MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme
Abstract
1. Introduction
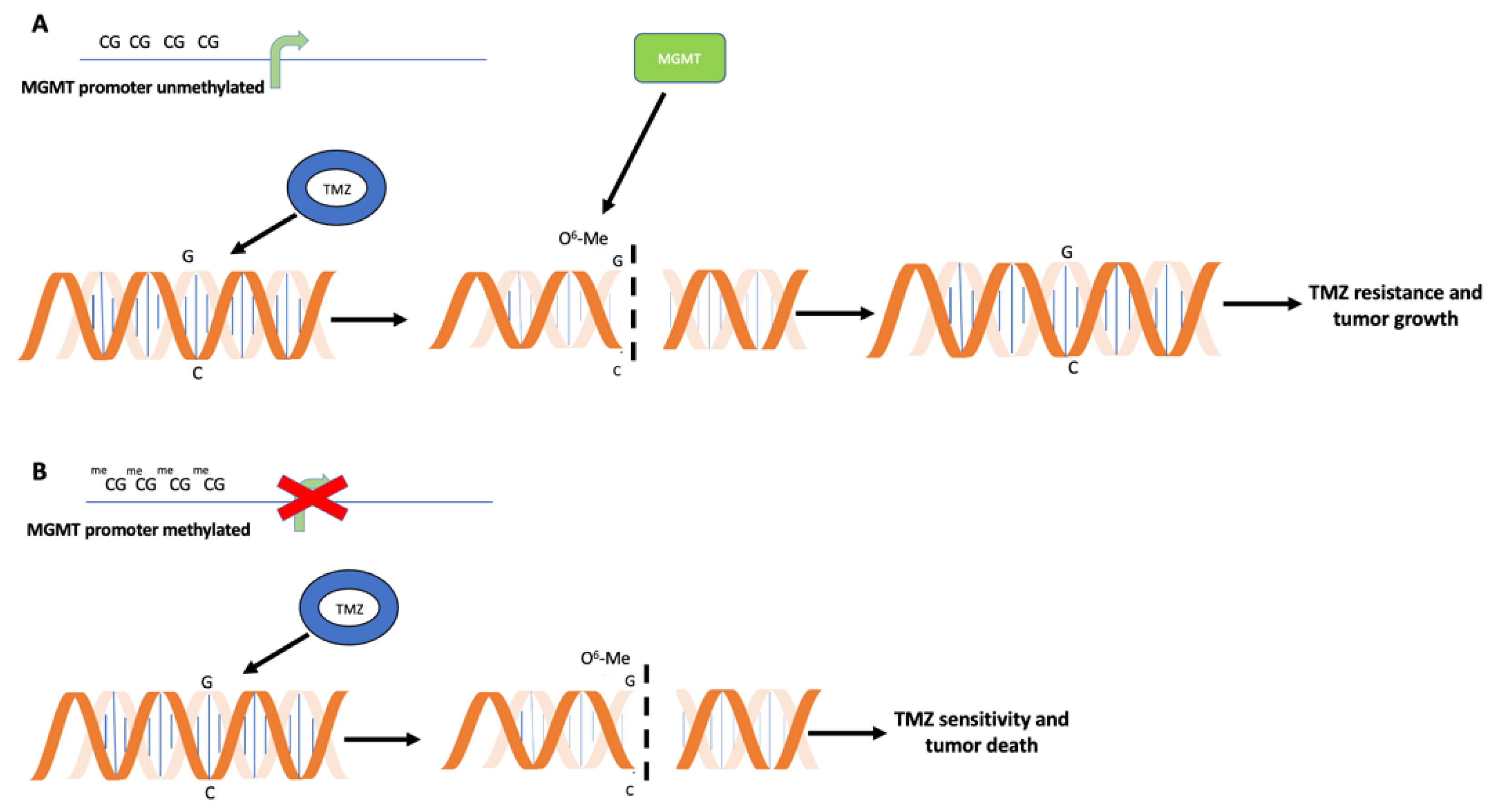
2. Relevance of MGMT Methylation Assessment for Glioblastoma Clinical Management
3. Techniques for DNA Methylation Testing in Glioblastomas
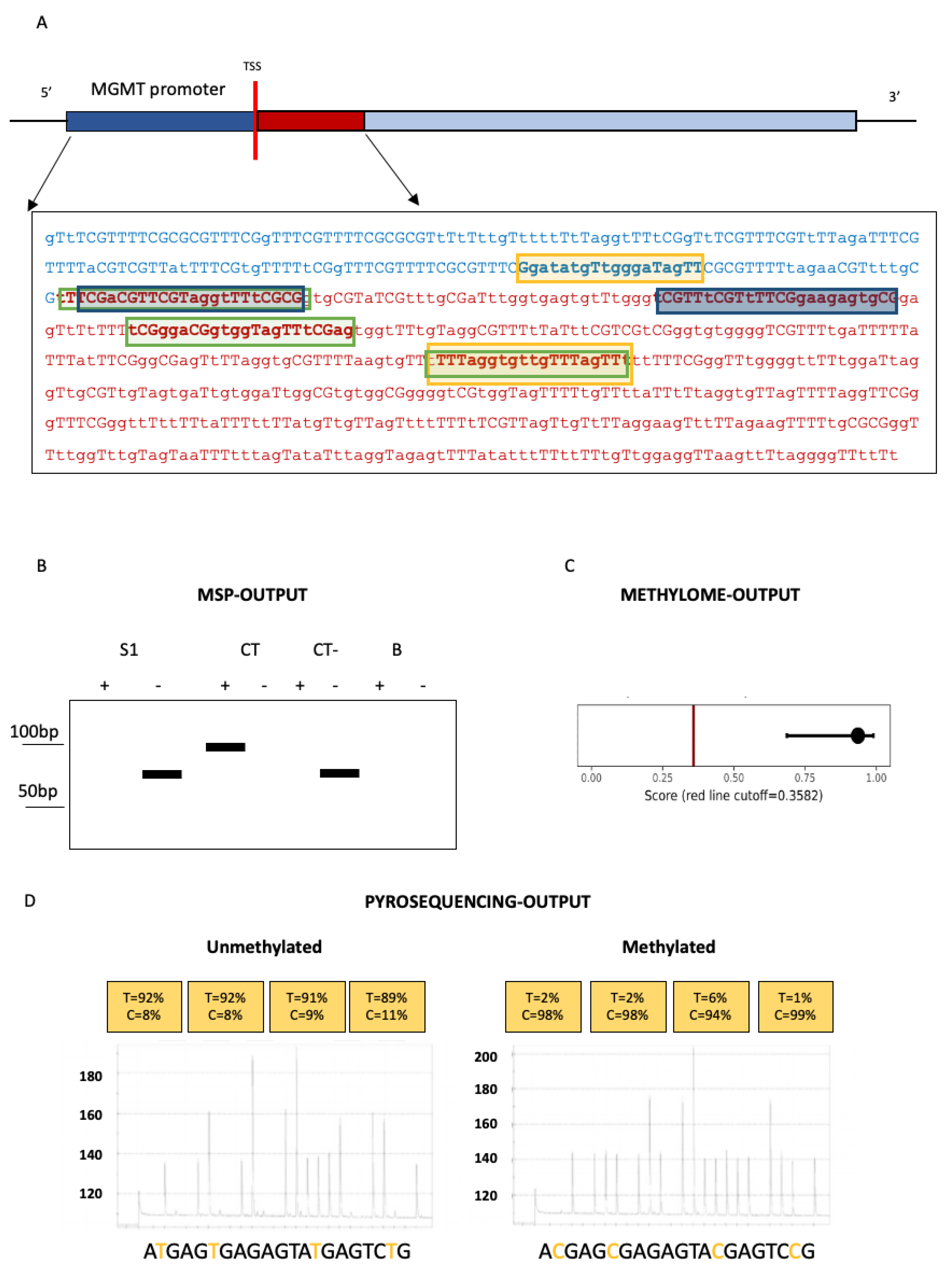
3.1. Techniques for MGMT Methylation Assessment in Glioblastomas
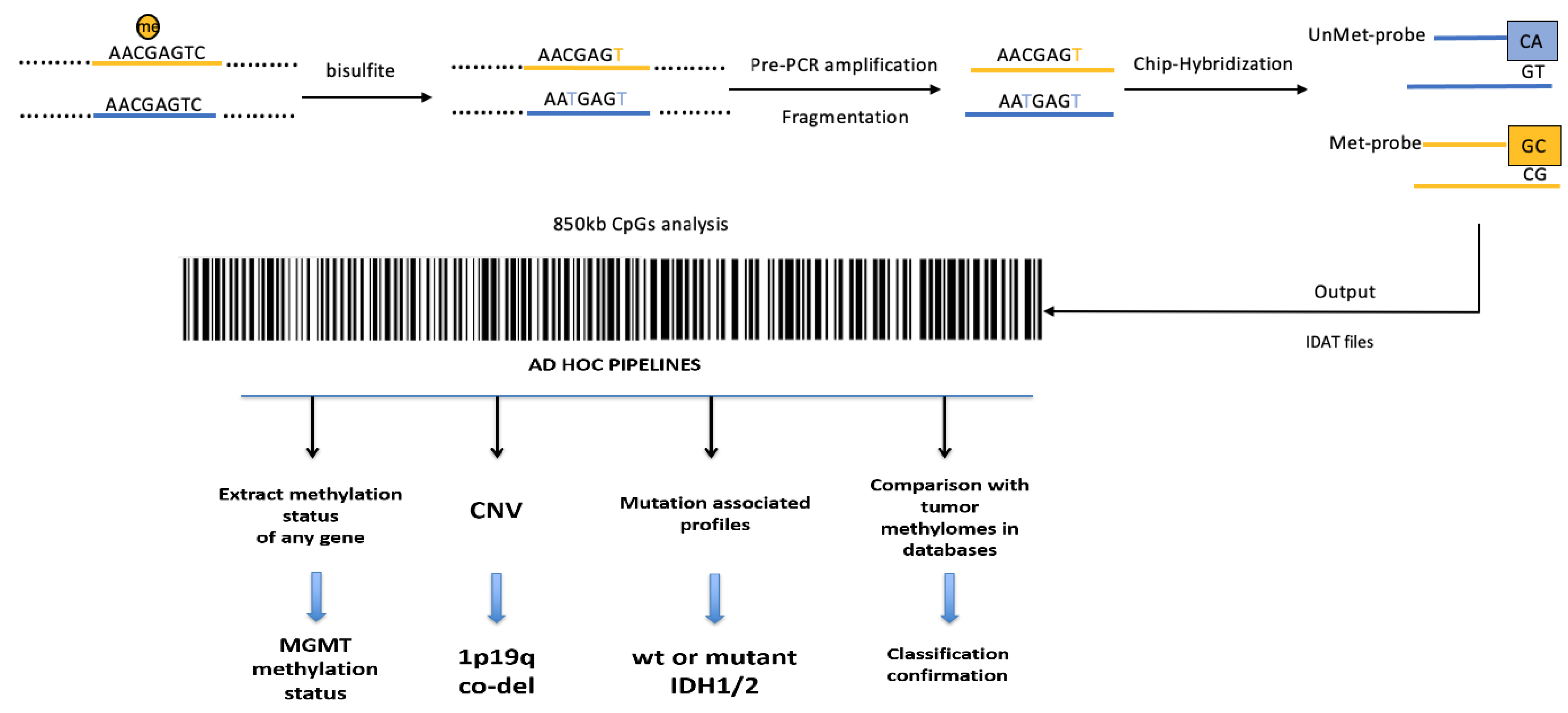
3.2. Whole-Genome Methylation Profiling (Methylome) of Glioblastomas
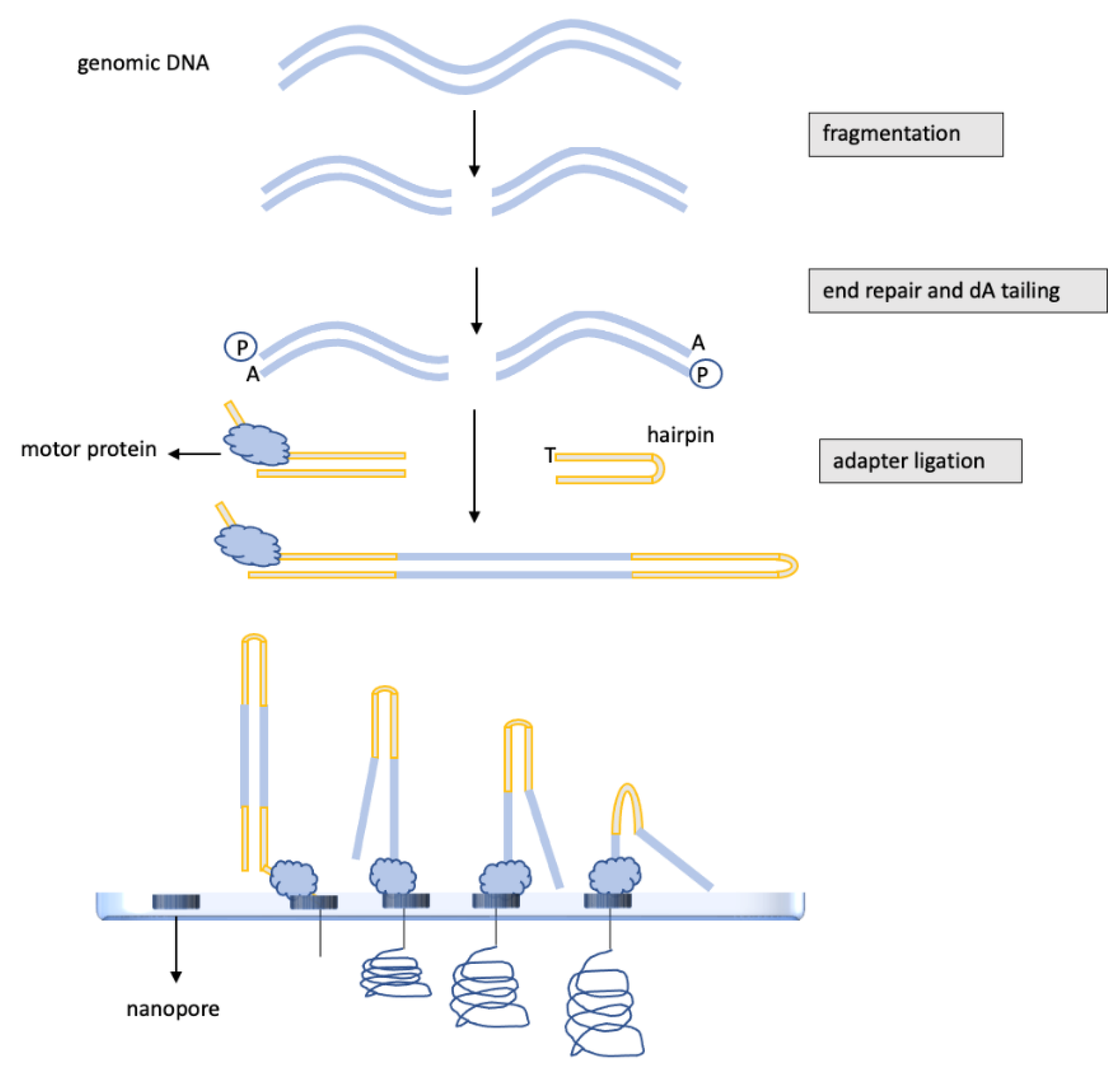
3.3. DNA Methylation Analysis of Glioblastomas by Nanopore, “Third-Generation” Sequencing
4. Glioblastoma DNA Methylation Assessment in Liquid Biopsies
5. IDH1/2 Mutations and the Methylator Phenotype of Glioblastomas: New Therapeutic Targets
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Vanyushin, B.F.; Mazin, A.L.; Vasilyev, V.K.; Belozersky, A.N. The content of 5-methylcytosine in animal DNA: The species and tissue specificity. Biochim. Biophys. Acta 1973, 299, 397–403. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Luo, H.; Krawczyk, M.; Wei, W.; Wang, W.; Wang, J.; Flagg, K.; Hou, J.; Zhang, H.; Yi, S.; et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Friedman, H.S.; Kerby, T.; Calvert, H. Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 2000, 6, 2585–2597. [Google Scholar] [PubMed]
- Bae, S.H.; Park, M.J.; Lee, M.M.; Kim, T.M.; Lee, S.H.; Cho, S.Y.; Kim, Y.H.; Kim, Y.J.; Park, C.K.; Kim, C.Y. Toxicity profile of temozolomide in the treatment of 300 malignant glioma patients in Korea. J. Korean Med. Sci. 2014, 29, 980–984. [Google Scholar] [CrossRef]
- Kuter, D.J. Treatment of chemotherapy-induced thrombocytopenia in patients with non-hematologic malignancies. Haematologica 2022, 107, 1243–1263. [Google Scholar] [CrossRef]
- Lam, F.C.; Morton, S.W.; Wyckoff, J.; Vu Han, T.L.; Hwang, M.K.; Maffa, A.; Balkanska-Sinclair, E.; Yaffe, M.B.; Floyd, S.R.; Hammond, P.T. Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 2018, 9, 1991. [Google Scholar] [CrossRef]
- Latancia, M.T.; Moreno, N.C.; Leandro, G.S.; Ribeiro, V.C.; de Souza, I.; Vieira, W.K.M.; Bastos, A.U.; Hoch, N.C.; Rocha, C.R.R.; Menck, C.F.M. DNA polymerase eta protects human cells against DNA damage induced by the tumor chemotherapeutic temozolomide. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2022, 878, 503498. [Google Scholar] [CrossRef]
- Han, S.; Meng, L.; Jiang, Y.; Cheng, W.; Tie, X.; Xia, J.; Wu, A. Lithium enhances the antitumour effect of temozolomide against TP53 wild-type glioblastoma cells via NFAT1/FasL signalling. Br. J. Cancer 2017, 116, 1302–1311. [Google Scholar] [CrossRef]
- Qian, X.C.; Brent, T.P. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997, 57, 3672–3677. [Google Scholar]
- Watts, G.S.; Pieper, R.O.; Costello, J.F.; Peng, Y.M.; Dalton, W.S.; Futscher, B.W. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol. Cell. Biol. 1997, 17, 5612–5619. [Google Scholar] [CrossRef] [PubMed]
- Danam, R.P.; Qian, X.C.; Howell, S.R.; Brent, T.P. Methylation of selected CpGs in the human O6-methylguanine-DNA methyltransferase promoter region as a marker of gene silencing. Mol. Carcinog. 1999, 24, 85–89. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Godard, S.; Dietrich, P.Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; de Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of 0-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Paz, M.F.; Yaya-Tur, R.; Rojas-Marcos, I.; Reynes, G.; Pollan, M.; Aguirre-Cruz, L.; García-Lopez, J.L.; Piquer, J.; Safont, M.J.; Balaña, C.; et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin. Cancer Res. 2004, 10, 4933–4938. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Brandes, A.A.; Tosoni, A.; Franceschi, E.; Sotti, G.; Frezza, G.; Amistà, P.; Morandi, L.; Spagnolli, F.; Ermani, M. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: Correlation with MGMT promoter methylation status. J. Clin. Oncol. 2009, 27, 1275–1279. [Google Scholar] [CrossRef]
- Perry, J.R.; Laperriere, N.; O’Callaghan, C.J.; Brandes, A.A.; Menten, J.; Phillips, C.; Fay, M.; Nishikawa, R.; Cairncross, J.G.; Roa, W.; et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N. Engl. J. Med. 2017, 376, 1027–1037. [Google Scholar] [CrossRef]
- Brandes, A.A.; Franceschi, E.; Tosoni, A.; Blatt, V.; Pession, A.; Tallini, G.; Bertorelle, R.; Bartolini, S.; Calbucci, F.; Andreoli, A.; et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J. Clin. Oncol. 2008, 26, 2192–2197. [Google Scholar] [CrossRef]
- Taal, W.; Brandsma, D.; de Bruin, H.G.; Bromberg, J.E.; Swaak-Kragten, A.T.; Sillevis Smitt, P.A.E.; van Es, C.A.; van den Bent, M.J. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer 2008, 113, 405–410. [Google Scholar] [CrossRef]
- Brandsma, D.; Stalpers, L.; Taal, W.; Sminia, P.; van den Bent, M.J. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008, 9, 453–461. [Google Scholar] [CrossRef]
- Philteos, J.; Karmur, B.S.; Mansouri, A. MGMT testing in glioblastomas: Pitfalls and opportunities. Am. J. Clin. Oncol. 2019, 42, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Graff, J.R.; Myöhänen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed]
- Della Monica, R.; Cuomo, M.; Visconti, R.; di Mauro, A.; Buonaiuto, M.; Costabile, D.; De Riso, G.; Di Risi, T.; Guadagno, E.; Tafuto, R.; et al. Evaluation of MGMT gene methylation in neuroendocrine neoplasms. Oncol. Res. 2022, 28, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797. [Google Scholar] [PubMed]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef] [PubMed]
- Vlassenbroeck, I.; Califice, S.; Diserens, A.C.; Migliavacca, E.; Straub, J.; Di Stefano, I.; Moreau, F.; Hamou, M.F.; Renard, I.; Delorenzi, M.; et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J. Mol. Diagn. 2008, 10, 332–337. [Google Scholar] [CrossRef]
- Wojdacz, T.K.; Dobrovic, A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007, 35, e41. [Google Scholar] [CrossRef]
- Switzeny, O.J.; Christmann, M.; Renovanz, M.; Giese, A.; Sommer, C.; Kaina, B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin. Epigenetics 2016, 8, 49. [Google Scholar] [CrossRef]
- Estival, A.; Sanz, C.; Ramirez, J.L.; Velarde, J.M.; Domenech, M.; Carrato, C.; de Las Peñas, R.; Gil-Gil, M.; Sepúlveda, J.; Armengol, R.; et al. Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 2019, 9, 11125. [Google Scholar] [CrossRef]
- Karayan-Tapon, L.; Quillien, V.; Guilhot, J.; Wager, M.; Fromont, G.; Saikali, S.; Etcheverry, A.; Hamlat, A.; Loussouarn, D.; Campion, L.; et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J. Neuro-Oncology 2010, 97, 311–322. [Google Scholar] [CrossRef]
- Christians, A.; Hartmann, C.; Benner, A.; Meyer, J.; von Deimling, A.; Weller, M.; Wick, W.; Weiler, M. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE 2012, 7, e33449. [Google Scholar] [CrossRef] [PubMed]
- Quillien, V.; Lavenu, A.; Karayan-Tapon, L.; Carpentier, C.; Labussière, M.; Lesimple, T.; Chinot, O.; Wager, M.; Honnorat, J.; Saikali, S.; et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012, 118, 4201–4211. [Google Scholar] [CrossRef] [PubMed]
- Jeuken, J.W.; Cornelissen, S.J.; Vriezen, M.; Dekkers, M.M.; Errami, A.; Sijben, A.; Boots-Sprenger, S.H.; Wesseling, P. MS-MLPA: An attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab. Investig. 2007, 87, 1055–1065. [Google Scholar] [CrossRef]
- Shah, N.; Lin, B.; Sibenaller, Z.; Ryken, T.; Lee, H.; Yoon, J.G.; Rostad, S.; Foltz, G. Comprehensive analysis of MGMT promoter methylation: Correlation with MGMT expression and clinical response in GBM. PLoS ONE 2011, 6, e16146. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, S.; Mama, N.; Ladib, M.; Karmeni, N.; Haddaji Mastouri, M.; Chourabi, M.; Mokni, M.; Tlili, K.; Krifa, H.; Yacoubi, M.T.; et al. MGMT methylation assessment in glioblastoma: MS-MLPA versus human methylation 450K beadchip array and immunohistochemistry. Clin. Transl. Oncol. 2016, 18, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Mancarella, D.; Plass, C. Epigenetic signatures in cancer: Proper controls, current challenges and the potential for clinical translation. Genome Med. 2021, 13, 23. [Google Scholar] [CrossRef]
- Orjuela, S.; Menigatti, M.; Schraml, P.; Kambakamba, P.; Robinson, M.D.; Marra, G. The DNA hypermethylation phenotype of colorectal cancer liver metastases resembles that of the primary colorectal cancers. BMC Cancer 2020, 20, 290. [Google Scholar] [CrossRef]
- Modhukur, V.; Sharma, S.; Mondal, M.; Lawarde, A.; Kask, K.; Sharma, R.; Salumets, A. Machine learning approaches to classify primary and metastatic cancers using tissue of origin-based DNA methylation profiles. Cancers 2021, 13, 3768. [Google Scholar] [CrossRef]
- Smit, K.N.; Boers, R.; Vaarwater, J.; Boers, J.; Brands, T.; Mensink, H.; Verdijk, R.M.; van Ijcken, W.F.J.; Gribnau, J.; de Klein, A.; et al. Genome-wide aberrant methylation in primary metastatic UM and their matched metastases. Sci. Rep. 2022, 12, 42. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. A methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Epigenetic Lab—CEINGE, Advanced Biotechnologies. Available online: https://www.ceinge.unina.it/en/epigenetic-lab (accessed on 9 May 2022).
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019, 178, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.; Ferreyra Vega, S.; Kling, T.; Bontell, T.O.; Jakola, A.S.; Carén, H. Intratumor DNA methylation heterogeneity in glioblastoma: Implications for DNA methylation-based classification. Neuro-Oncology 2019, 21, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.R.; Hudson, A.L.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Cheng, Z.J.; Vafaee, F.; Chen, J.; et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 2016, 6, 22477. [Google Scholar] [CrossRef]
- Akgül, S.; Patch, A.M.; D’Souza, R.C.J.; Mukhopadhyay, P.; Nones, K.; Kempe, S.; Kazakoff, S.H.; Jeffree, R.L.; Stringer, B.W.; Pearson, J.V.; et al. Intratumoural heterogeneity underlies distinct therapy responses and treatment resistance in glioblastoma. Cancers 2019, 11, 190. [Google Scholar] [CrossRef]
- Ramasamy, D.; Deva Magendhra Rao, A.K.; Rajkumar, T.; Mani, S. Non-CpG methylation—A key epigenetic modification in cancer. Brief. Funct. Genom. 2021, 20, 304–311. [Google Scholar] [CrossRef]
- Brigliadori, G.; Goffredo, G.; Bartolini, D.; Tosatto, L.; Gurrieri, L.; Mercatali, L.; Ibrahim, T. Influence of intratumor heterogeneity on the predictivity of MGMT gene promoter methylation status in glioblastoma. Front. Oncol. 2020, 10, 533000. [Google Scholar] [CrossRef]
- Chai, R.-C.; Liu, Y.-Q.; Zhang, K.-N.; Wu, F.; Zhao, Z.; Wang, K.-Y.; Jiang, T.; Wang, Y.-Z. A novel analytical model of MGMT methylation pyrosequencing offers improved predictive performance in patients with gliomas. Mod. Pathol. 2019, 32, 4–15. [Google Scholar] [CrossRef]
- Pezone, A.; Tramontano, A.; Scala, G.; Cuomo, M.; Riccio, P.; De Nicola, S.; Porcellini, A.; Chiariotti, L.; Avvedimento, E.V. Tracing and tracking epiallele families in complex DNA populations. NAR Genom. Bioinform. 2020, 2, lqaa096. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Patel, A.; Dogan, H.; Payne, A.; Krause, E.; Sievers, P.; Schoebe, N.; Schrimpf, D.; Blume, C.; Stichel, D.; Holmes, N.; et al. Rapid-CNS2: Rapid comprehensive adaptive nanopore-sequencing of CNS tumors, a proof-of-concept study. Acta Neuropathol. 2022, 143, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020, 38, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Wongsurawat, T.; Jenjaroenpun, P.; De Loose, A.; Alkam, D.; Ussery, D.W.; Nookaew, I.; Leung, Y.K.; Ho, S.M.; Day, J.D.; Rodriguez, A. A novel Cas9-targeted long-read assay for simultaneous detection of IDH1/2 mutations and clinically relevant MGMT methylation in fresh biopsies of diffuse glioma. Acta Neuropathol. Commun. 2020, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Djirackor, L.; Halldorsson, S.; Niehusmann, P.; Leske, H.; Capper, D.; Kuschel, L.P.; Pahnke, J.; Due-Tønnessen, B.J.; Langmoen, I.A.; Sandberg, C.J.; et al. Intraoperative DNA methylation classification of brain tumors impacts neurosurgical strategy. Neuro-Oncology Adv. 2021, 3, vdab149. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Lavon, I.; Refael, M.; Zelikovitch, B.; Shalom, E.; Siegal, T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro-Oncology 2012, 12, 173–180. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, W.; Wang, Y.; Guo, Y.; Cong, Z.; Du, F. MGMT promoter methylation in serum and cerebrospinal fluid as a tumor-specific biomarker of glioma. Biomed. Rep. 2015, 3, 543–548. [Google Scholar] [CrossRef][Green Version]
- Birkó, Z.; Nagy, B.; Klekner, Á.; Virga, J. Novel molecular markers in glioblastoma-benefits of liquid biopsy. Int. J. Mol. Sci. 2020, 21, 7522. [Google Scholar] [CrossRef]
- Chen, J.; Huan, W.; Zuo, H.; Zhao, L.; Huang, C.; Liu, X.; Hou, S.; Qi, J.; Shi, W. Alu methylation serves as a biomarker for non-invasive diagnosis of glioma. Oncotarget 2016, 7, 26099–26106. [Google Scholar] [CrossRef]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Maire, C.L.; Fuh, M.M.; Kaulich, K.; Fita, K.D.; Stevic, I.; Heiland, D.H.; Welsh, J.A.; Jones, J.C.; Görgens, A.; Ricklefs, T.; et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro-Oncology 2021, 23, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Persico, P.; Lorenzi, E.; Losurdo, A.; Dipasquale, A.; Di Muzio, A.; Navarria, P.; Pessina, F.; Politi, L.S.; Lombardi, G.; Santoro, A.; et al. Precision oncology in lower-grade gliomas: Promises and pitfalls of therapeutic strategies targeting IDH-mutations. Cancers 2022, 14, 1125. [Google Scholar] [CrossRef]
- Youssef, G.; Miller, J.J. Lower grade gliomas. Curr. Neurol. Neurosci. Rep. 2020, 20, 21. [Google Scholar] [CrossRef]
- Malta, T.M.; de Souza, C.F.; Sabedot, T.S.; Silva, T.C.; Mosella, M.S.; Kalkanis, S.N.; Snyder, J.; Castro, A.V.B.; Noushmehr, H. Glioma CpG island methylator phenotype (G-CIMP): Biological and clinical implications. Neuro-Oncology 2018, 20, 608–620. [Google Scholar] [CrossRef]
- Ichimura, K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012, 29, 131–139. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Borodovsky, A.; Salmasi, V.; Turcan, S.; Fabius, A.W.; Baia, G.S.; Eberhart, C.G.; Weingart, J.D.; Gallia, G.L.; Baylin, S.B.; Chan, T.A.; et al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 2013, 4, 1737–1747. [Google Scholar] [CrossRef]
- Rohle, D.; Popovici-Muller, J.; Palaskas, N.; Turcan, S.; Grommes, C.; Campos, C.; Tsoi, J.; Clark, O.; Oldrini, B.; Komisopoulou, E.; et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013, 340, 626–630. [Google Scholar] [CrossRef]
- Romani, M.; Pistillo, M.P.; Banelli, B. Epigenetic targeting of glioblastoma. Front. Oncol. 2018, 8, 448. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.M. IDH inhibitors in acute myeloid leukemia and myelodysplastic syndrome. Clin. Adv. Hematol. Oncol. 2021, 19, 556–558. [Google Scholar] [PubMed]
- Watts, J.M.; Baer, M.R.; Yang, J.; Prebet, T.; Lee, S.; Schiller, G.J.; Dinner, S.; Pigneux, A.; Montesinos, P.; Wang, E.S.; et al. Olutasidenib (FT-2102), an IDH1m inhibitor as a single agent or in combination with azacitidine, induces deep clinical responses with mutation clearance in patients with acute myeloid leukemia treated in a phase 1 dose escalation and expansion study. Blood 2019, 134 (Suppl. S1), 231. [Google Scholar] [CrossRef]
- A Study of FT 2102 in Participants with Advanced Solid Tumors and Gliomas with an IDH1 Mutation. Available online: https://clinicaltrials.gov/ct2/show/NCT03684811 (accessed on 9 May 2022).
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–663. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.; Zeng, T.; Pan, X.; Chen, L.; Liu, D.; Li, H.; Huang, T.; Cai, Y.D. Distinguishing glioblastoma subtypes by methylation signatures. Front. Genet. 2020, 11, 604336. [Google Scholar] [CrossRef]
- Sareen, H.; Garrett, C.; Lynch, D.; Powter, B.; Brungs, D.; Cooper, A.; Po, J.; Koh, E.S.; Vessey, J.Y.; McKechnie, S.; et al. The role of liquid biopsies in detecting molecular tumor biomarkers in brain cancer patients. Cancers 2020, 12, 1831. [Google Scholar] [CrossRef]
- Sharim, H.; Grunwald, A.; Gabrieli, T.; Michaeli, Y.; Margalit, S.; Torchinsky, D.; Arielly, R.; Nifker, G.; Juhasz, M.; Gularek, F.; et al. Long-read single-molecule maps of the functional methylome. Genome Res. 2019, 29, 646–656. [Google Scholar] [CrossRef]
- Rusk, N. Optically mapping methylation. Nat. Methods 2019, 16, 362. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Della Monica, R.; Cuomo, M.; Buonaiuto, M.; Costabile, D.; Franca, R.A.; Del Basso De Caro, M.; Catapano, G.; Chiariotti, L.; Visconti, R. MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme. Int. J. Mol. Sci. 2022, 23, 7148. https://doi.org/10.3390/ijms23137148
Della Monica R, Cuomo M, Buonaiuto M, Costabile D, Franca RA, Del Basso De Caro M, Catapano G, Chiariotti L, Visconti R. MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme. International Journal of Molecular Sciences. 2022; 23(13):7148. https://doi.org/10.3390/ijms23137148
Chicago/Turabian StyleDella Monica, Rosa, Mariella Cuomo, Michela Buonaiuto, Davide Costabile, Raduan Ahmed Franca, Marialaura Del Basso De Caro, Giuseppe Catapano, Lorenzo Chiariotti, and Roberta Visconti. 2022. "MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme" International Journal of Molecular Sciences 23, no. 13: 7148. https://doi.org/10.3390/ijms23137148
APA StyleDella Monica, R., Cuomo, M., Buonaiuto, M., Costabile, D., Franca, R. A., Del Basso De Caro, M., Catapano, G., Chiariotti, L., & Visconti, R. (2022). MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme. International Journal of Molecular Sciences, 23(13), 7148. https://doi.org/10.3390/ijms23137148








