc-AO-BBN, 99mTc-BBN and 99mTc-TPP-BBN. Synthesis in aqueous solution pH = 7, 30 min, 100 °C; [AO-Pz-BBN]= 10−4 M; [Pz-BBN]= 5 × 10−5 M; [TPP-Pz-BBN] = 10−4 M.

Table 1.
Radio-HPLC retention times and log Po/w values at pH 7.4 for complexes 99mTc-BBN, 99mTc-TPP-BBN and 99mTc-AO-BBN.
The in vitro stability of 99mTc-AO-BBN was evaluated in the presence of PBS pH 7.4 and cell culture medium (DMEM) upon incubation of the complex at 37 °C for 24 h, corresponding to the maximum incubation time used in the biological assays. Aliquots of the mixtures were analyzed by RP-HPLC, and representative examples of the respective radiochromatograms are shown in Figure 2. As we previously reported for 99mTc-BBN [27], the complex 99mTc-AO-BBN remained stable in the presence of PBS and the cell culture medium.
2.2. Cellular Uptake and Binding Studies
The cellular uptake of 99mTc-AO-BBN was studied in PC3 cells that overexpress GRPR, aiming to evaluate the ability of this dual-targeted complex to bind and internalize into the cells and to compare with the results previously reported by us for 99mTc-BBN and 99mTc-TPP-BBN [27]. The study was performed by incubating the PC3 cells with 99mTc-AO-BBN for different time intervals (5–180 min). For each time point, the overall radioactivity associated with the cells (cellular uptake) and that associated with the membrane, as well as that internalized in the cells, were determined by gamma-counting measurements. The results are presented in Figure 3 in comparison with those reported previously for 99mTc-BBN and 99mTc-TPP-BBN [27].
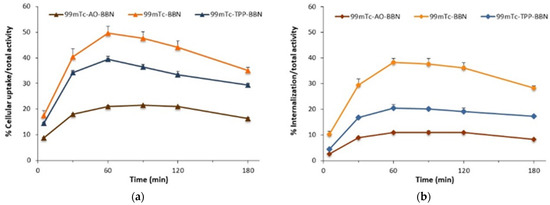
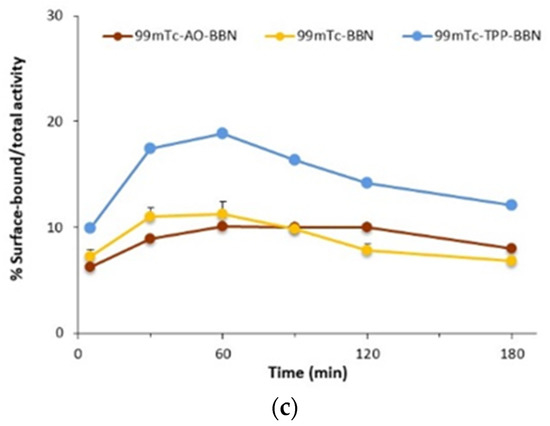
Figure 3.
Time-dependent cellular uptake (a), internalization (b) and surface-bound (c) 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN in PC3 cells at 37 °C. The results are expressed as a percentage of the total (applied) activity and calculated from independent biological replicates (mean ± SEM, n = 4).
As shown in Figure 3, the cellular uptake (Figure 3a) and internalization (Figure 3b) curves obtained for 99mTc-AO-BBN have a similar profile to those that we previously reported for 99mTc-BBN and 99mTc-TPP-BBN [27], where the maximum values (per applied radioactivity) were observed for the 1-h incubation time point. The maximum uptake and internalized values spanned the ranges 21.1–49.7% and 11.0–38.4%, respectively, a trend observed in both cases: 99mTc-BBN > 99mTc-TPP-BBN > 99mTc-AO-BBN. Furthermore, a different trend was observed for the cell membrane binding (per applied activity) with 99mTc-TPP-BBN, showing the highest values (Figure 3c). However, considering the percentage of the total cell-associated activity that attached to the membrane, the dual-targeted complexes presented a higher tendency to bind to the cell membrane when compared with the single-targeted congener. For instance, at the 60-min time point, for which a maximum uptake value was observed for all radiocomplexes, roughly 50% of the total cell-associated 99mTc-AO-BBN and 99mTc-TPP-BBN were bound to the cell membrane against only ca. 20% for 99mTc-BBN. This behavior is certainly related to the larger overall positive charge of the dual-targeted complexes and/or presence of the intrinsically positive pharmacophores, favoring electrostatic interactions with the negatively charged cell membrane.
Thereafter, we performed blockade assays with the GRPR agonist [Tyr4]-BBN to evaluate if the internalization of 99mTc-AO-BBN is mediated by the GRPR. For that, 99mTc-AO-BBN was incubated with PC3 cells for different time points (30 and 60 min) in the presence of a fixed concentration of cold [Tyr4]-BBN, as we previously described for 99mTc-BBN and 99mTc-TPP-BBN. The percentages of inhibition found for 99mTc-AO-BBN were 78% for both time points, corresponding to values that were intermediate between those determined for 99mTc-BBN (70–72%) and 99mTc-TPP-BBN (88–89%) under the same conditions (Figure S3) [27]. These data indicate that the interaction with the GRPR seems to be the dominant process in the internalization of the different radiocomplexes, including for the dual-targeted ones.
The receptor-binding affinities of both dual-targeting complexes for the human GRPR were determined by competition-binding assays at 4 °C in prostate adenocarcinoma PC3 cells against [125I-Tyr4]-BBN. The IC50 values were determined from the competitive receptor-binding curves represented in Figure S4. All BBN derivatives displaced the radioligand from the GRPR-binding sites in a dose-dependent way. The sub-nanomolar IC50 value of Re-TPP-BBN (0.38 ± 0.14 nM) was similar to the one obtained for the [Tyr4]-BBN reference peptide (0.56 ± 0.16 nM), the latest being very close to the nanomolar value reported in the literature (1.8 ± 0.2 nM) [32]. These results indicate that neither the introduction of the TPP moiety and complexation with Re reduced the binding affinity to the receptor. In contrast, a lower binding affinity was observed for the Re-AO-BBN complex (IC50 = 26.5 ± 6.8 nM) that could, in part, be responsible for the lowest level of cell internalization (Figure 3b).
2.3. Nuclear and Mitochondrial Uptake
Besides the ability of the radiocomplexes to be internalized by PC3 cells, their subcellular localization can influence the potential radiobiological effects—in particular, those induced by the AE emitted by 99mTc. For the designed dual-targeted complexes, the cell nucleus and the mitochondria were identified as their putative radiosensitive subcellular targets. Thus, the nuclear and mitochondrial uptake of the different radiocomplexes under study were assessed using the appropriate commercial subcellular fractionation kits, as we described elsewhere [27,29].
As shown in Figure 4a, the AO-containing radiopeptide 99mTc-AO-BBN presented a fast and consistent nuclear uptake that spanned a very narrow range (30.5–32.4% of the radioactivity associated to the cells), significantly higher than that found for 99mTc-BBN (8.7–16.9%) and 99mTc-TPP-BBN (9.1–16.2%) (p < 0.05 when 99mTc-AO-BBN is compared with both 99mTc-BBN and 99mTc-TPP-BBN for all time points, except when 99mTc-AO-BBN is compared with 99mTc-BBN for 120 min).
Figure 4.
Nuclear uptake (a) and mitochondrial uptake (b) of 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN in PC3 cells at 37 °C expressed as a percentage of the cell-associated activity (* p < 0.05).
The mitochondrial uptake values measured for 99mTc-AO-BBN (0.50 and 0.62% of the radioactivity associated with the cells for 60 and 120 min of incubation, respectively) are slightly lower than the corresponding values that we previously reported for 99mTc-TPP-BBN (0.54 and 0.84%) but higher than those found for 99mTc-BBN (0.24 and 0.19%) (Figure 4b) [27].
Fractionation assays can leave some organelles largely intact, namely the nucleus, while others such as the mitochondria can be damaged [5] with their contents released into other subcellular components. As a consequence, there is a cross-contamination between the mitochondrial and the cytosolic fractions based on the fractionation protocol, as we discussed elsewhere [27,33]. For these reasons, the mitochondrial uptake values measured have to be seen as relative values useful to rank the mitochondria-tropism of the radiocomplexes, which follows the order 99mTc-TPP-BBN > 99mTc-AO-BBN > 99mTc-BBN. This trend can be accounted for by the presence of the TPP pharmacophore, which has well-recognized mitochondrion tropic properties [34,35]. On the other hand, the AO group can act as a DNA intercalator with the ability to carry its compounds to the cell nucleus, as we previously showed based on fluorescence microscopy studies [30,36]. However, other authors also reported that some compounds carrying AO pharmacophores are able to accumulate in the mitochondria, with or without simultaneous localization in the nucleus [37,38,39]. The results obtained for 99mTc-AO-BBN seem to be in line with this possibility, as this radiocomplex showed some accumulation in the mitochondria, and the highest nuclear uptake that varied in the order 99mTc-AO-BBN > 99mTc-BBN > 99mTc-TPP-BBN.
2.4. Fluorescence Microscopy Studies
To evaluate the subcellular localization of the complex carrying the AO moiety and, in particular, the possibility of mitochondrial accumulation, we performed fluorescence microscopy studies with Re-AO-BBN, taking advantage of the fluorescent properties of the AO moiety. First, we performed a live cell wide field microscopy assay to assess the uptake kinetics of this complex in PC3 cells. As can be seen in Figure 5a, at 30 min, Re-AO-BBN was already accumulated in the nucleus, particularly in the nucleoli of the cells, while lower amounts were detected in the cytoplasm despite the cell-to-cell variations. This nuclear localization is in accordance with our previous studies [30,36] and with the high nuclear uptake of the radioactive congener 99mTc-AO-BBN (Figure 4a).
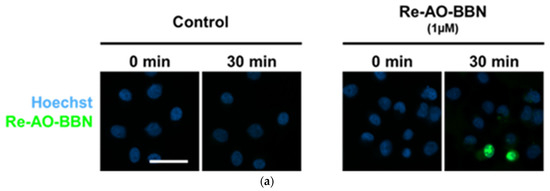
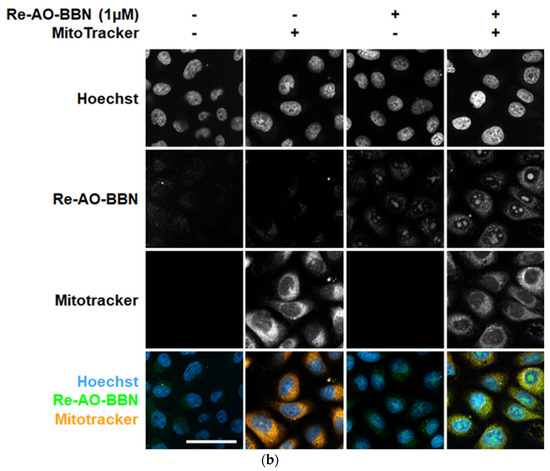
Figure 5.
The cellular uptake and localization of Re-AO-BBN in PC3 evaluated by fluorescence microscopy. (a) Live cell uptake of Re-AO-BBN (1 µM). (b) Intracellular localization of Re-AO-BBN and MitoTracker. Confocal slices show nucleolar and cytoplasmic accumulation of Re-AO-BBN. Colocalization of Re-AO-BBN and MitoTracker fluorescence is quantified at Pearson’s r = 0.31. Mitochondrial morphology is not changed by the presence of Re-AO-BBN. Scale bar = 50 µm.
We then performed the incubation of cells with Re-AO-BBN and Mitotracker Deep Red FM, a cell-permeable dye that stains the mitochondria, followed by fixation and confocal microscopy analysis (Figure 5b).
When the cells are fixed, we can observe that Re-AO-BBN shows a pronounced nuclear accumulation (as in the case of live cells) but, also, a diffuse cytoplasmic localization. Some colocalization is detected with the Mitotracker dye (Pearson’s Correlation Coefficient, PCC = 0.31), suggesting that, indeed, Re-AO-BBN is able to accumulate in the mitochondria of PC3 cells. This observation is consistent with the mitochondria uptake detected for 99mTc-AO-BBN (Figure 4b). However, unlike the extreme values, the meaning of the intermediate PCC values, as the ones obtained in our work, is often difficult to interpret. Dunn et al. outlined a consensus for PCC interpretation [40] in which colocalization was classified as high (PCC > 0.9), reasonably strong (PCC ~0.6), poor (PCC ~0.16) and not significant (PCC ~0.07). Horton et al. defined mitochondria localization mitochondria-penetrating peptides as very high (PCC ~0.5–0.6) or low (PCC ~0.2) [41], and similar values were reported for peptides with (PCC ~0.54–0.94) and without (PCC ~0.15) mitotropic features [42].
2.5. Evaluation of Radiobiological Effects
2.5.1. Clonogenic Assays
We proceeded with the evaluation of the radiocytotoxicity of 99mTc-AO-BBN in PC3 cells, using the clonogenic assay upon exposure of the cells to increasing activities of the compound, as we previously described for 99mTc-BBN and 99mTc-TPP-BBN [27]. The clonogenic assay evaluates the ability of a single cell to grow into a colony, i.e., to undergo continuous proliferation, and is often used to study the effect of IR on the survival of cancer cells, either for EBRT or for TRT with medical radionuclides [43]. The results obtained for 99mTc-AO-BBN are presented in Figure 6a in comparison with 99mTc-BBN and 99mTc-TPP-BBN [27]. For all the tested activities, the dual-targeted 99mTc-AO-BBN induced a much stronger reduction in cell survival than the single-targeted 99mTc-BBN, although weaker than that caused by the other dual-targeted complex, 99mTc-TPP-BBN.
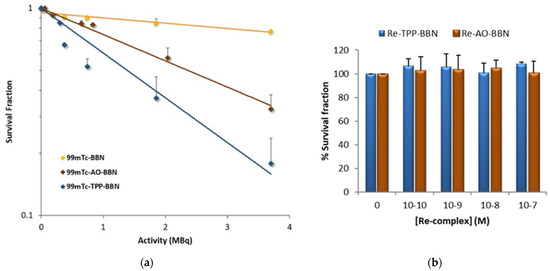
Figure 6.
Survival fraction from clonogenic assay experiments. (a) PC3 cells were incubated with 0–3.7 MBq of HPLC-purified 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN for 24 h at 37 °C. The lethal dose (LD50) was >5, 2.92 and 1.39 MBq for 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN, respectively. (b) PC3 cells were incubated with Re-AO-BBN and Re-TPP-BBN complexes (0, 10−7, 10−8, 10−9 and 10−10 M) for 24 h at 37 °C. The data correspond to the mean ± SEM of 3 replicates.
The clonogenic assays were performed with HPLC-purified radiocomplexes, in which the excess of the respective ligands was removed and did not contribute to the observed cytotoxicity. The total molar concentration of Tc (99mTc and 99Tc) involved in the clonogenic assays was of the order of 10−9 M, even for the highest tested dose of 3.7 MBq, since the 99mTcO4− used to prepare the radioconjugates was obtained by the elution of a 99Mo/99mTc generator submitted 24 h before to a preventive elution. This preventive elution was performed to minimize the accumulation of reactive oxygen species (ROS) and 99Tc inside the generator column, which could introduce some bias on the observed radiobiological effects.
We used the complexes Re-AO-BBN and Re-TPP-BBN as surrogates of the 99mTc congeners and evaluated their influence in the survival of PC3 cells in the concentration range 10−10–10−7 M. Re complexes are commonly used as surrogates for 99mTc congeners, as described above for the chemical identification of 99mTc-AO-BBN by HPLC in comparison with Re-AO-BBN, due to the physicochemical similarities of Tc and Re that form isostructural complexes with similar biological properties [44,45]. Even for the PC3 cells treated with the highest concentration of Re complexes (10−7 M), which is ca. 100-fold superior to that used in the clonogenic assays with the 99mTc complexes, there was no effect on their survival when compared with the control (Figure 6b). Therefore, the cytotoxicity observed for 99mTc-AO-BBN and 99mTc-TPP-BBN is most probably the result of biological effects due to the radiation emitted by 99mTc.
2.5.2. γ-H2AX and Micronucleus Assays
The phosphorylation of the Ser-139 residue of the histone variant H2AX, forming γ-H2AX, is an early cellular response to the induction of DSBs. The detection of this phosphorylation event has emerged as a highly specific and sensitive molecular marker for monitoring DNA damage [46]. The formation of a γ-H2AX foci can act as a molecular marker of cellular radiosensitivity in order to evaluate the efficacy of new therapeutic approaches and, also, to predict individual responses to IR even in clinical settings [47]. For this study, PC3 cells were incubated with 1.85 MBq of the different radiocomplexes under study for 24 h at 37 °C. 99mTcO4−, which is not internalized by the cells, was also evaluated under the same conditions, and basal control assays were also performed in parallel without any radioactive compounds. Several randomly selected images were collected for each condition, using a fluorescence microscope, which were then analyzed with Cellprofiler55® [48]. The results, represented as the average number of foci per nuclei, are presented in Figure 7a.
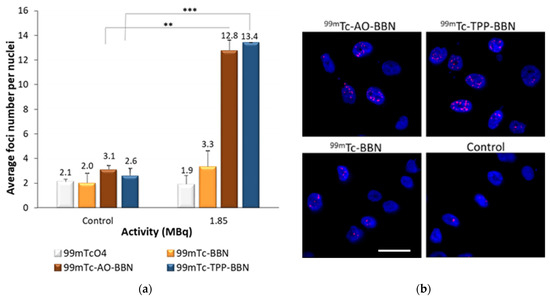
Figure 7.
DNA damage induced by HPLC-purified 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN and 99mTcO4 (1.85 MBq) in PC3 cells after 24 h of incubation at 37 °C. (a) The average number of foci per nuclei in PC3 cells, the data corresponding to the mean ± SEM of 3 replicates (** p < 0.01 and *** p < 0.005). (b) Fluorescence images of PC3 cells exposed to 1.85 MBq of 99mTc complexes. Cells were immunostained for γ-H2AX, and DAPI was used to visualize the nuclei. Scale bar = 50 µm.
PC3 cells not exposed to the radioactive compounds presented a low average number of foci, very similar among the different assays, corresponding to the basal “background” DNA damage that naturally occurs. 99mTcO4− did not cause any increase in the foci number when compared with the respective control. The 99mTc-BBN complex also did not significantly induce more DNA damage relative to the control. By contrast, the dual-targeted complexes 99mTc-AO-BBN and 99mTc-TPP-BBN led to a significant increase of the foci number in the PC3 cells, with the average number of foci increasing almost 4–5 times when compared to cells not exposed to the radiocomplexes. Altogether, these results show that the intracellular localization of the 99mTc complexes plays a crucial role in the induction of DNA damage, which is more significant for the complexes preferentially located in the nucleus and/or the mitochondria.
Another biomarker that can be used to evaluate the effects of IR in cells is the formation of micronuclei. A micronucleus (MN) is an extranuclear body that forms during anaphase, as a result of unrepaired chromosome breaks, DNA misrepair or the formation of acentric chromosome fragments, namely upon the cell exposure to IR. The formation of micronuclei in the cells treated with complexes 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN was evaluated by the cytokinesis block micronucleus assay (CBMN) [49]. The cells were exposed to the radiocomplexes for 24 h in the activity range 0.185–3.7 MBq, and the results were expressed as the MN yield corresponding to the distribution of the MN number per 1000 binucleated (BN) cells normalized to the control (Figure 8).
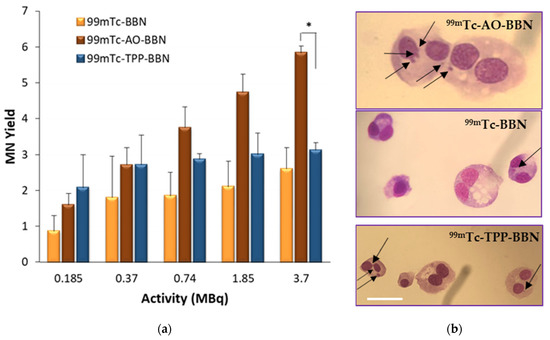
Figure 8.
Micronucleus formation in PC3 cells exposed to the HPLC-purified 99mTc-BBN, 99mTc-AO-BBN and 99mTc-TPP-BBN (0.185–3.7 MBq) after 24 h of incubation with the compounds at 37 °C. (a) Activity-dependent MN yield normalized to the control. The data correspond to the mean ± SEM of 3 replicates (* p < 0.05). (b) Binucleated cells without and with MN observed at a x43-fold magnification. Scale bar = 35 µm.
The dual-targeted complexes 99mTc-AO-BBN and 99mTc-TPP-BBN were more able to trigger the formation of the micronuclei compared with the single-targeted counterpart 99mTc-BBN, the respective MN yields appearing in the ranges 1.6–5.8, 2.1–3.1 and 0.9–2.6, respectively. For the highest tested activity (3.7 MBq), 99mTc-AO-BBN induced a significantly higher number of micronuclei when compared with 99mTc-TPP-BBN (p < 0.05). These results are contrary to those observed in the γ-H2AX assay, in which both compounds exhibited a similar behaviour. Compared with the γ-H2AX foci, MN formation can be seen as a late biological effect that is observed in cells that undergo cellular division and escape the DNA repair mechanisms. Thus, these trends seem to indicate that the DNA lesions induced by 99mTc-AO-BBN are less prone to be repaired than those induced by 99mTc-TPP-BBN.
DNA lesions, such as DSBs, can be produced by direct radiation hits like those resulting from DNA interactions with AEs emitted by 99mTc [18]. However, despite their high LET, AEs can also contribute indirectly to the formation of DNA lesions through the formation of reactive oxygen species (ROS) generated in cells during oxidative stress [7,50,51,52,53]. As discussed above, 99mTc-TPP-BBN induced the formation of a significant number of γ-H2AX foci rather comparable to that induced by 99mTc-AO-BBN, even if 99mTc-TPP-BBN was the tested complex with the lowest uptake in the nucleus of the target PC3 cells. This result seems to indicate a substantial involvement of indirect processes in the DNA damage induced by 99mTc-TPP-BBN. 99mTc-TPP-BBN has led to a lower efficiency of MN formation when compared to 99mTc-AO-BBN but was able to affect the cellular proliferation of PC3 cells to a larger extent than the latter, probably due to its mitotropic nature.
2.5.3. Effects on Mitochondrial Function
Mitochondria have a central role in cellular metabolism and are the principal player in the production of cellular energy through the formation of ATP by oxidative phosphorylation (OXPHOS) processes within the Krebs cycle and also being the main source of endogenous ROS. Consequently, mitochondria are strongly involved in the biological response of tumor cells to IR exposure, therefore considered radiosensitive organelles. In fact, similar to nuclear DNA, mitochondrial DNA (mtDNA) can also be damaged by direct radiation effects or by processes mediated by ROS. Most relevantly, mitochondrial DNA is not protected by histones and is less prone to undergo efficient repair mechanisms when compared to nuclear DNA [51,54]. As a consequence, the irradiation of mitochondria triggers radiation-induced cell signaling pathways such as apoptosis [51,55,56].
The subcellular localization studies showed that the complexes 99mTc-TPP-BBN and 99mTc-BBN have the highest and lowest mitochondrial uptakes, respectively. Most relevantly, the clonogenic assay results demonstrated that 99mTc-TPP-BBN reduces much more strongly the survival of PC3 cells than 99mTc-BBN for the same range of applied activities and in a dose-dependent manner. Thus, we selected these complexes to evaluate how the different mitochondrial uptakes influence the mitochondrial function of PC3 cells exposed to the compounds. For this study, we evaluated the effect of the exposure of the cells to 99mTc-TPP-BBN and 99mTc-BBN on the mtDNA copy number and on the ATP production.
To assess the effect on the mtDNA copy number, PC3 cells were incubated with 9.25 MBq/mL of each complex for 2 h at 37 °C. The applied activity used in this study was the same as that used in the γ-H2AX assay, being the same order of magnitude as the LD50 obtained for 99mTc-TPP-BBN in the clonogenic assay. After exposure to the compounds, the total DNA was extracted based on a commercial kit, and the mtDNA copy number was determined using a qPCR technique, as detailed in the experimental section. The obtained results, relative to the control cells incubated only with the cell medium, are presented in Figure 9a. It was found that the mtDNA copy number of the control cells and PC3 cells treated with 99mTc-BBN were not statistically different. On the contrary, 99mTc-TPP-BBN led to an approximate 50% reduction of the mtDNA content when compared to the control cells, indicating that this mitotropic complex might have the ability to induce mitochondrial dysfunction with an impact on the mtDNA content.
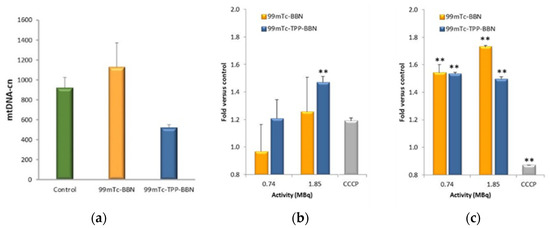
Figure 9.
Evaluation of the mitochondrial effect. (a) Mitochondrial DNA copy number (mtDNA-cn) of PC3 cells after 2 h of incubation with 9.25 MBq/mL of 99mTc-BBN and 99mTc-TPP-BBN at 37 °C. Cells only with the regular medium were used as the control. (b) Membrane integrity and (c) ATP levels for PC3 cells treated for 2 h at 37 °C with 0.74 and 1.85 MBq of 99mTc-BBN and 99mTc-TPP-BBN. Cells incubated with the assay medium (HBSS supplemented with galactose and 1% BSA) were used as a basal control, and cells incubated with CCCP (10 μM) as a positive control for mitochondrial inhibition. The data are represented as the fold versus the basal control. The data correspond to the mean ± SEM of 3 replicates (** p < 0.01 versus the control).
To the best of our knowledge, no studies have been previously reported to evaluate the effect of radionuclides internalized by tumor cells on their mtDNA copy number. In contrast with the results obtained for 99mTc-TPP-BBN, the external irradiation of cells with IR (e.g., X-ray or α-particles) often leads to an increased mtDNA copy number, eventually compensating the poor repair capability of mtDNA and acting as a self-protective mechanism of tumor cells to prevent apoptosis [56,57,58]. Interestingly, cytotoxic drugs that are able to accumulate in the mitochondria and bind to mtDNA, such as the classical chemotherapeutic drugs cisplatin and doxorubicin or a RuII–PtII bimetallic complex, inhibit mtDNA synthesis and cause its depletion [59,60,61], as we observed for 99mTc-TPP-BBN. It is possible to hypothesize that the localization of 99mTc-TPP-BBN in the mitochondria could favor the occurrence of direct mtDNA damage induced by the AEs emitted by the radionuclide.
Finally, we studied the metabolic activity of PC3 cells by measuring their ATP production following the treatment of the cells with different activities of 99mTc-TPP-BBN and 99mTc-BBN for 2 h at 37 °C. The study was performed using the commercially available kit “Mitochondrial ToxGlo™ Assay” that quantifies biomarkers associated with changes in the cell membrane integrity and cellular ATP levels. It first measures the cell membrane integrity with a fluorescent probe that cannot cross the membrane of viable cells. An increase in fluorescence of the cells exposed to a toxic agent in comparison with a control reflects the loss of membrane integrity and cytotoxicity. Next, cellular ATP production is measured in a luminescent assay. By using a galactose-containing medium (versus glucose), the oxidative phosphorylation pathway to generate ATP is favored over glycolysis, and this leads to the best conditions to evaluate the effects on mitochondrial ATP production. The combination of the two sets of data (cell death and ATP production) can give hints about the involvement of mitochondrial dysfunction or non-mitochondrial-associated cytotoxic mechanisms. This study was also conducted for cyanide-m-chlorophenylhydrazone (CCCP) [35], which is a mitochondria toxin that does not compromise the cell viability at the concentration tested. The results are presented in Figure 9.
As expected, the exposure of PC3 cells to CCCP did not compromise their viability (Figure 9b) but significantly reduced the ATP production when compared to the control cells, as the compound acts as an OXPHOS uncoupler (Figure 9c). 99mTc-TPP-BBN and 99mTc-BBN caused similar effects in the metabolism of PC3 cells, i.e., a significant increase of the ATP levels (Figure 9c).
In which concerns the membrane integrity, the effects were more pronounced for 99mTc-TPP-BBN, which caused a significant loss of membrane integrity at the highest activity tested (p < 0.01 vs control), while the effect of 99mTc-BBN was not significant (Figure 9b). This result indicates that 99mTc-TPP-BBN seems to be more effective in the induction of cell death than 99mTc-BBN, which is in accordance with the greater decrease in survival observed in the clonogenic assay (Figure 6a).
Unlike 99mTc-BBN, mitotropic 99mTc-TPP-BBN decreases the expression of the mtDNA of PC3 cells in the same way as described for different cytotoxic drugs, possibly due to localized mitochondria-targeted effects involving the AEs emitted by 99mTc. However, the decrease in mtDNA copy number did not imply a reduction of the ATP production in the mitochondria. On the contrary, there was an augmented ATP production upon exposure of the cells to the compound, as was also observed for the cells treated with 99mTc-BBN.
Other authors reported a similar effect following the external irradiation of tumor cells with X-rays, which was attributed to OXPHOS enhancement as a response to cellular stress induced by the radiation [62]. By its nature, external X-ray irradiation does not allow the selective irradiation of mitochondria, such as 99mTc-BBN due to its subcellular distribution. The enhanced ATP production observed for the PC3 cells treated with 99mTc-BBN and 99mTc-TPP-BBN certainly reflected the overall irradiation of the cells by the AEs, X-ray and γ-photons emitted by the complexes, and not only the mitochondria-targeted effects, and might be interpreted as an initial sign of apoptosis, which is an energetically demanding cell death mechanism. In summary, 99mTc-TPP-BBN reduced the mtDNA copy number of PC3 cells, but apparently, the cells still retained a high mitochondrial ATP production after exposure to the compound. To better understand this result, further studies are needed. Namely, these studies should involve the evaluation of the influence of different times of exposure to 99mTc-TPP-BBN on the relationship between the mtDNA copy number and ATP production, as well as different time points after the incubation period. Furthermore, additional studies should be extended to other tumoral cell lines, as the radiobiological effects might be cell line-dependent. This could include other human tumoral cells with different levels of GRPR expression and normal cells from the same tissue. However, these more detailed studies are out of the scope of the present contribution.
3. Materials and Methods
Unless otherwise stated, all chemicals and solvents were of reagent grade and used without further purification. Compound 1, ethyl 4-((2-(tert-butoxycarbonylamino)ethyl)(2-(4-(2-(2,5-dioxopyrrolidinyloxy)-2-oxoethyl)-3,5-dimethylpyrazol)ethyl)amino) butanoate, was prepared as described in the literature [63]. Compound 2, 3,6-bis(dimethylamino)-10-(3-(1,3-dioxoisoindolin-2-yl)-propyl)acridinium iodide (2), was prepared according to the published methods [64]. The starting material fac-[Re(H2O)3(CO)3]Br was synthesized according to the literature [65]. The G3-BBN [7-14] peptide was synthesized by a standard Fmoc strategy, as described in the literature [35]. The G3-BBN [7-14] derivatives Pz-BBN, TPP-BBN, Re-BBN and Re-TPP-BBN were prepared as previously described [27,30]. Na[99mTcO4] was eluted from a commercial 99Mo/99mTc generator using a 0.9% saline solution. The radioactive precursor fac-[99mTc(CO)3(H2O)3]+ was prepared as we described earlier [18,30].
1H-, 13C- and 31P-NMR spectra were recorded on a Bruker Avance III 400 MHz or 300 MHz spectrometers. The chemical shifts (δ) are given in ppm and were referenced to the residual solvent resonances relative to tetramethylsilane (SiMe4). Coupling constants (J) are given in Hz.
Mass spectra were acquired in an electrospray ionization/quadrupole ion trap (ESI/QITMS) Bruker HCT mass spectrometer (Bruker, Billerica, MA, USA). Samples were injected in mixtures of water:acetonitrile or water:methanol and injected at a flow rate of 150 µL·h−1.
Column chromatography was performed with silica gel 60 (Merck). HPLC (Perkin Elmer, Waltham, MA, USA) analysis of the Re and 99mTc complexes was performed on a Perkin–Elmer LC pump 200 coupled to a LC 290 tunable UV–Vis detector and to a Berthold LB-509 radiometric detector, using an analytic Macherey-Nagel C18 reversed-phase column (Nucleosil 100–10, 250 × 4 mm) with a flow rate of 1 mL/min. HPLC solvents consisted of 0.1% CF3COOH in H2O (eluent A) and 0.1% CF3COOH in acetonitrile (eluent B), and two different gradients were used: HPLC (method I): the gradient was: t = 0–25 min, 10–90% eluent B; 25–27 min,10–100% eluent B; 27–30 min, 100% eluent B; 30–31 min, 100–10% eluent B; 31–35 min, 10% eluent B. HPLC (method II): the gradient was: t = 0–3 min, 0% eluent B; 3–3.1 min, 0–25% eluent B; 3.1–9 min, 25% eluent B; 9–9.1 min, 25–34% eluent B; 9.1–25.1 min, 34–54% eluent B; 25.1–26 min, 54-0% eluent B; 26–30 min, 0% eluent B.
3.1. Synthesis of Acridine Orange Derivatives
3.1.1. Ethyl 4-((2-(4-(2-(10-4-(Propylamino)-3,6-bis(dimethylamino)acridinium)-2-oxoethyl)-3,5-dimethylpyrazol)ethyl) (2-(Tert-butoxycarbonylamino) thyl)amino) Butanoate (3)
DIPEA (0.355 mmol) was added to a solution of 3,6-bis(dimethylamino)-10-(3-(1,3-dioxoisoindolin-2-yl)-propyl)acridinium iodide (0.160 g, 0.355 mmol) in dry DMF (5 mL), and the mixture was stirred for 1 h. Then, compound 1, ethyl 4-((2-(tert-butoxycarbonylamino)ethyl)(2-(4-(2-(2,5-dioxopyrrolidinyloxy)-2-oxoethyl)-3,5-dimethylpyrazol)ethyl)amino) butanoate (0.220 g, 0.40 mmol), in DMF (5 mL) was added, and the mixture was stirred for 3 days at room temperature. Then, the solvent was evaporated, and the residue was purified with silica gel column chromatography (eluent: EtOH, 100→50/CHCl3, 0→50/NH3, 5→20. Rf (NH3, 0.1 mL/EtOH, 7 mL/CHCl3, 3 mL) = 0.14. Yield: 0.103 g; 30 %.
1H NMR (300 MHz, CD3OD): δ (ppm) 1.21 (t, 3H, CH3); 1.28 (s br, 2H, CH2); 1.41 (s, 9H, 3CH3, BOC); 1.59 (m, 2H, CH2); 2.24–2.15 (m, 6H + 4H, CH3/CH2); 2.43 (t, 3H, CH3); 2.49 (t, 3H, CH3); 2.74 (t, 3H, CH3); 3.01 (m, 2H, CH2); 3.32 (s, 12H, CH3); 3.50 (t, 2H, CH2); 4.06 (m, 4H, CH2); 4.69 (t, 2H, CH2); 6.66 (s, 2H, 2CH-Ar); 7,27 (dd, 2H, 2CH-Ar); 7.90 (d, 2H, 2CH-Ar); 8,64 (s, H, CH-Ar).
13C NMR (75 MHz, CD3OD): δ (ppm) 9.96 (CH3), 11.90 (CH3), 14.59 (CH3-Et), 23.68, 27.02, 28.80 (3CH3-BOC), 30.68, 31.91, 38.32, 29.50, 40.87 (4CH3), 46.53, 54.52, 54.81, 54.95, 61.37 (CH2-Et), 79.97 (C-BOC), 93.45 (2CH-Ar), 111.20 (C(4)pz), 115.54 (2CH-Ar), 118.50 (C-Ar), 134.42 (2CH-Ar), 139.65 (C3/5-pz), 144.08 (CH-Ar), 144.37 (C-Ar), 147.65 (C3/5-pz), 157.45 (C=O), 174.35 (C=O), 175.37 (C=O).
ESI-MS (+) m/z calcd for [C42H63N8O5]+ = 759.49; found 759.5.
HPLC (method I): Rt = 17.2 min.
3.1.2. 4-((2-(4-(2-(10-4-(Propylamino)-3,6-bis(dimethylamino)acridinium)-2-oxoethyl)-3,5-dimethylpyrazol)ethyl) (2-(Tert-butoxycarbonylamino) ethyl)amino) Butanoic Acid (4)
To a solution of 1 (0.060g; 0.068 mmol) in distilled H2O (3 mL) were added 0.5 mL of 1 M NaOH aqueous solution. This mixture was heated at 75 °C overnight. The next day, a solution of 3 M aqueous HCl was added dropwise to the mixture until reaching a final pH of about 7. The product was extracted with chloroform, dried with MgSO4 and the solvent was removed under vacuum. The compound was recovered after purification with Sep Pak OASIS MAX cartridges (Waters Co, 6 cc/150 mg 30 μm, Part No. 186000369) preconditioned according to the manufacturer’s protocol using a MeOH–H2O gradient. After washing with 5 mL of 5% NH4OH in H2O, the product was eluted with 5 mL of methanol. Yield: 32.5 mg; 56.1%.
1H NMR (300 MHz, CD3OD): δ (ppm) 1.37 (s, 9H, CH3); 1.63 (m br, 2H, CH2); 2.13–2.22 (m, 10H, 2CH3 + 2CH2); 2.53–2.63 (m, 4H, CH2); 2.89 (t, 2H, CH2, J = 6.0); 3.05 (m, 2H, CH2); 3.31 (m, 14H, CH2 + CH3); 3.49 (t, 2H, CH2, J = 7.26); 4.07 (t, 2H, CH2, J = 5.90); 7.72–4.63 (m, 2H, CH2); 6.65 (s, 2H, CH-Ar); 7.25 (dd, 2H, CH-Ar, J = 9.28, J = 1.98); 7.88 (d, 2H, CH-Ar, J = 9.34); 8,63 (s, H, CH-Ar).
13C NMR (75 MHz, CD3OD): δ (ppm) 9.96 (CH3), 11.90 (CH3), 23.84 (CH2), 27.02 (CH2), 28.77 (3CH3, BOC), 31.98 (CH2), 34.82 (CH2), 38.35 (CH2), 38.96 (CH2), 40.89 (4CH3), 46.55 (CH2), 47.53 (CH2), 48.15 (CH2), 54.76 (CH2), 54.83 (CH2), 55.44 (CH2), 80.07 (C-BOC), 93.49 (2CH-Ar), 111.19 (C(4) pz), 115.54 (2CH-Ar), 118.50 (4C-Ar), 134.41 (2CH-Ar), 139.89 (C3/C5 pz), 144.08 (2C-Ar), 144.35 (CH-Ar), 147.93 (C3/C5 pz), 157.46 (2CH-Ar), 158.28 (C=O, BOC), 174.43 (C=O), 179.72 (C=O).
ESI-MS (+) m/z calcd for [C40H59N8O5]+ = 731.46; found: 731.8.
HPLC (method I): Rt = 16.5 min.
3.1.3. 4-((2-(4-(2-(10-(4-Butylamino)-3,6-bis(dimethylamino)acridinium)-2-oxoethyl)-3,5-dimethylpyrazol)ethyl)(2-aminoethyl)amino) Butanoic Acid (L1)
A solution of 4 (12.1 mg; 0.0141 mmol) and trifluoroacetic acid (1 mL) in 1 mL of dichloromethane was reacted at room temperature over five hours. The product obtained was pure after evaporating the solvent under reduced pressure. Yield: 10.5 mg; 98%.
1H NMR (300 MHz, CD3OD): δ (ppm) 1.76–1.62 (m, 2H, CH2); 2.25–2.09 (m, 10H, 2CH3 + 2CH2); 2.78 (t, 2H, CH2, J = 6.87); 3.05–3.01 (m, 2H, CH2); 3.14–3.09 (m, 6H, CH2); 3.31 (m, 14H, CH2 + CH3); 4.48 (t, 2H, CH2, J = 7.25); 4.20 (t, 2H, CH2, J = 5.80); 4.69 (m, 2H, CH2); 6.66 (s, 2H, CH-Ar); 7.25 (dd, 2H, CH-Ar, J = 9.31, J = 1.99); 7.88 (d, 2H, CH-Ar, J = 9.36); 8,63 (s, H, CH-Ar).
13C NMR (75 MHz, CD3OD): δ (ppm) 9.59 (CH3), 11.74 (CH3), 21.94, 27.06, 30.76, 31.51, 37.60, 38.33, 40.86 (4CH3), 46.12, 46.47, 51.77, 54.00, 54.35, 93.48 (2CH-Ar), 112.02 (C(4)pz), 115.56 (2CH-Ar), 118.53 (2C-Ar), 134.44 (2CH-Ar), 140.04 (C3/5pz), 144.40 (CH-Ar), 148.11 (C3/5pz), 157.49 (C-Ar), 174.13 (C=O ), 176.82 (C=O).
ESI-MS (+) m/z calcd for [C35H51N8O3+] = 631.41; found: 632.5. calcd for [M+H]2+ = 316.7; found: 317.1.
HPLC (method II): Rt = 15.4 min.
3.1.4. Synthesis of the Re(I) Complex
fac-[Re(CO)3(k3-L1)]2+(CF3CO2Na) (Re-AO-COOH) was obtained by a reaction of [Re(H2O)3(CO)3]Br with an equimolar amount of L1 in refluxing H2O (12 h). The solvent was removed under reduced pressure, and the product was extracted with methanol to give Re-AO-COOH. Yield: 18.7 mg; 90%.
1H NMR (300 MHz, CD3OD): δ (ppm) 1.93–2.05 (m, 1H, CH2), 2.07–2.27 (m, 2H, CH2), 2.31 (s, 3H, CH3), 2.39 (s, 3H, CH3), 2.42 (m, 2H, CH2), 2.58–2.68 (m, 1H, CH2), 2.71–2.78 (m, 1H, CH), 2.87 (m, 2H, CH2), 3.09–3.19 (m, 2H, CH2), 3.32 (m, CH3, 12H), 3.42 (s, CH2, 2H), 3.52 (m, 4H, CH2), 3.68 (m, 1H, CH), 4.04 (s br, 1H, NH), 4.22 (m, 1H, CH), 4.54 (m, 1H, CH), 4.73 (t, 2H, CH2), 5.51 (q br, 1H, NH), 6.68 (s, 2H, CH-Ar); 7.26 (dd, 2H, CH-Ar, J = 9.29, J = 1.80); 7.89 (d, 2H, CH-Ar, J = 9.34); 8.64 (s, H, CH-Ar).
13C NMR (75 MHz, MeOD) δ (ppm) 194.95 (C≡O), 194.88 (C≡O), 193.82 (C≡O), 176.16 (C=O), 173.17 (C=O), 157.47 (2C-Ar), 153.97 (C3/C5 pz), 144.38 (CH-Ar), 144.10 (2C-Ar), 144.03 (C3/C5 pz), 134.43 (2CH-Ar), 118.51 (2C-Ar), 115.54 (2CH-Ar), 114.06 (C(4)pz), 93.53 (2CH-Ar), 66.94 (CH2), 62.40 (CH2), 53.69 (CH2), 48.43 (CH2 under residual MeOH, as demonstrated by the HSQC experiment), 46.42 (CH2), 43.60 (CH2), 40.91 (4CH3), 38.43 (CH2), 31.51 (CH2), 31.34 (CH2), 27.01 (CH2), 20.49 (CH2), 14.71 (CH3) and 10.37 (CH3).
ESI-MS (+) calcd for [C38H51N8O6Re + CF3COONa]2+ = 519.16; found: 519.9.
HPLC (method II): Rt = 20.4 min.
3.1.5. Synthesis of the Peptide Conjugate (AO-Pz-BBN)
Compound 4 was coupled to the N-terminal of the G3-BBN [7-14] peptide (2.5 equiv) in DMF, in the presence of O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU: 2.5 equiv) and DIPEA (4.6 equiv). The reaction was run for 4 h at room temperature, under a nitrogen atmosphere. After this time, the solvent was evaporated to dryness and the residue was treated with a mixture of TFA and dichloromethane (1:1) for 2 h at room temperature, under a nitrogen atmosphere, to remove the Boc protecting group from the terminal amine of the pyrazole-diamine framework. Thereafter, the solution was concentrated under a nitrogen stream and precipitated in Et2O, after standing in the freezer for 1 h. The resulting precipitate was washed two times with cold Et2O, dried and purified by RP-HPLC to yield the peptide conjugate AO-Pz-BBN, which was characterized by analytical HPLC and ESI-MS.
ESI-MS m/z calcd for [C84H123N24O14S+2H]3+ = 575.65; found 575.7.
HPLC (method II): Rt = 17.5 min.
3.1.6. Synthesis of the Re(I) Conjugate (Re-AO-BBN)
The complex Re-AO-COOH was coupled to the N-terminal of the G3-BBN [7-14] peptide (1.1 equiv) using HBTU (1.2 equiv) and DIPEA (4.6 equiv) in the DMF solution and reacted for 3 h at room temperature, under a nitrogen atmosphere. The resulting mixture was dried under vacuum, purified by RP-HPLC and the recovered conjugate characterized by ESI-MS.
ESI-MS m/z calcd for [C87H123N24O17SRe + H]3+ = 665.29; found [M+H]3+ 665.7; calcd for [C87H123N24O17SRe]2+ = 997.44; found [M]2+ 997.9.
HPLC ((method II): Rt = 24.4 min.
3.2. Synthesis and In Vitro Evaluation of 99mTc(I) Complexes
3.2.1. Radiolabeling Procedure
The radiocomplexes 99mTc-BBN and 99mTc-TPP-BBN were obtained by the reactions of the respective ligands with the organometallic precursor fac-[99mTc(CO)3(H2O)3]+, as we described elsewhere [27]; 99mTc-AO-BBN was obtained using the same procedure: to a nitrogen-purged vial containing the AO-Pz-BBN ligand (using concentrations as low as 10−4 M) was added the organometallic precursor fac-[99mTc(CO)3(H2O)3]+ (2–5 mCi) in aqueous solution at pH 7. The reaction mixture was then heated at 100 °C for 30 min to give the desired 99mTc complex (99mTc-AO-BBN). After cooling to room temperature, the radiochemical purity (RCP) was determined by RP-HPLC analysis. 99mTc-AO-BBN was purified by HPLC to remove the excess of the respective ligand. The purified 99mTc complex was collected in 0.1 M PBS pH 7.4 containing 0.2% of BSA to avoid adsorption to the vials and the organic solvent was removed under a nitrogen stream. The chemical identity of 99mTc-AO-BBN was established by comparison of its HPLC profile with that of the corresponding rhenium complex.
99mTc-AO-BBN: HPLC: Rt = 25.0 min (γ detection; method II); RCP > 98% (after purification by HPLC).
3.2.2. In Vitro Stability Studies
Radiochemical stability of 99mTc-AO-BBN was assessed by RP-HPLC analysis at several time points (2 h, 4 h and 24 h) following the incubation of the compound in PBS pH 7.4 and in DMEM cell culture medium at 37 °C, respectively.
3.2.3. Lipophilicity Determination
The lipophilicity of 99mTc-AO-BBN was evaluated by the “shake flask” method [31]. Briefly, 25 µL of the radiocomplex solution were added to a mixture of n-octanol (1 mL) and PBS pH = 7.4 (1 mL), previously saturated in each other by stirring the mixture. This mixture was vortexed and centrifuged (5000 rpm, 10 min, room temperature) to allow phase separation. Aliquots of 50 µL of both octanol and PBS were counted in a gamma counter. The partition coefficient (Po/w) was calculated by dividing the counts in the octanol phase by those in the buffer, and the results were expressed as Log Po/w. Log Po/w= −0.84 (99mTc-AO-BBN).
3.3. Cellular Studies
3.3.1. Cell Culture
PC3 human prostate cancer cells (ECACC 90112714, England, UK) were grown in DMEM containing GlutaMax supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin antibiotic solution (all from Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in a humidified atmosphere of 95% air and 5% CO2 at 37 °C (Heraeus, Hanau, Germany).
3.3.2. Internalization and Cellular Uptake
Time-dependent accumulation of 99mTc-AO-BBN in PC3 cells was studied, as we have previously described for 99mTc-BBN and 99mTc-TPP-BBN [27]. PC3 cells were seeded at a density of 0.125 million per well in 24-well plates and allowed to attach overnight. The cells were incubated at 37 °C for a period of 5 min to 3 h with about 7.4 kBq (0.2 µCi) of the HPLC-purified radiocompound in 0.5 mL of culture medium. After each incubation time, the unbound radiocomplex was removed and the cells washed with ice-cold DMEM medium. Cell surface-bound radiocompound was removed by two steps of acid wash (50 mM glycine-HCl/100 mM NaCl, pH 2.8) at room temperature for 4 min. The pH was neutralized with cold PBS with 0.2% BSA, and subsequently, the cells were lysed with 1 M NaOH for 10 min at 37 °C to determine the internalized radiocompound. The activity in both cell surface-bound and internalized fractions was measured using a gamma counter (LB 2111, Berthold) and is reported as a proportion to the total applied radioactivity, together representing the cellular uptake of the radiocomplex. Assays for each time point were performed in quadruplicate, and data were presented as average ± SEM of typically three independent experiments.
For assessing the specific GRPR-mediated cellular uptake and internalization of 99mTc-AO-BBN, a similar study was performed in which the radiocomplex was incubated for 0.5 h and 1 h, with or without the potent GRPR agonist [Tyr4] Bombesin (0.25 µg/0.5 mL/well).
3.3.3. Competitive Radioligand Binding Assay
The in vitro cell-binding assays were performed with the prostate carcinoma cell line PC3. Briefly, cells were seeded in 24-well plates (150,000 cells per well) and allowed to attach overnight. On the day of the experiment, cells were rinsed with ice-cold binding assay medium (RPMI 1640 medium supplemented with 1% (v/v) FBS, 25 mM HEPES and 1% (w/v) BSA). Competition was conducted by incubation of [125I-Tyr4]BBN (15,000 cpm, 0.2 mL, PerkinElmer, Inc., Boston, MA, USA) in the presence of increasing concentrations (10−13 to 10−5 M) of BBN-bearing Re-complexes in the binding buffer (0.1 mL, total volume per well 0.3 mL) for 90 min at 4 °C. The competition was interrupted by discarding supernatants and washing the cells twice with ice-cold PBS with 0.2% BSA. Cells were then lysed with 1 M NaOH treatment (2 × 0.4 mL, 10 min at 37 °C). Lysates were collected and counted for their radioactivity content in an automated γ–counter (HIDEX AMG, Hidex, Turku, Finland). IC50 values (concentration of competitor required to inhibit 50% of the radioligand binding) were calculated by nonlinear regression according to a one-site model using GraphPad PRISM 5 software (San Diego, CA, USA) and are the average of three independent experiments. Nonspecific binding was defined as the amount of activity bound in the presence of 10−5 M of the respective BBN-bearing Re-complex or [Tyr4]BBN.
3.3.4. Nuclear Uptake
PC3 cells were seeded at a density of 50,000 cells per well in a 24-well plate and allowed to attach overnight. The cells were incubated with different activities (0.185–7.4 MBq) of HPLC-purified 99mTc-BBN, 99mTc-TPP-BBN and 99mTc-AO-BBN in 0.5 mL of culture medium for 24 h at 37 °C. Incubation was terminated by washing the cells with ice-cold PBS. Cells were detached from the plates using trypsin/EDTA solution (Gibco-Invitrogen). The unbound radioactive compound was removed by centrifugation of the cell suspension, followed by washing the cellular pellet with ice-cold PBS with 0.2% BSA. The activity of cellular pellet was measured, using a gamma counter, to quantify the total cellular uptake of the radiocompound. The pellet was then resuspended in 2 mL of ice-cold cell lysis buffer (10 mM Tris, 1.5 mM MgCl2 and 140 mM NaCl) containing 0.1% of IGEPAL-ca 630 (Sigma) and incubated on ice for 10 min to disrupt the cell membrane. After the lysis, the suspension was centrifuged at 1300× g for 2 min at 4 °C, the supernatant (cytoplasm) was separated from the pellet (nuclei), and the activity in both fractions measured. The nuclear uptake was expressed in a percentage of internalized activity.
3.3.5. Mitochondrial Uptake
The mitochondrial uptake of 99mTc-AO-BBN was assessed based on compartmental fractionation studies in PC3 cells, as we previously described for 99mTc-BBN and 99mTc-TPP-BBN [27]. Adherent and confluent cells (T75 culture flask) were incubated with about 7.4 MBq (100 µCi) of the HPLC purified radiocompounds in 2 mL of culture medium for 1 h and 2 h at 37 °C and 5% CO2. The harvested cell suspension with 10–20 × 106 cells was split into 3 fractions (3 replicates, 2 mL each), centrifuged at 850× g for 2 min at 4 °C (Centrifuge 5804R, Eppendorf) and cells washed twice with cold PBS to remove the unbound radiocomplex and the activity of whole-cell fraction was measured (cellular uptake determination). To obtain the mitochondrial fraction, the cells were treated with the “Mitochondria Isolation Kit for Cultured cells” (Thermo Scientific, Walthman, MA, USA) according to the manufacturer’s protocol. Briefly, the pellet (one-third of total pellet) was resuspended with reagent A (0.8 mL/2 × 107 cells) supplemented with a cocktail of protease inhibitors (Roche, Basel, Switzerland), vortexed at medium speed for 5 s and incubated on ice for exactly 2 min. Reagent B (10 µL/2 × 107 cells) was added and the samples were vortexed at maximum speed for 5 s, incubated on ice, and vortexed every minute for 5 min. Then, reagent C (0.8 mL/2 × 107 cells), supplemented with a cocktail of protease inhibitors, was added, and the cells suspension centrifuged for 10 min at 700× g at 4 °C (Centrifuge 5417R, Eppendorf, Hamburg, Germany). The nucleus isolation steps were monitored by trypan blue staining under a phase contrast microscope. The resulting pellet corresponds to the nuclear fraction. The supernatant, transferred to a new tube, was centrifuged for 15 min at 3000× g at 4 °C to remove the lysosomal and peroxisomal contaminants. The pellet, containing the isolated mitochondria, was again treated with reagent C (0.5 mL/2 × 107 cells) and centrifuged for 5 min at 12,000× g. The activity in all fractions collected were directly counted in a dose calibrator (Berthold, LB2111, Bad Wildbad, BW, Germany).
3.3.6. Cellular Uptake by Fluorescence Microscopy
PC3 cells seeded at a density of 5000 cells per well in microscopy-grade 96 well plate (Greiner, 655090, Frickenhausen, Germany) and allowed to attach for 24 to 48 h.
Re-AO-BBN uptake was assessed through live cell wide-field fluorescence microscopy. Initially, cells were washed with PBS and stained with Hoechst 33342 (0.2 µg/mL) prepared in culture medium for 30 min. Then, the culture media was replaced by FluoroBrite™ DMEM with high D-glucose (Gibco A18967-01, Grand Island, NY, USA) containing 1 µM Re-AO-BBN or a compound-free control. Time-lapse imaging started immediately on a Leica DMI6000 B widefield fluorescence system equipped with a HC PL APO 40 × /1.10 water immersion objective, EL6000 mercury metal halide light source, Hamamatsu Orca-Flash4.0 CMOS camera (2048 × 2048 pixels, pixel size: 6.5 × 6.5 μm) and bandpass filter cubes. 37 °C and 5% CO2 were maintained throughout. Time resolution was 10 min. Images were background corrected using the flat field/dark frame algorithm.
To study the intracellular location of uptaken Re-AO-BBN, cells were washed with PBS, incubated with MitoTracker™ Deep Red FM (1:1000, Invitrogen, Bleiswijk, The Netherlands) and Re-AO-BBN (1 µM) in FluoroBrite™ DMEM with high D-glucose for 30 min, fixed with 4% paraformaldehyde (in PBS) at 4 °C for 20 min, washed with PBS, incubated with Hoechst (0.2 µg/mL) for 1 h, washed and left submerged in PBS. Cells were imaged on a Leica TCS SP8 confocal microscope, equipped with a HC Plan Apo 20 × /0.75 dry objective, 405, 488 and 638 nm laser lines, PMT (MitoTracker) and HyD (Hoechst, Re-AO-BBN) detectors. The pinhole was set to 1 airy unit and Nyquist sampling was met in xyz (132 × 132 × 685 nm). Mitochondria accumulation was assessed by the Pearson coefficient between Re-AO-BBN and MitoTracker fluorescence intensities, considering an appropriate threshold.
3.3.7. Clonogenic Assay
The clonogenic assay was performed for 99mTc-AO-BBN and for the rhenium conjugates Re-TPP-BBN and Re-AO-BBN, as we have previously described for 99mTc-BBN, 99mTc-TPP-BBN [27]. PC3 cells were seeded at a density of 50000 cells per well in a 24 well-plate and allowed to attach overnight. Cells were incubated with several activities (0–3.7 MBq) of 99mTc-AO-BBN and several concentrations (10−10–10−7 M) of the rhenium conjugates in 0.5 mL of culture medium, for 24 h at 37 °C. Immediately, after incubation, cells were seeded out in appropriate dilution to form colonies, in 2 weeks, with at least 50 cells. Colonies were fixed with methanol: glacial acetic acid (3:1) and stained with Giemsa (4%).
The Plating Efficiency (PE), ratio of the number of colonies to the number of cells seeded, and the Survival Fraction (SF), number of colonies that arise after treatment of cells, expressed in terms of PE, were obtained following the methodology described in [66], where:
3.3.8. γ-H2AX Assay and Foci Analysis
PC3 cells were seeded at a density of 10000 cells per well in an eight-well chamber slide and allowed to attach overnight. Cells were incubated with several activities (0.37, 0.74, 1.85 and 3.7 MBq) of HPLC purified 99mTc-BBN, 99mTc-TPP-BBN and 99mTc-AO-BBN in 0.3 mL of culture medium, for 24 h at 37 °C. [99mTcO4]− was used as a negative control, at the same activities, since it does not efficiently internalize into cells [21] and consequently should not target the DNA to induce radiotoxic effects. PC3 cells were washed three times with PBS, and fixed with 4% formaldehyde in PBS for 15 min. After washing with PBS, cells were permeabilized with Triton X-100 (0.5%) at room temperature for 5 min followed by two washing steps with 1% BSA in PBS. Then, cells were incubated with an anti-γ-H2AX primary antibody (mouse anti-γ-H2AX (ser139), Stressgen) at 2 μg/mL for 1 h. After being washed twice with 1% BSA in PBS, cells were incubated with a Texas Red-X-conjugated anti-mouse secondary antibody at 1mg/mL for 1 h, followed by three washing steps with PBS. Cells were finally mounted in anti-fade mounting media with DAPI (Vectashield H-1200, Vector Laboratories).
Cells were analyzed under 64x magnification. Several high-quality 2D images were randomly collected in each slide and analyzed using the pipeline Speckle Count from the freeware CellProfiler [48]. At least 200 nuclei were analyzed per experiment per dose. Statistical analysis was performed with Origin 7.5 software.
3.3.9. Micronuclei Assay
PC3 cells were seeded at a density of 50000 cells per well in a 24 well-plate and allowed to attach overnight. Cells were incubated with several activities (0, 0.185, 0.37, 0.74, 1.85, 3.7 MBq) of HPLC purified 99mTc-BBN, 99mTc-TPP-BBN and 99mTc-AO-BBN in 0.5 mL of culture medium, for 24 h at 37 °C. The cytokinesis-block micronucleus (CBMN) assay allows better precision because the data obtained are not confounded by altered cell division kinetics caused by cytotoxicity of agents tested or sub-optimal cell culture conditions [67]. Cytochalasin B (Sigma-Aldrich) in a final concentration of 2 µg/mL was added to the culture medium 44 h after initial incubation to inhibit cytokinesis, allowing cells to be binucleated. After 24 h of incubation with cytochalasin B (time needed to have the majority of cells binucleated), cells were harvested by centrifugation and submitted to a mild hypotonic shock (0.075 mol/L KCl solution) to enlarge the cellular cytoplasm. The cells were then smeared onto clean glass slides, allowed to dry, fixed with methanol:glacial acetic acid (3:1) and finally stained with 4% (w/v) Giemsa (Merck; Darmstadt, Germany).
3.3.10. Assessment of mtDNA Copy Number
Cells were grown close to confluence in T25 flasks before being incubated (or not as a control) for 2 h with 9.25 MBq/mL of 99mTc-BBN and 99mTc-TPP-BBN. After incubation, the cells were detached, washed once with PBS and resuspended in 200 µL PBS. Then, total DNA was isolated using the “DNeasy Blood and Tissue Kit” (Qiagen), according to the manufacturer’s instructions. DNA concentration and purity were determined using a μDrop™ plate in a Varioskan™ LUX microplate reader (Thermo Fisher Scientific), and the samples were stored at −20 °C until further use. Only samples with high purity were used for the qPCR. qPCR was carried out in 96-well plates using the qTower3 system (Analytik Jena GmbH). Each well contained a reaction volume of 20 μL, which was composed of: 6.25 ng of total DNA, 1 × KAPA2G Fast HotStart ReadyMix (Roche), 1 × fluorescent dye EvaGreen (Biotium) and 500 nM of the forward and reverse primers (StabVida). PCR efficiencies were calculated using the qPCRsoft 4.1 (Analytik Jena GmbH) by preparing template dilutions corresponding to 12.5, 6.25, 3.125 and 1.5625 ng of total cellular DNA. The presence of the specific amplification products was confirmed through agarose gel electrophoresis. The results were used to calculate the number of copies of mitochondrial DNA as previously described [68], assuming a ploidy of 3.3 for the PC3 cell line [69].
3.3.11. Determination of Cell Membrane Integrity and Cellular ATP Levels
PC3 cells were seeded at a density of 10,000 cells per well in a 96 well-plate and allowed to attach overnight. Cells were incubated with several activities (0.74 and 1.85 MBq) of HPLC purified 99mTc-BBN and 99mTc-TPP-BBN in 0.1 mL of HBSS supplemented with galactose and 1% BSA culture medium, for 2 h at 37 °C. Cells incubated only with this medium were used as a basal control and cells incubated with CCCP (10 μM) as a positive control for mitochondrial inhibition. After the 2 h, the Mitochondrial Toxglo assay was performed according to the manufacturer’s instructions (Promega). Assays were performed in triplicate.
3.3.12. Statistical Analysis
All data were shown as the mean values ± standard error of the mean (S.E.M.). Statistical analysis was carried out using OriginLab software. Statistical differences between treatment and control samples were assessed using two-tailed Student’s t-test. Differences among more than two groups were assessed by one-way ANOVA followed by Tukey’s test. The threshold for statistical significance was set to p = 0.05.
4. Conclusions
To our knowledge, this work represents the first contribution reporting a comparative study of the radiobiological effects induced by structurally related targeted radioconjugates, 99mTc-TPP-BBN and 99mTc-AO-BBN, showing preferential accumulation in the mitochondria or nucleus of tumor cells. These dual-targeted radioconjugates caused a remarkably high reduction of the survival of PC3 cells when compared with the single-targeted congener 99mTc-BBN. Moreover, the dual-targeted radioconjugates led to an augmented formation of γ−H2AX foci and micronuclei. 99mTc-TPP-BBN also caused the reduction of mtDNA copy number, although enhancing the ATP production by PC3 cells. These differences can be attributed to the augmented uptake of 99mTc-TPP-BBN in the mitochondria and enhanced uptake of 99mTc-AO-BBN in the nucleus, which promoted the irradiation of these radiosensitive organelles with the short path-length AEs emitted by 99mTc. In particular, the results obtained for 99mTc-TPP-BBN reinforce the relevance of targeting the mitochondria to promote stronger radiobiological effects by AE-emitting radioconjugates. Nonetheless, these effects are not necessarily localized only in the mitochondria since there is an intricate crosstalk between mitochondrial dysfunction, nuclear DNA damage and different cell death mechanisms.
In brief, our study showed that mitochondria targeting is as effective as the nuclear targeting to induce lethal radiobiological effects in tumor cells, thus deserving further attention in the design of radioconjugates for Auger therapy of cancer. These results are inspiring for us and for other researchers from this field, opening the way to apply our approach to more efficient Auger emitting radiometals (e.g., 111In, 161Tb or 191Pt), using other clinically relevant targeting vectors (e.g., somatostatin analogs or PSMA inhibitors) in combination with alternative cleavable linkers and/or mitotropic moieties.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23137238/s1.
Author Contributions
Conceptualization, A.P. and P.R.; chemical synthesis, C.F., F.S. and E.P.; radiosynthesis, C.F. and E.P.; cellular studies, P.R., J.F.G., C.I.G.P., F.M. and A.B.; microscopy studies, F.M and H.M.B.; writing—original draft preparation, A.P. and P.R. and writing—review and editing, all authors. Funding acquisition, A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundação para a Ciência e Tecnologia, Portugal (projects UID/Multi/04349/2019,PTDC/QUI-NUC/30147/2017, PTDC/BTM-TEC/29256/2017 and PTDC/MED-QUI/1554/2020 to C2TN and UIDB/04046/2020 and UIDP/04046/2020 to BioISI). C. Pinto acknowledges FCT for the PhD fellowship 2020.07119.BD. Microscopy studies were performed at the Faculty of Sciences of the University of Lisboa Microscopy Facility, a node of the Portuguese Platform of BioImaging, reference PPBI-POCI-01-0145-FEDER-022122.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ruigrok, E.A.; van Weerden, W.M.; Nonnekens, J.; de Jong, M. The Future of PSMA-Targeted Radionuclide Therapy: An Overview of Recent Preclinical Research. Pharmaceutics 2019, 11, 560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasul, S.; Hacker, M.; Kretschmer-Chott, E.; Leisser, A.; Grubmüller, B.; Kramer, G.; Shariat, S.; Wadsak, W.; Mitterhauser, M.; Hartenbach, M.; et al. Clinical outcome of standardized (177)Lu-PSMA-617 therapy in metastatic prostate cancer patients receiving 7400 MBq every 4 weeks. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 713–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathekge, M.; Bruchertseifer, F.; Knoesen, O.; Reyneke, F.; Lawal, I.; Lengana, T.; Davis, C.; Mahapane, J.; Corbett, C.; Vorster, M.; et al. (225)Ac-PSMA-617 in chemotherapy-naive patients with advanced prostate cancer: A pilot study. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 129–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, A.; Facca, V.J.; Cai, Z.; Reilly, R.M. Auger electrons for cancer therapy—A review. EJNMMI Radiopharm. Chem. 2019, 4, 27. [Google Scholar] [CrossRef] [Green Version]
- Bavelaar, B.M.; Lee, B.Q.; Gill, M.R.; Falzone, N.; Vallis, K.A. Subcellular Targeting of Theranostic Radionuclides. Front. Pharmacol. 2018, 9, 996. [Google Scholar] [CrossRef]
- Rosenkranz, A.A.; Slastnikova, T.A.; Georgiev, G.P.; Zalutsky, M.R.; Sobolev, A.S. Delivery systems exploiting natural cell transport processes of macromolecules for intracellular targeting of Auger electron emitters. Nucl. Med. Biol. 2020, 80–81, 45–56. [Google Scholar] [CrossRef]
- Paillas, S.; Ladjohounlou, R.; Lozza, C.; Pichard, A.; Boudousq, V.; Jarlier, M.; Sevestre, S.; Le Blay, M.; Deshayes, E.; Sosabowski, J.; et al. Localized Irradiation of Cell Membrane by Auger Electrons Is Cytotoxic Through Oxidative Stress-Mediated Nontargeted Effects. Antioxid. Redox Signal. 2016, 25, 467–484. [Google Scholar] [CrossRef] [Green Version]
- Costa, I.M.; Siksek, N.; Volpe, A.; Man, F.; Osytek, K.M.; Verger, E.; Schettino, G.; Fruhwirth, G.O.; Terry, S.Y. Relationship of In Vitro Toxicity of Technetium-99m to Subcellular Localisation and Absorbed Dose. Int. J. Mol. Sci. 2021, 22, 13466. [Google Scholar] [CrossRef]
- Rigby, A.; Blower, J.E.; Blower, P.J.; Terry, S.Y.; Abbate, V. Targeted Auger electron-emitter therapy: Radiochemical approaches for thallium-201 radiopharmaceuticals. Nucl. Med. Biol. 2021, 98–99, 1–7. [Google Scholar] [CrossRef]
- Kortylewicz, Z.P.; Kimura, Y.; Inoue, K.; Mack, E.; Baranowska-Kortylewicz, J. Radiolabeled cyclosaligenyl monophosphates of 5-iodo-2′-deoxyuridine, 5-iodo-3′-fluoro-2′,3′-dideoxyuridine, and 3′-fluorothymidine for molecular radiotherapy of cancer: Synthesis and biological evaluation. J. Med. Chem. 2012, 55, 2649–2671. [Google Scholar] [CrossRef] [Green Version]
- Kiess, A.P.; Minn, I.; Chen, Y.; Hobbs, R.; Sgouros, G.; Mease, R.C.; Pullambhatla, M.; Shen, C.J.; Foss, C.A.; Pomper, M.G. Auger Radiopharmaceutical Therapy Targeting Prostate-Specific Membrane Antigen. J. Nucl. Med. 2015, 56, 1401–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.J.; Minn, I.; Hobbs, R.F.; Chen, Y.; Josefsson, A.; Brummet, M.; Banerjee, S.R.; Brayton, C.F.; Mease, R.C.; Pomper, M.G.; et al. Auger radiopharmaceutical therapy targeting prostate-specific membrane antigen in a micrometastatic model of prostate cancer. Theranostics 2020, 10, 2888–2896. [Google Scholar] [CrossRef] [PubMed]
- Matthias, J.; Engelhardt, J.; Schäfer, M.; Bauder-Wüst, U.; Meyer, P.T.; Haberkorn, U.; Eder, M.; Kopka, K.; Hell, S.W.; Eder, A.C. Cytoplasmic Localization of Prostate-Specific Membrane Antigen Inhibitors May Confer Advantages for Targeted Cancer Therapies. Cancer Res. 2021, 81, 2234–2245. [Google Scholar] [CrossRef] [PubMed]
- Howell, R.W.; Kassis, A.I.; Adelstein, S.J.; Rao, D.V.; Wright, H.A.; Hamm, R.N.; Turner, J.E.; Sastry, K.S. Radiotoxicity of Platinum-195m-Labeled trans-Platinum (II) in Mammalian Cells. Radiat. Res. 1994, 140, 55–62. [Google Scholar] [CrossRef]
- Obata, H.; Tsuji, A.B.; Kumata, K.; Sudo, H.; Minegishi, K.; Nagatsu, K.; Takakura, H.; Ogawa, M.; Kurimasa, A.; Zhang, M.R. Development of Novel 191Pt-Labeled Hoechst33258: 191Pt Is More Suitable than 111In for Targeting DNA. J. Med. Chem. 2022, 65, 5690–5700. [Google Scholar] [CrossRef]
- bin Othman, M.F.; Mitry, N.R.; Lewington, V.J.; Blower, P.J.; Terry, S.Y. Re-assessing gallium-67 as a therapeutic radionuclide. Nucl. Med. Biol. 2017, 46, 12–18. [Google Scholar] [CrossRef] [Green Version]
- Verger, E.; Cheng, J.; De Santis, V.; Iafrate, M.; Jackson, J.A.; Imberti, C.; Fruhwirth, G.O.; Blower, P.J.; Ma, M.T.; Burnham, D.R.; et al. Validation of the plasmid study to relate DNA damaging effects of radionuclides to those from external beam radiotherapy. Nucl. Med. Biol. 2021, 100–101, 36–43. [Google Scholar] [CrossRef]
- Pereira, E.; Do Quental, L.; Palma, E.; Oliveira, M.C.; Mendes, F.; Raposinho, P.; Correia, I.; Lavrado, J.; Di Maria, S.; Belchior, A.; et al. Evaluation of Acridine Orange Derivatives as DNA-Targeted Radiopharmaceuticals for Auger Therapy: Influence of the Radionuclide and Distance to DNA. Sci. Rep. 2017, 7, 42544. [Google Scholar] [CrossRef]
- Reissig, F.; Mamat, C.; Steinbach, J.; Pietzsch, H.J.; Freudenberg, R.; Navarro-Retamal, C.; Caballero, J.; Kotzerke, J.; Wunderlich, G. Direct and Auger Electron-Induced, Single- and Double-Strand Breaks on Plasmid DNA Caused by 99mTc-Labeled Pyrene Derivatives and the Effect of Bonding Distance. PLoS ONE 2016, 11, e0161973. [Google Scholar] [CrossRef]
- Kotzerke, J.; Punzet, R.; Runge, R.; Ferl, S.; Oehme, L.; Wunderlich, G.; Freudenberg, R. 99mTc-labeled HYNIC-DAPI causes plasmid DNA damage with high efficiency. PLoS ONE 2014, 9, e104653. [Google Scholar] [CrossRef]
- Imstepf, S.; Pierroz, V.; Raposinho, P.; Bauwens, M.; Felber, M.; Fox, T.; Shapiro, A.B.; Freudenberg, R.; Fernandes, C.; Gama, S.; et al. Nuclear Targeting with an Auger Electron Emitter Potentiates the Action of a Widely Used Antineoplastic Drug. Bioconjugate Chem. 2015, 26, 2397–2407. [Google Scholar] [CrossRef] [PubMed]
- Häfliger, P.; Agorastos, N.; Spingler, B.; Georgiev, O.; Viola, G.; Alberto, R. Induction of DNA-Double-Strand Breaks by Auger Electrons from 99mTc Complexes with DNA-Binding Ligands. ChemBioChem 2005, 6, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Idrissou, M.B.; Pichard, A.; Tee, B.; Kibedi, T.; Poty, S.; Pouget, J.P. Targeted Radionuclide Therapy Using Auger Electron Emitters: The Quest for the Right Vector and the Right Radionuclide. Pharmaceutics 2021, 13, 980. [Google Scholar] [CrossRef] [PubMed]
- Vallis, K.A.; Reilly, R.M.; Scollard, D.; Merante, P.; Brade, A.; Velauthapillai, S.; Caldwell, C.; Chan, I.; Freeman, M.; Lockwood, G.; et al. Phase I trial to evaluate the tumor and normal tissue uptake, radiation dosimetry and safety of (111)In-DTPA-human epidermal growth factor in patients with metastatic EGFR-positive breast cancer. Am. J. Nucl. Med. Mol. Imaging 2014, 4, 181–192. [Google Scholar]
- Rosenkranz, A.A.; Slastnikova, T.A.; Karmakova, T.A.; Vorontsova, M.S.; Morozova, N.B.; Petriev, V.M.; Abrosimov, A.S.; Khramtsov, Y.V.; Lupanova, T.N.; Ulasov, A.V.; et al. Antitumor Activity of Auger Electron Emitter (111)In Delivered by Modular Nanotransporter for Treatment of Bladder Cancer with EGFR Overexpression. Front. Pharmacol. 2018, 9, 1331. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Gao, P.; Wu, T.; Pan, W.; Li, N.; Tang, B. Organelle-localized radiosensitizers. Chem. Commun. 2020, 56, 10621–10630. [Google Scholar] [CrossRef]
- Figueiredo, D.; Fernandes, C.; Silva, F.; Palma, E.; Raposinho, P.; Belchior, A.; Vaz, P.; Paulo, A. Synthesis and Biological Evaluation of 99mTc(I) Tricarbonyl Complexes Dual-Targeted at Tumoral Mitochondria. Molecules 2021, 26, 441. [Google Scholar] [CrossRef]
- Maucksch, U.; Runge, R.; Wunderlich, G.; Freudenberg, R.; Naumann, A.; Kotzerke, J. Comparison of the radiotoxicity of the 99mTc-labeled compounds 99mTc-pertechnetate, 99mTc-HMPAO and 99mTc-MIBI. Int. J. Radiat. Biol. 2016, 92, 698–706. [Google Scholar] [CrossRef]
- Belchior, A.; Di Maria, S.; Fernandes, C.; Vaz, P.; Paulo, A.; Raposinho, P. Radiobiological and dosimetric assessment of DNA-intercalated 99mTc-complexes bearing acridine orange derivatives. EJNMMI Res. 2020, 10, 79. [Google Scholar] [CrossRef]
- Esteves, T.; Marques, F.; Paulo, A.; Rino, J.; Nanda, P.; Smith, C.J.; Santos, I. Nuclear targeting with cell-specific multifunctional tricarbonyl M(I) (M is Re, 99mTc) complexes: Synthesis, characterization, and cell studies. JBIC J. Biol. Inorg. Chem. 2011, 16, 1141–1153. [Google Scholar] [CrossRef]
- Troutner, D.E.; Volkert, W.A.; Hoffman, T.J.; Holmes, R.A. A neutral lipophilic complex of 99mTc with a multidentate amine oxime. Int. J. Appl. Radiat. Isot. 1984, 35, 467–470. [Google Scholar] [CrossRef]
- Marsouvanidis, P.J.; Nock, B.A.; Hajjaj, B.; Fehrentz, J.A.; Brunel, L.; M’kadmi, C.; van der Graaf, L.; Krenning, E.P.; Maina, T.; Martinez, J.; et al. Gastrin releasing peptide receptor-directed radioligands based on a bombesin antagonist: Synthesis, (111)in-labeling, and preclinical profile. J. Med. Chem. 2013, 56, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Mendes, F.; Gano, L.; Fernandes, C.; Paulo, A.; Santos, I. Studies of the myocardial uptake and excretion mechanisms of a novel 99mTc heart perfusion agent. Nucl. Med. Biol. 2012, 39, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef] [PubMed]
- Moura, C.; Mendes, F.; Gano, L.; Santos, I.; Paulo, A. Mono- and dicationic Re(I)/(99m)Tc(I) tricarbonyl complexes for the targeting of energized mitochondria. J. Inorg. Biochem. 2013, 123, 34–45. [Google Scholar] [CrossRef]
- Esteves, T.; Xavier, C.; Gama, S.; Mendes, F.; Raposinho, P.D.; Marques, F.; Paulo, A.; Pessoa, J.C.; Rino, J.; Viola, G.; et al. Tricarbonyl M(I) (M = Re, 99mTc) complexes bearing acridine fluorophores: Synthesis, characterization, DNA interaction studies and nuclear targeting. Org. Biomol. Chem. 2010, 8, 4104–4116. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Kim, Y.S.; Shi, J.; Jacobson, O.; Chen, X.; Liu, S. Evaluation of 64Cu-Labeled Acridinium Cation: A PET Radiotracer Targeting Tumor Mitochondria. Bioconjugate Chem. 2011, 22, 700–708. [Google Scholar] [CrossRef] [Green Version]
- Ngen, E.J.; Rajaputra, P.; You, Y. Evaluation of delocalized lipophilic cationic dyes as delivery vehicles for photosensitizers to mitochondria. Bioorg. Med. Chem. 2009, 17, 6631–6640. [Google Scholar] [CrossRef]
- Rodriguez, M.E.; Azizuddin, K.; Zhang, P.; Chiu, S.M.; Lam, M.; Kenney, M.E.; Burda, C.; Oleinick, N.L. Targeting of mitochondria by 10-N-alkyl acridine orange analogues: Role of alkyl chain length in determining cellular uptake and localization. Mitochondrion 2008, 8, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol.-Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [Green Version]
- Horton, K.L.; Stewart, K.M.; Fonseca, S.B.; Guo, Q.; Kelley, S.O. Mitochondria-Penetrating Peptides. Chem. Biol. 2008, 15, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Q.; Dong, X.; Yang, F.; Zhou, S.; Xiang, M.; Lou, L.; Yao, S.Q.; Gao, L. Engineered Cell-Penetrating Peptides for Mitochondrion-Targeted Drug Delivery in Cancer Therapy. Chem.—A Eur. J. 2021, 27, 14721–14729. [Google Scholar] [CrossRef] [PubMed]
- Buch, K.; Peters, T.; Nawroth, T.; Sänger, M.; Schmidberger, H.; Langguth, P. Determination of cell survival after irradiation via clonogenic assay versus multiple MTT Assay—A comparative study. Radiat. Oncol. 2012, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abram, U.; Alberto, R. Technetium and rhenium: Coordination chemistry and nuclear medical applications. J. Braz. Chem. Soc. 2006, 17, 1486–1500. [Google Scholar] [CrossRef]
- Morais, G.R.; Paulo, A.; Santos, I. Organometallic Complexes for SPECT Imaging and/or Radionuclide Therapy. Organometallics 2012, 31, 5693–5714. [Google Scholar] [CrossRef]
- Mah, L.J.; El-Osta, A.; Karagiannis, T.C. γH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Lobachevsky, P.; Leong, T.; Daly, P.; Smith, J.; Best, N.; Tomaszewski, J.; Thompson, E.R.; Li, N.; Campbell, I.G.; Martin, R.F.; et al. Compromized DNA repair as a basis for identification of cancer radiotherapy patients with extreme radiosensitivity. Cancer Lett. 2016, 383, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [Green Version]
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007, 2, 1084–1104. [Google Scholar] [CrossRef] [Green Version]
- Srinivas, U.S.; Tan, B.W.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Pouget, J.P.; Lozza, C.; Deshayes, E.; Boudousq, V.; Navarro-Teulon, I. Introduction to radiobiology of targeted radionuclide therapy. Front. Med. 2015, 2, 12. [Google Scholar] [CrossRef] [Green Version]
- Pouget, J.P.; Santoro, L.; Piron, B.; Paillas, S.; Ladjohounlou, R.; Pichard, A.; Poty, S.; Deshayes, E.; Constanzo, J.; Bardiès, M. From the target cell theory to a more integrated view of radiobiology in Targeted radionuclide therapy: The Montpellier group’s experience. Nucl. Med. Biol. 2022, 104–105, 53–64. [Google Scholar] [CrossRef]
- Reuvers, T.G.A.; Kanaar, R.; Nonnekens, J. DNA Damage-Inducing Anticancer Therapies: From Global to Precision Damage. Cancers 2020, 12, 2098. [Google Scholar] [CrossRef]
- Averbeck, D.; Rodriguez-Lafrasse, C. Role of Mitochondria in Radiation Responses: Epigenetic, Metabolic, and Signaling Impacts. Int. J. Mol. Sci. 2021, 22, 11047. [Google Scholar] [CrossRef]
- Richardson, R.B.; Harper, M.E. Mitochondrial stress controls the radiosensitivity of the oxygen effect: Implications for radiotherapy. Oncotarget 2016, 7, 21469–21483. [Google Scholar] [CrossRef] [Green Version]
- Kobashigawa, S.; Kashino, G.; Suzuki, K.; Yamashita, S.; Mori, H. Ionizing radiation-induced cell death is partly caused by increase of mitochondrial reactive oxygen species in normal human fibroblast cells. Radiat. Res. 2015, 183, 455–464. [Google Scholar] [CrossRef]
- Zhou, X.; Li, N.; Wang, Y.; Wang, Y.; Zhang, X.; Zhang, H. Effects of X-irradiation on mitochondrial DNA damage and its supercoiling formation change. Mitochondrion 2011, 11, 886–892. [Google Scholar] [CrossRef]
- Zhang, S.; Wen, G.; Huang, S.X.; Wang, J.; Tong, J.; Hei, T.K. Mitochondrial alteration in malignantly transformed human small airway epithelial cells induced by α-particles. Int. J. Cancer 2013, 132, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Zaidieh, T.; Smith, J.R.; Ball, K.E.; An, Q. ROS as a novel indicator to predict anticancer drug efficacy. BMC Cancer 2019, 19, 1224. [Google Scholar] [CrossRef] [Green Version]
- Ashley, N.; Poulton, J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene 2009, 28, 3880–3891. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Zhang, D.Y.; Zhang, H.; Cao, J.J.; Tan, C.P.; Ji, L.N.; Mao, Z.W. Photodamaging of Mitochondrial DNA to Overcome Cisplatin Resistance by a Ru(II) -Pt(II) Bimetallic Complex. Chemistry 2018, 24, 18971–18980. [Google Scholar] [CrossRef]
- Deguchi, T.; Hosoya, K.; Kim, S.; Murase, Y.; Yamamoto, K.; Bo, T.; Yasui, H.; Inanami, O.; Okumura, M. Metformin preferentially enhances the radio-sensitivity of cancer stem-like cells with highly mitochondrial respiration ability in HMPOS. Mol. Ther. Oncolytics 2021, 22, 143–151. [Google Scholar] [CrossRef]
- Moura, C.; Esteves, T.; Gano, L.; Raposinho, P.D.; Paulo, A.; Santos, I. Synthesis, characterization and biological evaluation of tricarbonyl M(i) (M = Re, 99mTc) complexes functionalized with melanin-binding pharmacophores. New J. Chem. 2010, 34, 2564–2578. [Google Scholar] [CrossRef]
- Vultos, F.; Fernandes, C.; Mendes, F.; Marques, F.; Correia, J.D.; Santos, I.; Gano, L. A Multifunctional Radiotheranostic Agent for Dual Targeting of Breast Cancer Cells. ChemMedChem 2017, 12, 1103–1107. [Google Scholar] [CrossRef]
- Lazarova, N.; James, S.; Babich, J.; Zubieta, J. A convenient synthesis, chemical characterization and reactivity of [Re(CO)3(H2O)3]Br: The crystal and molecular structure of [Re(CO)3(CH3CN)2Br]. Inorg. Chem. Commun. 2004, 7, 1023–1026. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Fenech, M. The in vitro micronucleus technique. Mutat. Res. 2000, 455, 81–95. [Google Scholar] [CrossRef]
- Dannenmann, B.; Lehle, S.; Lorscheid, S.; Huber, S.M.; Essmann, F.; Schulze-Osthoff, K. Simultaneous quantification of DNA damage and mitochondrial copy number by long-run DNA-damage quantification (LORD-Q). Oncotarget 2017, 8, 112417–112425. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Q.; Kyle, E.; Patel, S.; Housseau, F.; Hakim, F.; Lieberman, R.; Pins, M.; Blagosklonny, M.V.; Bergan, R.C. Prostate cancer chemoprevention agents exhibit selective activity against early stage prostate cancer cells. Prostate Cancer Prostatic Dis. 2001, 4, 81–91. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).