LRF Promotes Indirectly Advantageous Chromatin Conformation via BGLT3-lncRNA Expression and Switch from Fetal to Adult Hemoglobin
Abstract
:1. Introduction
2. Results
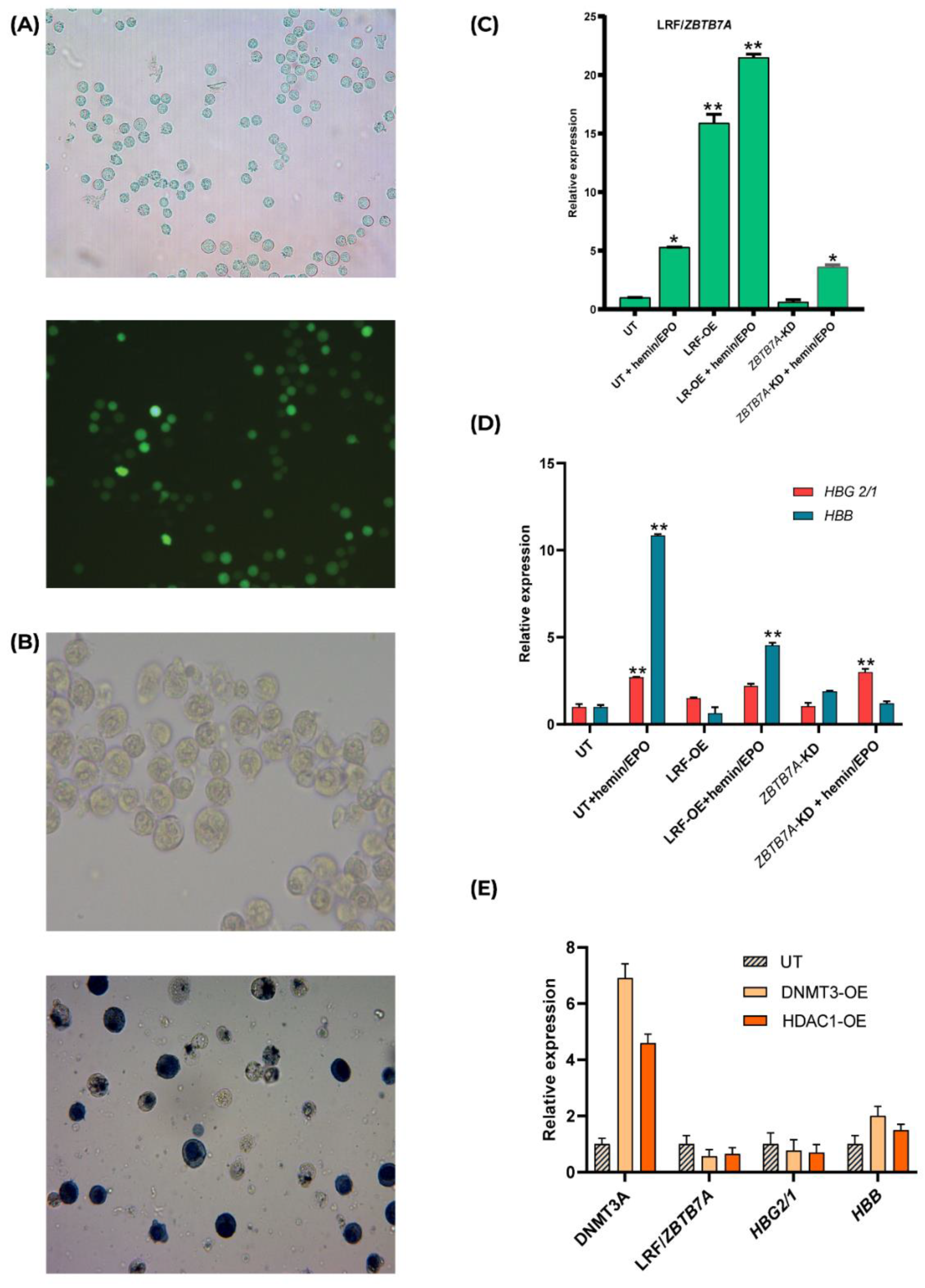
2.1. LRF/ZBTB7A Overexpression in Transfected K562 Clones and Hemin/EPO Treatment Induces Preferentially β- Than γ-Globin Expression
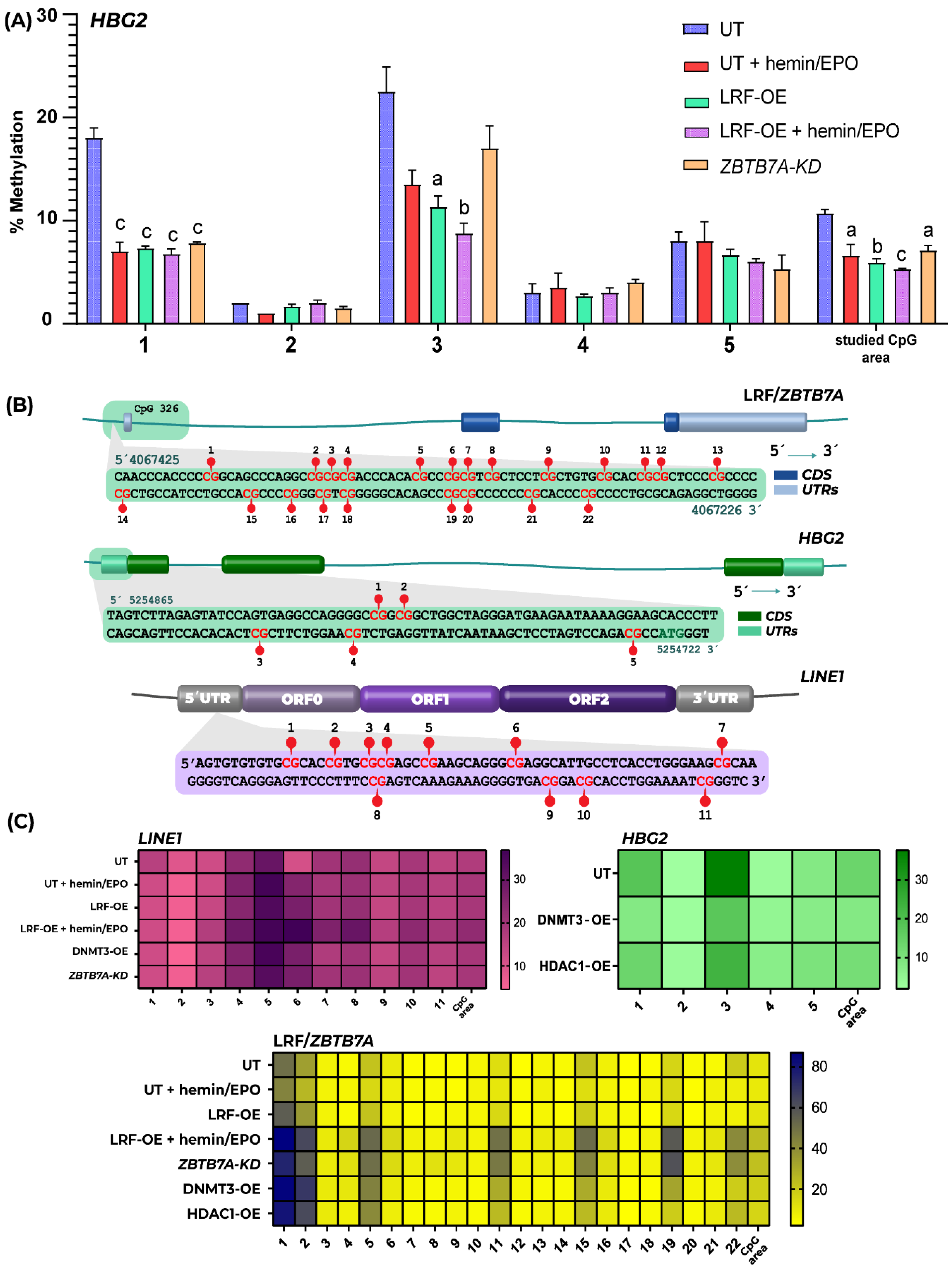
2.2. Epigenetic Events Promoted by Overexpression of DNMT3A and HDAC1 Leads to Insubstantial Changes in γ- and β-Globin Expression Levels
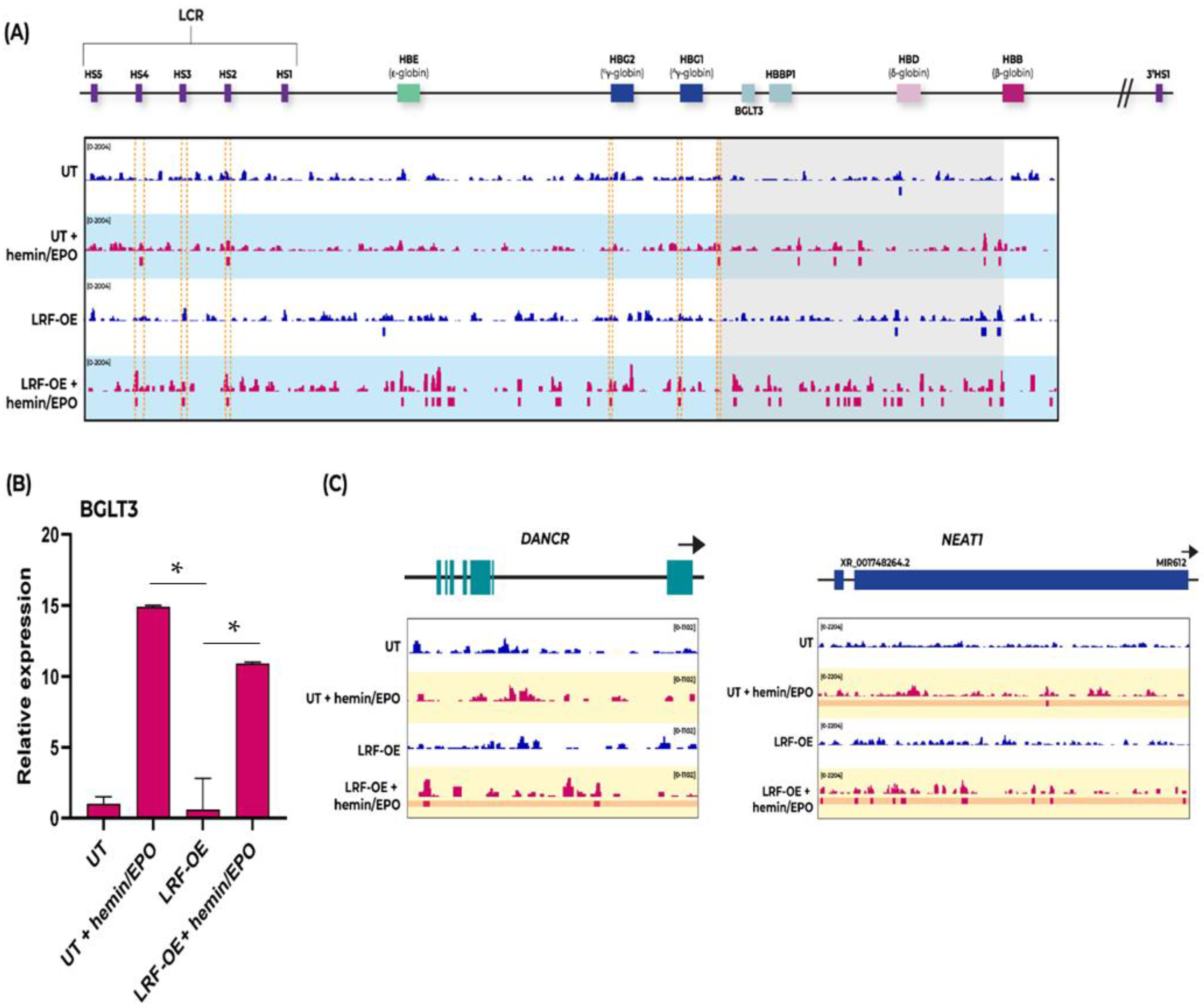
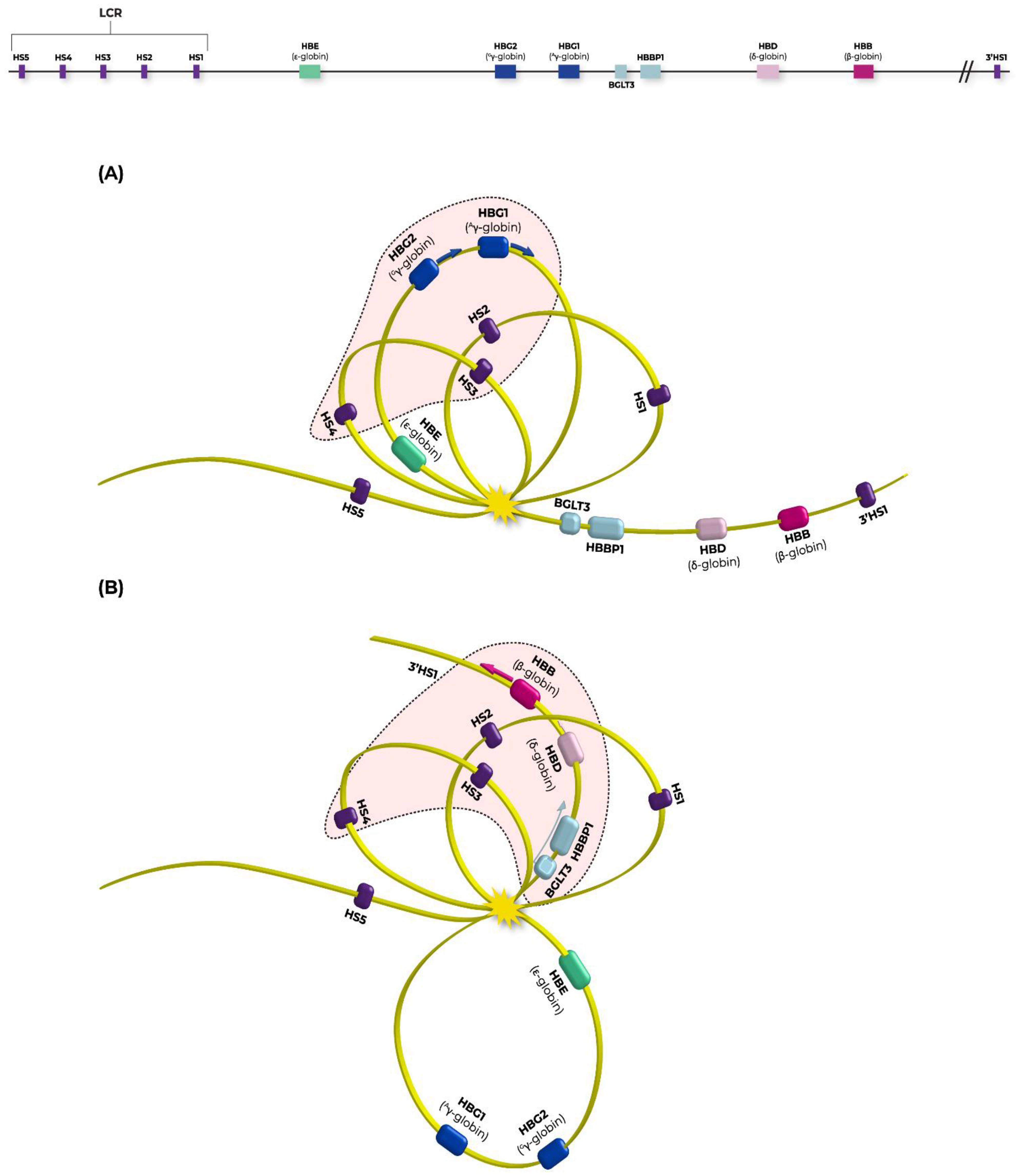
2.3. ChIP-Seq Analysis Reveals a Novel Function of LRF’s Regulatory Properties on BGLT3 Gene
3. Discussion
4. Materials and Methods
4.1. Cell Culture
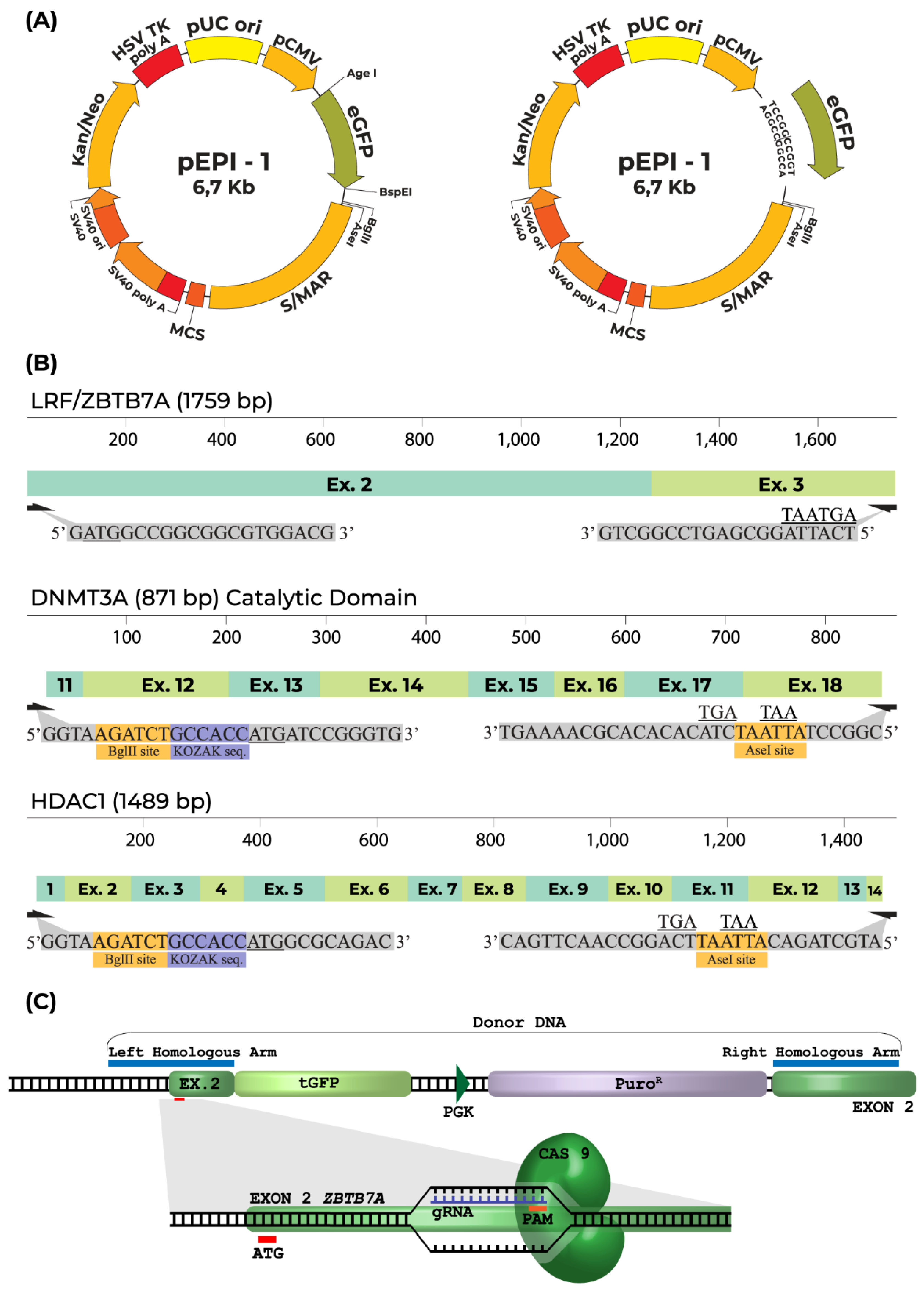
4.2. Plasmid Construction
4.3. LRF/ZBTB7A Knockout Using CRISPR/Cas9 System
4.4. RT-qPCR
4.5. Chromatin Immunoprecipitation
4.6. ChIP-Seq Library Construction and Next-Generation Sequencing
4.7. Analysis, Peak Calling and Visualization of ChIP-Seq Data
4.8. Methylation Analysis/Pyrosequencing
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giardine, B.; Borg, J.; Viennas, E.; Pavlidis, C.; Moradkhani, K.; Joly, P.; Bartsakoulia, M.; Riemer, C.; Miller, W.; Tzimas, G.; et al. Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 2014, 42, D1063–D1069. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, N.; Zuccato, C.; Finotti, A.; Lampronti, I.; Borgatti, M.; Gambari, R. Involvement of miRNA in erythroid differentiation. Epigenomics 2012, 4, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Rupon, J.W.; Krivega, I.; Breda, L.; Motta, I.; Jahn, K.S.; Reik, A.; Gregory, P.D.; Rivella, S.; Dean, A.; et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 2014, 158, 849–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Chen, Y.; Zhang, Y.; Liu, Y.; Liu, N.; Botten, G.A.; Cao, H.; Orkin, S.H.; Zhang, M.Q.; Xu, J. Multiplexed capture of spatial configuration and temporal dynamics of locus-specific 3D chromatin by biotinylated dCas9. Genome Biol. 2020, 21, 59. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, V.G.; Xu, J.; Byron, R.; Greisman, H.A.; Fisher, C.; Weatherall, D.J.; Sabath, D.E.; Groudine, M.; Orkin, S.H.; Premawardhena, A.; et al. A functional element necessary for fetal hemoglobin silencing. N. Engl. J. Med. 2011, 365, 807–814. [Google Scholar] [CrossRef] [Green Version]
- Lulli, V.; Romania, P.; Morsilli, O.; Cianciulli, P.; Gabbianelli, M.; Testa, U.; Giuliani, A.; Marziali, G. MicroRNA-486-3p regulates gamma-globin expression in human erythroid cells by directly modulating BCL11A. PLoS ONE 2013, 8, e60436. [Google Scholar] [CrossRef] [Green Version]
- Masuda, T.; Wang, X.; Maeda, M.; Canver, M.C.; Sher, F.; Funnell, A.P.; Fisher, C.; Suciu, M.; Martyn, G.E.; Norton, L.J.; et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 2016, 351, 285–289. [Google Scholar] [CrossRef] [Green Version]
- Ivaldi, M.S.; Diaz, L.F.; Chakalova, L.; Lee, J.; Krivega, I.; Dean, A. Fetal gamma-globin genes are regulated by the BGLT3 long noncoding RNA locus. Blood 2018, 132, 1963–1973. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Xu, S.; Yao, Q.; Zhu, Q.; Kai, Y.; Hsu, J.Y.; Sakon, P.; Pinello, L.; Yuan, G.C.; Bauer, D.E.; et al. Transcription factor competition at the gamma-globin promoters controls hemoglobin switching. Nat. Genet. 2021, 53, 511–520. [Google Scholar] [CrossRef]
- Bianchi, N.; Zuccato, C.; Lampronti, I.; Borgatti, M.; Gambari, R. Fetal Hemoglobin Inducers from the Natural World: A Novel Approach for Identification of Drugs for the Treatment of β-Thalassemia and Sickle-Cell Anemia. Evid. Based Complement. Altern. Med. 2009, 6, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Musallam, K.M.; Taher, A.T.; Cappellini, M.D.; Sankaran, V.G. Clinical experience with fetal hemoglobin induction therapy in patients with beta-thalassemia. Blood 2013, 121, 2199–2212; quiz 2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, R.B.; Mitchener, J.W.; Huisman, T.H. Studies on the fetal hemoglobin in the persistent high Hb-F anomaly. Blood 1961, 18, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Chondrou, V.; Stavrou, E.F.; Markopoulos, G.; Kouraklis-Symeonidis, A.; Fotopoulos, V.; Symeonidis, A.; Vlachaki, E.; Chalkia, P.; Patrinos, G.P.; Papachatzopoulou, A.; et al. Impact of ZBTB7A hypomethylation and expression patterns on treatment response to hydroxyurea. Hum. Genom. 2018, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Chumchuen, S.; Pornsukjantra, T.; Khamphikham, P.; Anurathapan, U.; Sripichai, O.; Songdej, D.; Hongeng, S. Downregulation of Transcription Factor LRF/ZBTB7A Increases Fetal Hemoglobin Expression in β-Thalassemia/Hemoglobin E Erythroid Cells. Blood 2019, 134 (Suppl. S1), 3549. [Google Scholar] [CrossRef]
- Fibach, E.; Kollia, P.; Schechter, A.N.; Noguchi, C.T.; Rodgers, G.P. Hemin-induced acceleration of hemoglobin production in immature cultured erythroid cells: Preferential enhancement of fetal hemoglobin. Blood 1995, 85, 2967–2974. [Google Scholar] [CrossRef]
- Green, A.R.; Rockman, S.; DeLuca, E.; Begley, C.G. Induced myeloid differentiation of K562 cells with downregulation of erythroid and megakaryocytic transcription factors: A novel experimental model for hemopoietic lineage restriction. Exp. Hematol. 1993, 21, 525–531. [Google Scholar]
- Naumann, S.; Reutzel, D.; Speicher, M.; Decker, H.J. Complete karyotype characterization of the K562 cell line by combined application of G-banding, multiplex-fluorescence in situ hybridization, fluorescence in situ hybridization, and comparative genomic hybridization. Leuk. Res. 2001, 25, 313–322. [Google Scholar] [CrossRef]
- Sasaki, D.; Kosunago, S.; Mikami, T.; Matsumoto, T.; Suzuki, M. Growth-inhibition by hemin in K562 human leukemic cells is related to hemoglobin-producing activity. Biol. Pharm. Bull. 1994, 17, 586–590. [Google Scholar] [CrossRef] [Green Version]
- Finotti, A.; Gasparello, J.; Breveglieri, G.; Cosenza, L.C.; Montagner, G.; Bresciani, A.; Altamura, S.; Bianchi, N.; Martini, E.; Gallerani, E.; et al. Development and characterization of K562 cell clones expressing BCL11A-XL: Decreased hemoglobin production with fetal hemoglobin inducers and its rescue with mithramycin. Exp. Hematol. 2015, 43, 1062–1071.e3. [Google Scholar] [CrossRef] [Green Version]
- Lessard, S.; Beaudoin, M.; Benkirane, K.; Lettre, G. Comparison of DNA methylation profiles in human fetal and adult red blood cell progenitors. Genome Med. 2015, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Hashimoto, H.; Horton, J.R.; Zhang, X. Chapter 2—Mechanisms of DNA Methylation, Methyl-CpG Recognition, and Demethylation in Mammals. In Handbook of Epigenetics; Tollefsbol, T., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 9–24. [Google Scholar]
- Ren, W.; Gao, L.; Song, J. Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation. Genes 2018, 9, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.D.; Kim, J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: A meta-analysis. PLoS ONE 2012, 7, e34615. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Verboon, J.M.; Zhang, Y.; Liu, N.; Kim, Y.J.; Marglous, S.; Nandakumar, S.K.; Voit, R.A.; Fiorini, C.; Ejaz, A.; et al. A unified model of human hemoglobin switching through single-cell genome editing. Nat. Commun. 2021, 12, 4991. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sun, M.Z.; Lv, X.; Guo, C.; Liu, S. ETV6 Regulates Hemin-Induced Erythroid Differentiation of K562 Cells through Mediating the Raf/MEK/ERK Pathway. Biol. Pharm. Bull. 2022, 45, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Woessmann, W.; Zwanzger, D.; Borkhardt, A. ERK signaling pathway is differentially involved in erythroid differentiation of K562 cells depending on time and the inducing agent. Cell Biol. Int. 2004, 28, 403–410. [Google Scholar] [CrossRef]
- Tsolaki, V.C.; Georgiou-Siafis, S.K.; Tsamadou, A.I.; Tsiftsoglou, S.A.; Samiotaki, M.; Panayotou, G.; Tsiftsoglou, A.S. Hemin accumulation and identification of a heme-binding protein clan in K562 cells by proteomic and computational analysis. J. Cell. Physiol. 2022, 237, 1315–1340. [Google Scholar] [CrossRef] [PubMed]
- Antoniani, C.; Meneghini, V.; Lattanzi, A.; Felix, T.; Romano, O.; Magrin, E.; Weber, L.; Pavani, G.; El Hoss, S.; Kurita, R.; et al. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human beta-globin locus. Blood 2018, 131, 1960–1973. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; Kang, Q.; Zhu, X.; Chen, Q.; Wang, X.; Chen, Y.; Ouyang, J.; Zhang, L.; Tan, H.; Chen, R.; et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2015, 34, 1768–1779. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [Green Version]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Liu, K.; Sun, C.W.; Pawlik, K.M.; Townes, T.M. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat. Genet. 2010, 42, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Borg, J.; Papadopoulos, P.; Georgitsi, M.; Gutierrez, L.; Grech, G.; Fanis, P.; Phylactides, M.; Verkerk, A.J.; van der Spek, P.J.; Scerri, C.A.; et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 2010, 42, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Norton, L.J.; Funnell, A.P.W.; Burdach, J.; Wienert, B.; Kurita, R.; Nakamura, Y.; Philipsen, S.; Pearson, R.C.M.; Quinlan, K.G.R.; Crossley, M. KLF1 directly activates expression of the novel fetal globin repressor ZBTB7A/LRF in erythroid cells. Blood Adv. 2017, 1, 685–692. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.; Williamson, I.; Bickmore, W.A.; Dostie, J. An Overview of Genome Organization and How We Got There: From FISH to Hi-C. Microbiol. Mol. Biol. Rev. MMBR 2015, 79, 347–372. [Google Scholar] [CrossRef] [Green Version]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Cuddapah, S.; Jothi, R.; Schones, D.E.; Roh, T.Y.; Cui, K.; Zhao, K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009, 19, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K.; Kas, E.; Poljak, L.; Adachi, Y. Scaffold-associated regions: Cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 1992, 2, 275–285. [Google Scholar] [CrossRef]
- Heng, H.H.; Goetze, S.; Ye, C.J.; Liu, G.; Stevens, J.B.; Bremer, S.W.; Wykes, S.M.; Bode, J.; Krawetz, S.A. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004, 117, 999–1008. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.B.; Kramer, J.A.; Krawetz, S.A. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997, 25, 1419–1425. [Google Scholar] [CrossRef]
- Mathai, J.; Mittal, S.P.; Alam, A.; Ranade, P.; Mogare, D.; Patel, S.; Saxena, S.; Ghorai, S.; Kulkarni, A.P.; Chattopadhyay, S. SMAR1 binds to T(C/G) repeat and inhibits tumor progression by regulating miR-371-373 cluster. Sci. Rep. 2016, 6, 33779. [Google Scholar] [CrossRef] [Green Version]
- Narwade, N.; Patel, S.; Alam, A.; Chattopadhyay, S.; Mittal, S.; Kulkarni, A. Mapping of scaffold/matrix attachment regions in human genome: A data mining exercise. Nucleic Acids Res. 2019, 47, 7247–7261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Keller, C.A.; Giardine, B.; Grevet, J.D.; Davies, J.O.J.; Hughes, J.R.; Kurita, R.; Nakamura, Y.; Hardison, R.C.; Blobel, G.A. Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Dev. 2017, 31, 1704–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Di, L.J.; Lv, X.; Zheng, W.; Xue, Z.; Guo, Z.C.; Liu, D.P.; Liang, C.C. Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster. PLoS ONE 2009, 4, e4629. [Google Scholar]
- de Laat, W.; Grosveld, F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003, 11, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, Y.; Si, Y.; Wang, F.; Yu, J. Long non-coding RNAs in hematopoietic regulation. Cell Regen. 2018, 7, 27–32. [Google Scholar] [CrossRef]
- Ren, Y.; Zhu, J.; Han, Y.; Li, P.; Wu, J.; Qu, H.; Zhang, Z.; Fang, X. Regulatory association of long noncoding RNAs and chromatin accessibility facilitates erythroid differentiation. Blood Adv. 2021, 5, 5396–5409. [Google Scholar] [CrossRef]
- Baliga, B.S.; Mankad, M.; Shah, A.K.; Mankad, V.N. Mechanism of differentiation of human erythroleukaemic cell line K562 by hemin. Cell Prolif. 1993, 26, 519–529. [Google Scholar] [CrossRef]
- Wang, D.; Si, S.; Wang, Q.; Luo, G.; Du, Q.; Liang, Q.; Guo, X.; Zhang, G.; Feng, J.; Leng, Z. MiR-27a Promotes Hemin-Induced Erythroid Differentiation of K562 Cells by Targeting CDC25B. Cell. Physiol. Biochem. 2018, 46, 365–374. [Google Scholar] [CrossRef] [Green Version]
- Rowley, P.T.; Ohlsson-Wilhelm, B.M.; Farley, B.A. K562 human erythroleukemia cells demonstrate commitment. Blood 1985, 65, 862–868. [Google Scholar] [CrossRef] [Green Version]
- Piechaczek, C.; Fetzer, C.; Baiker, A.; Bode, J.; Lipps, H.J. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999, 27, 426–428. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Dastani, Z.; Hivert, M.F.; Timpson, N.; Perry, J.R.; Yuan, X.; Scott, R.A.; Henneman, P.; Heid, I.M.; Kizer, J.R.; Lyytikainen, L.P.; et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: A multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012, 8, e1002607. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albarino, C.G.; Romanowski, V. Phenol extraction revisited: A rapid method for the isolation and preservation of human genomic DNA from whole blood. Mol. Cell. Probes 1994, 8, 423–427. [Google Scholar] [CrossRef]
| Assay | Gene | Sequence | Primer |
|---|---|---|---|
| Pyrosequencing CpG assay | LRF/ZBTB7A | GTAGATTTTTTTGTGTTAAGGA | Forward |
| AACAAACCCCCAACCTCTAC | 5′ biotinylated Reverse | ||
| GGGATTTTTATAGTTTTATTTTTAA | Forward Pyrosequencing | ||
| HBG2 | 5′ ATGGTGGGAGAAGAAAATTAGTT 3′ | Forward | |
| 5′ TCTCCTCCAACATCTTCCACATTCA 3′ | 5′ biotinylated Reverse | ||
| 5′ ATAGTTTTAGAGTATTTAGTGA 3′ | Forward Pyrosequencing | ||
| LINE1 | 5′ AAGTAAGTTTGGGTAATGG 3′ | Forward | |
| 5′ AACAACTCCCATCTACAACTCCC 3′ | 5′ biotinylated Reverse | ||
| 5′ GGGTGGGAGTGAT 3′ | Forward Pyrosequencing | ||
| qPCR | GAPDH | 5′ AGGGCTGCTTTTAACTCTGGT 3′ | Forward |
| 5′ CCCCACTTGATTTTGGAGGGA 3′ | Reverse | ||
| LRF/ZBTB7A | 5′ GAAGCCCTACGAGTGCAACATC 3′ | Forward | |
| 5′ GTGGTTCTTCAGGTCGTAGTTGTG 3′ | Reverse | ||
| HBG2/1 | 5′ AACCCCAAAGTCAAGGCACA 3′ | Forward | |
| 5′ GATTGCCAAAACGGTCACCA 3′ | Reverse | ||
| HBB | 5′ CTGAGTGAGCTGCACTGTGA 3’ | Forward | |
| 5′ ATTGGACAGCAAGAAAGCGA 3′ | Reverse | ||
| DNMT3A | 5′ GGTGCTGTCTCTCTTTGATGGA 3′ | Forward | |
| 5′ GGATATGCTTCTGTGTGACGC 3′ | Reverse | ||
| BGLT3 | 5′ TCAGGGGTAACACACAAACCA 3′ | Forward | |
| 5′ GTCTCATGTGCTGCACGTCT 3′ | Reverse | ||
| HDAC1 | 5′ CCCAATGAAGCCTCACCGAA 3′ | Forward | |
| 5′ CGGATGGAGCGCAAGAATTT 3′ | Reverse | ||
| Cloning | LRF/ZBTB7A | 5′ GATGGCCGGCGGCGTGGACG 3′ | Forward |
| 5′ TCATTAGGCGAGTCCGGCTGTGAAG 3′ | Reverse | ||
| DNMT3A | 5′ GGTAAGATCTGCCACCATGATCCGGGTG 3′ | Forward | |
| 5′ CGGCCTATTAATCTACACACACGAAAGT 3′ | Reverse | ||
| HDAC1 | 5′ GGTAAGATCTGCCACCATGGCGCAGAC 3′ | Forward | |
| 5′ TAGCCTAGTTATGCTAGCATTAATTCAGG 3′ | Reverse |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chondrou, V.; Shaukat, A.-N.; Psarias, G.; Athanasopoulou, K.; Iliopoulou, E.; Damanaki, A.; Stathopoulos, C.; Sgourou, A. LRF Promotes Indirectly Advantageous Chromatin Conformation via BGLT3-lncRNA Expression and Switch from Fetal to Adult Hemoglobin. Int. J. Mol. Sci. 2022, 23, 7025. https://doi.org/10.3390/ijms23137025
Chondrou V, Shaukat A-N, Psarias G, Athanasopoulou K, Iliopoulou E, Damanaki A, Stathopoulos C, Sgourou A. LRF Promotes Indirectly Advantageous Chromatin Conformation via BGLT3-lncRNA Expression and Switch from Fetal to Adult Hemoglobin. International Journal of Molecular Sciences. 2022; 23(13):7025. https://doi.org/10.3390/ijms23137025
Chicago/Turabian StyleChondrou, Vasiliki, Athanasios-Nasir Shaukat, Georgios Psarias, Katerina Athanasopoulou, Evanthia Iliopoulou, Ariadne Damanaki, Constantinos Stathopoulos, and Argyro Sgourou. 2022. "LRF Promotes Indirectly Advantageous Chromatin Conformation via BGLT3-lncRNA Expression and Switch from Fetal to Adult Hemoglobin" International Journal of Molecular Sciences 23, no. 13: 7025. https://doi.org/10.3390/ijms23137025
APA StyleChondrou, V., Shaukat, A.-N., Psarias, G., Athanasopoulou, K., Iliopoulou, E., Damanaki, A., Stathopoulos, C., & Sgourou, A. (2022). LRF Promotes Indirectly Advantageous Chromatin Conformation via BGLT3-lncRNA Expression and Switch from Fetal to Adult Hemoglobin. International Journal of Molecular Sciences, 23(13), 7025. https://doi.org/10.3390/ijms23137025







