Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake
Abstract
1. Introduction
2. Results and Discussion
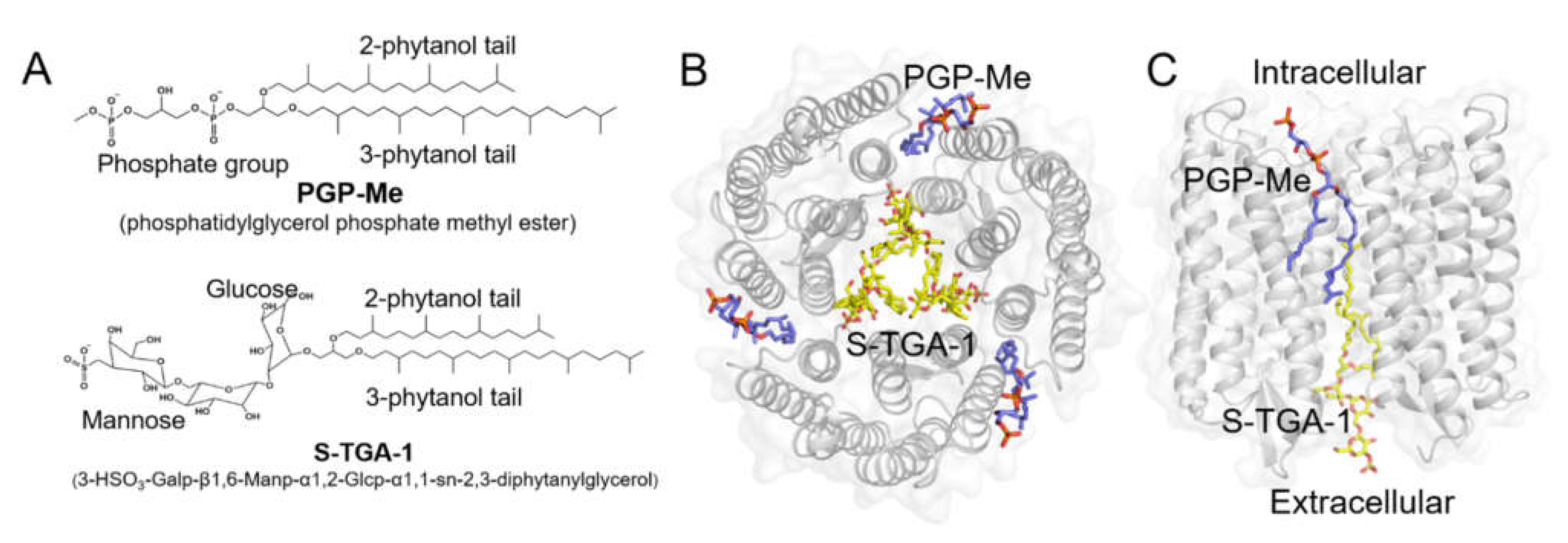
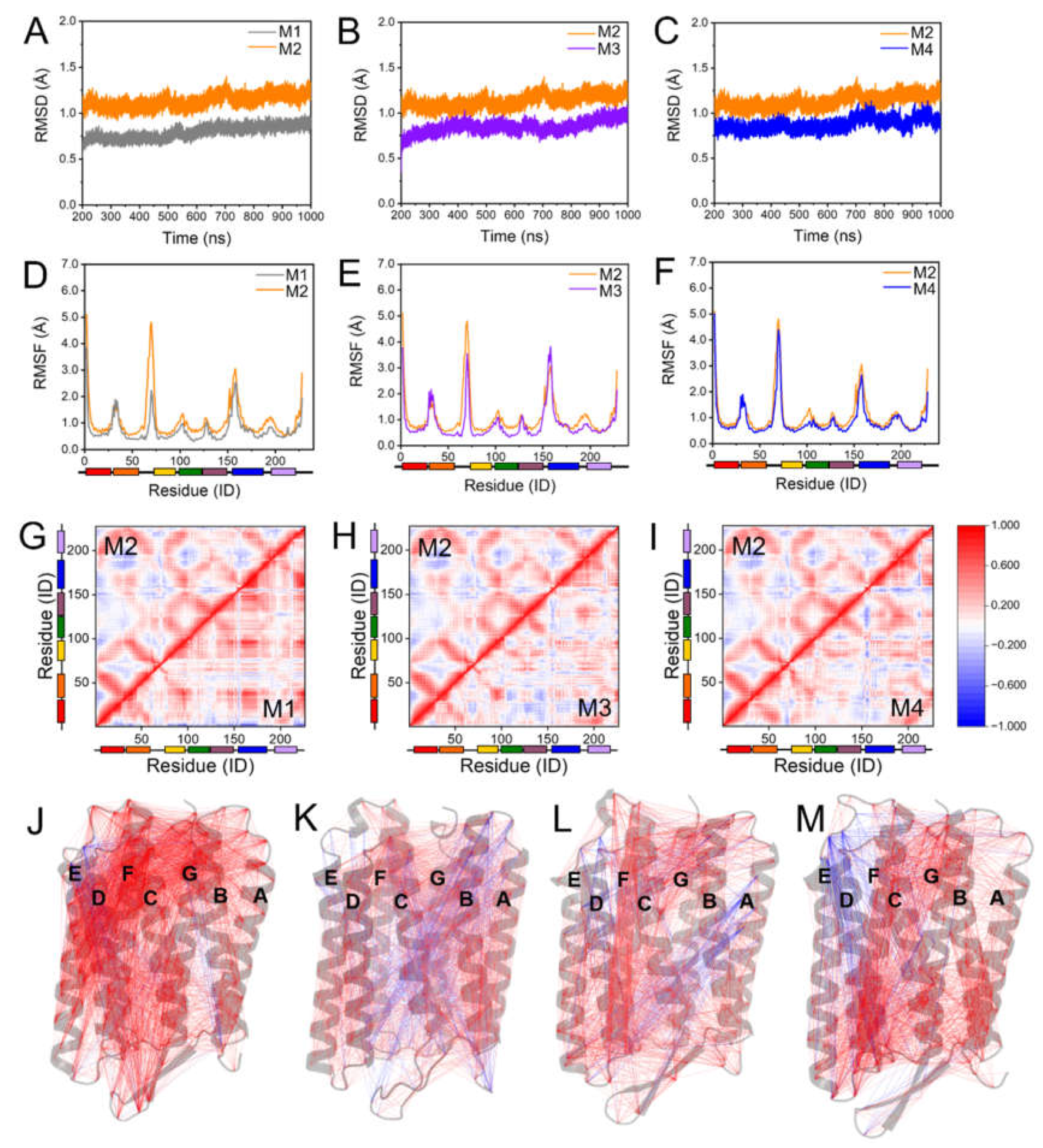
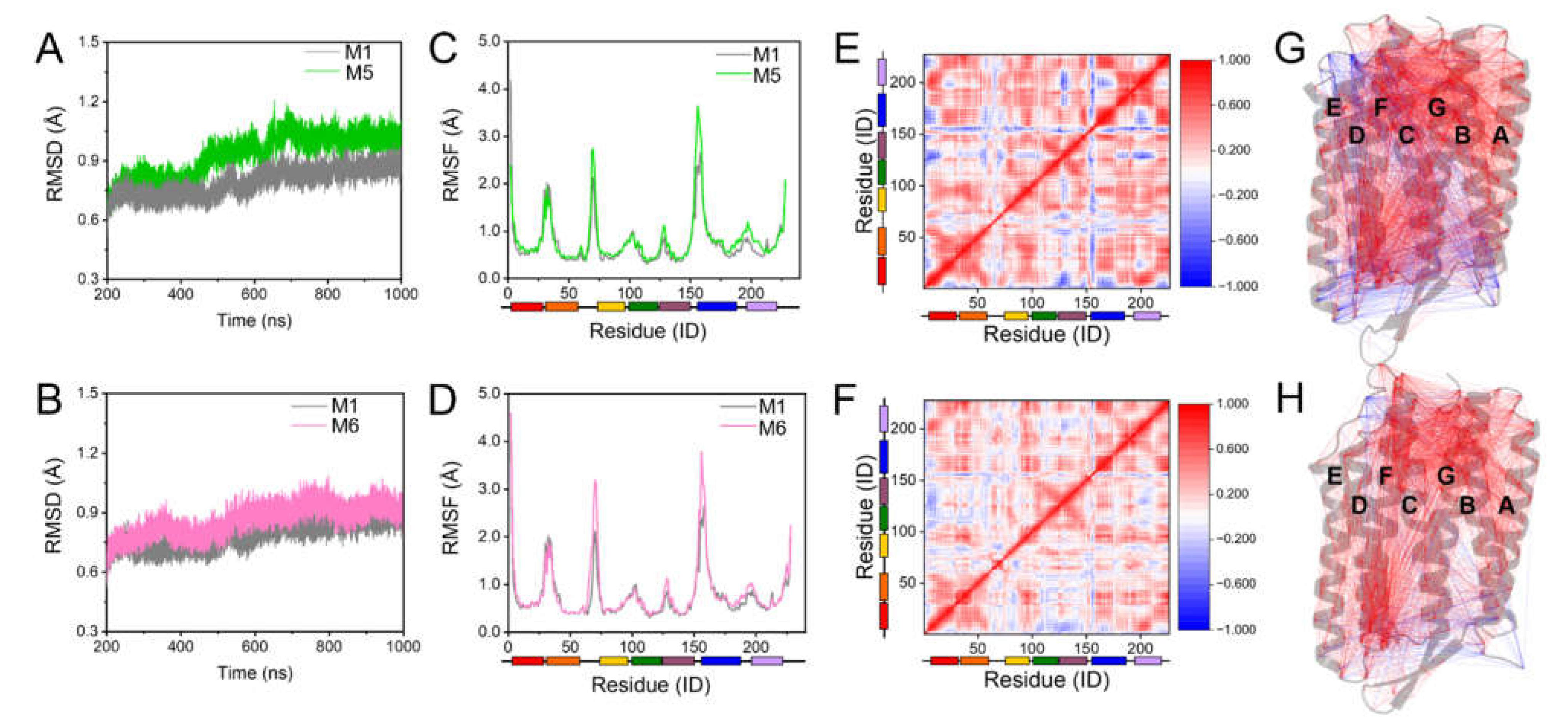
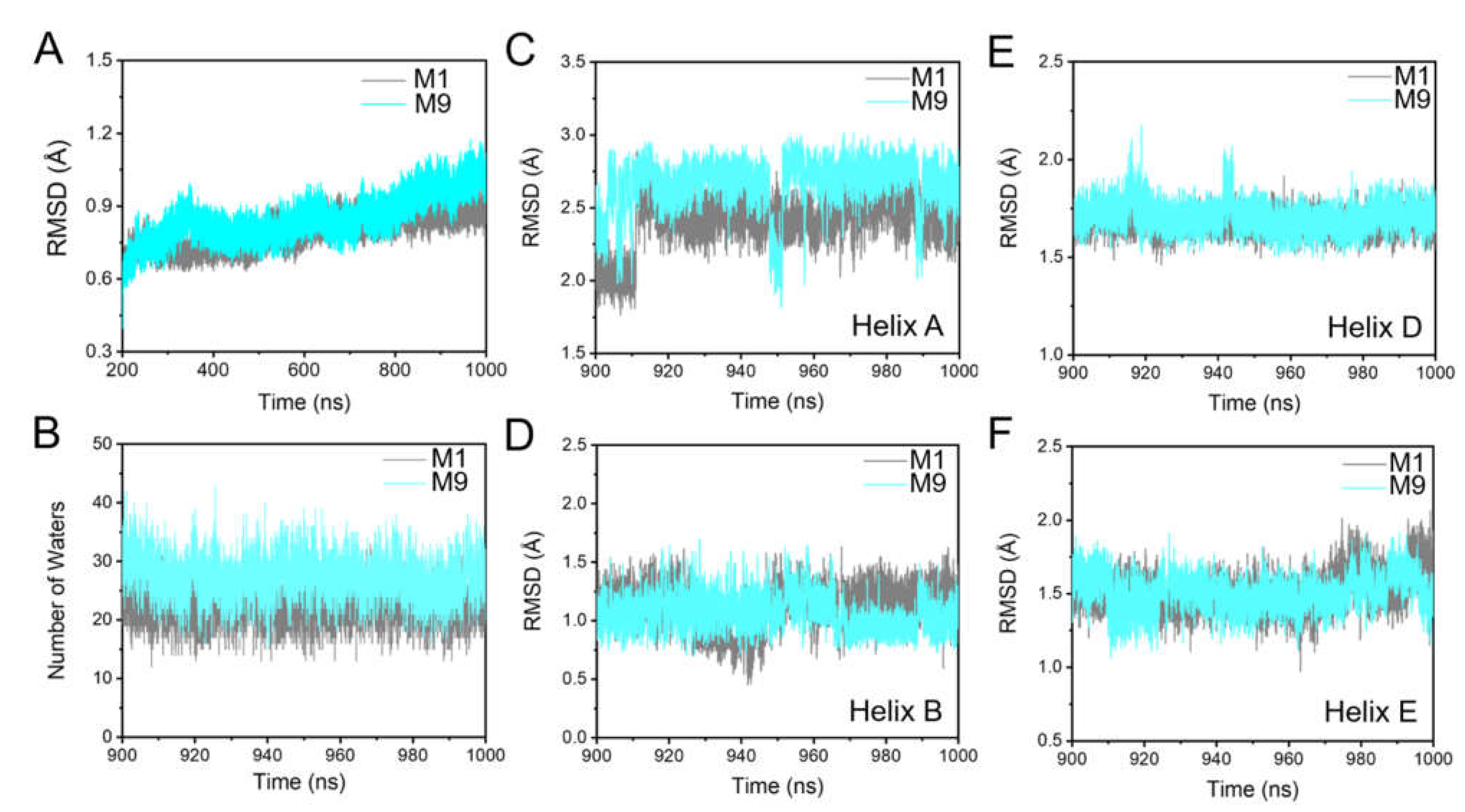
2.1. Maintaining the Overall Coherent Dynamics by S-TGA-1 and PGP-Me Binding
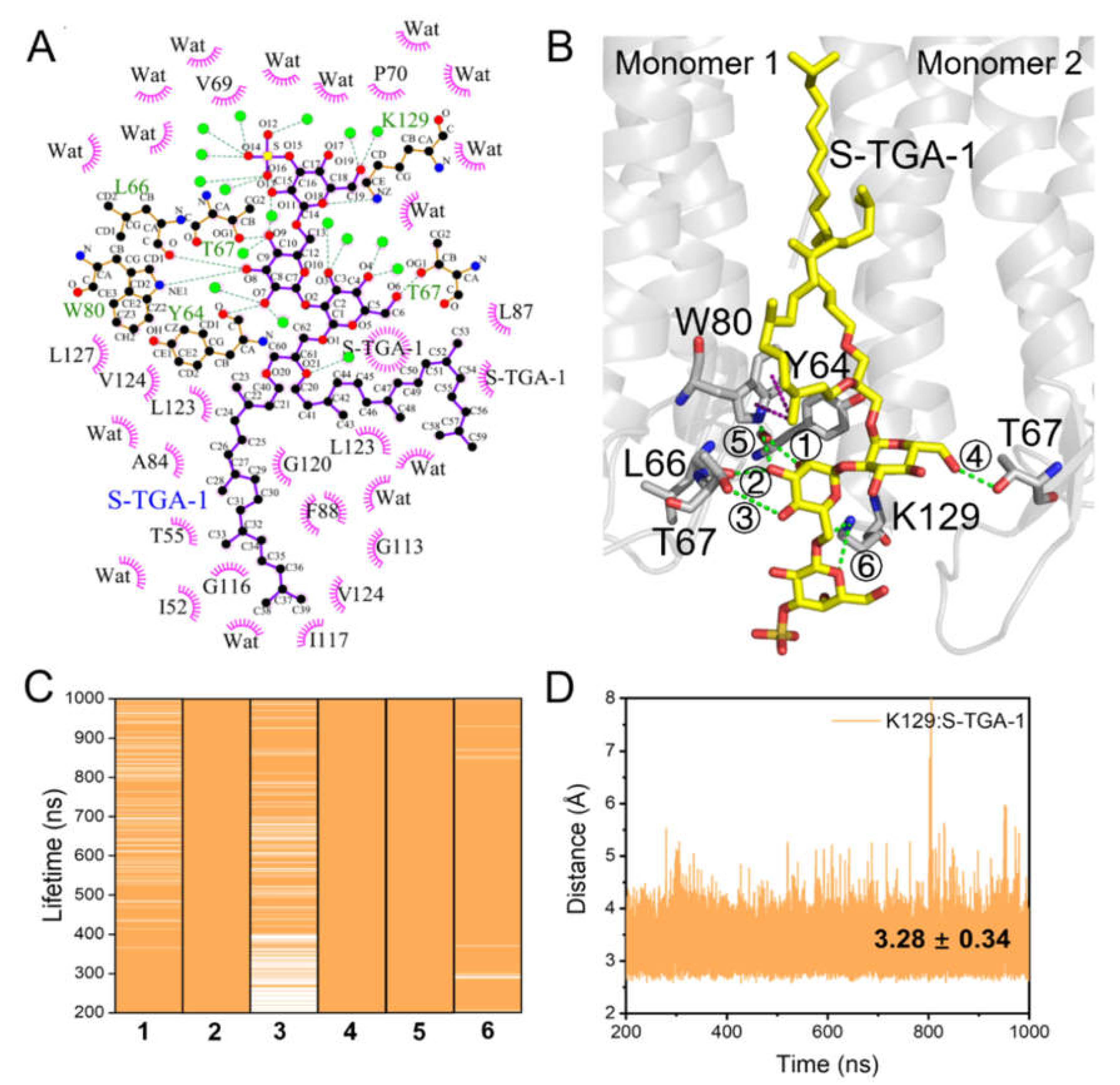
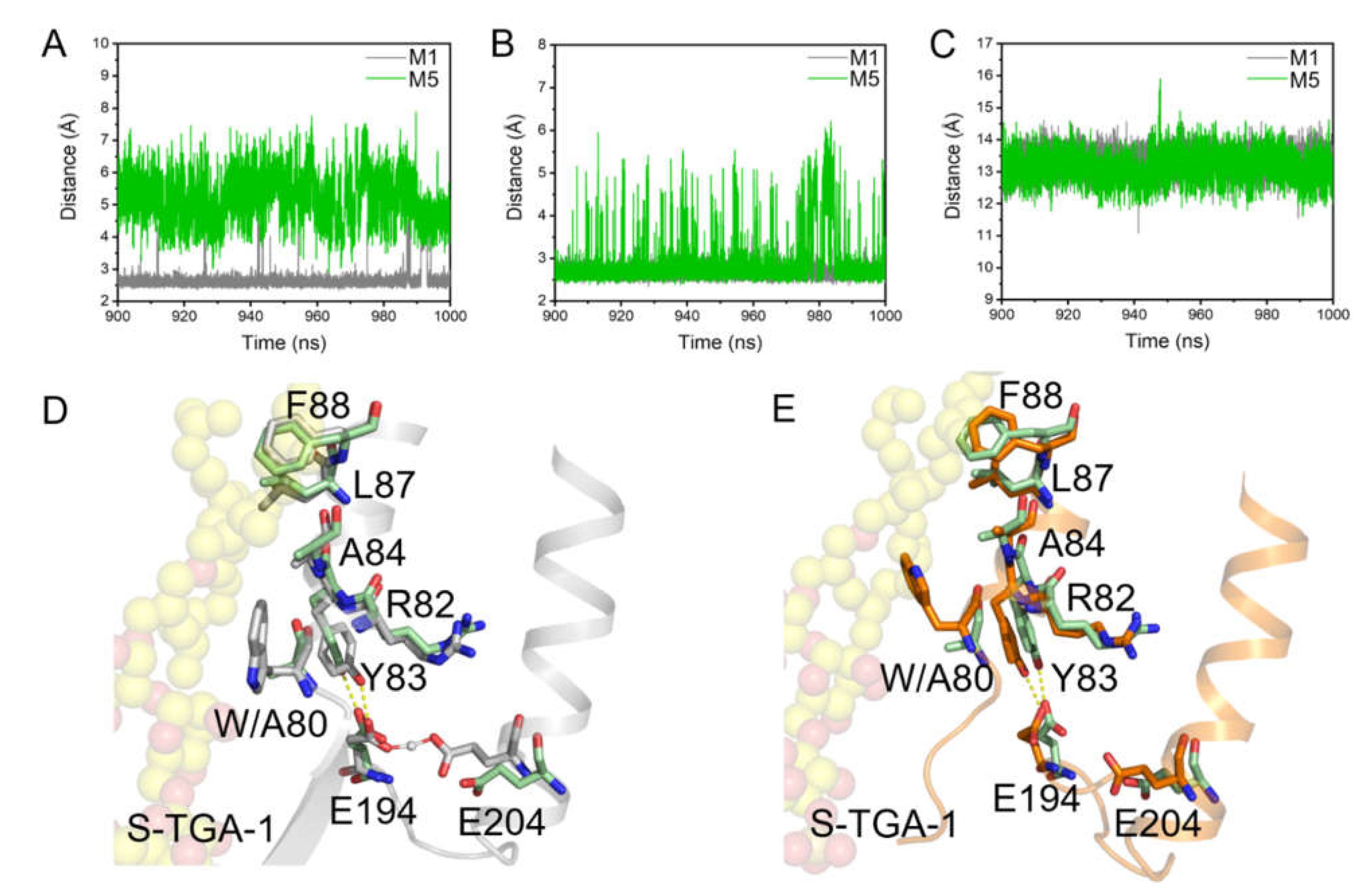
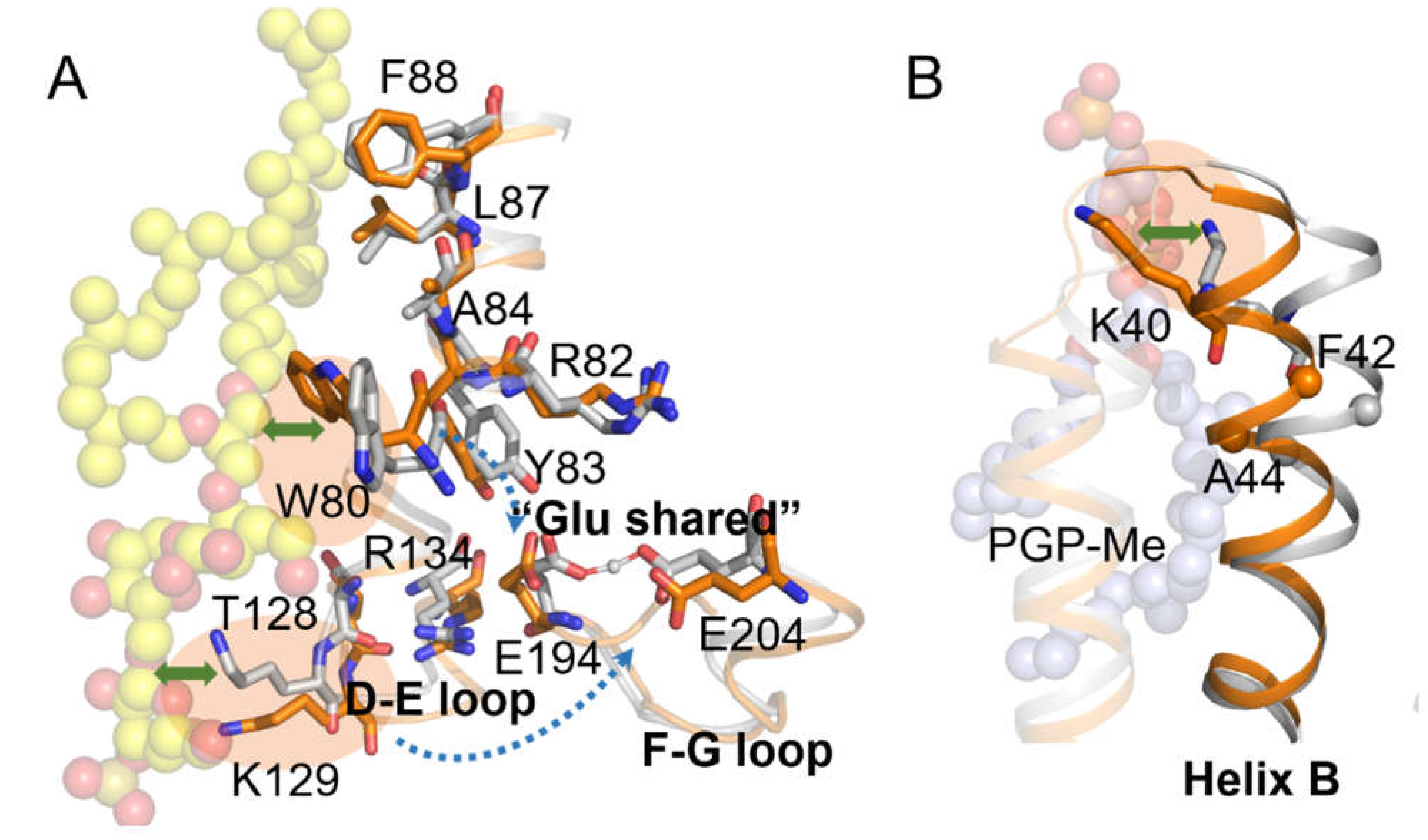
2.2. Coupling of S-TGA-1 with the Extracellular Surface of bR for Proton Release
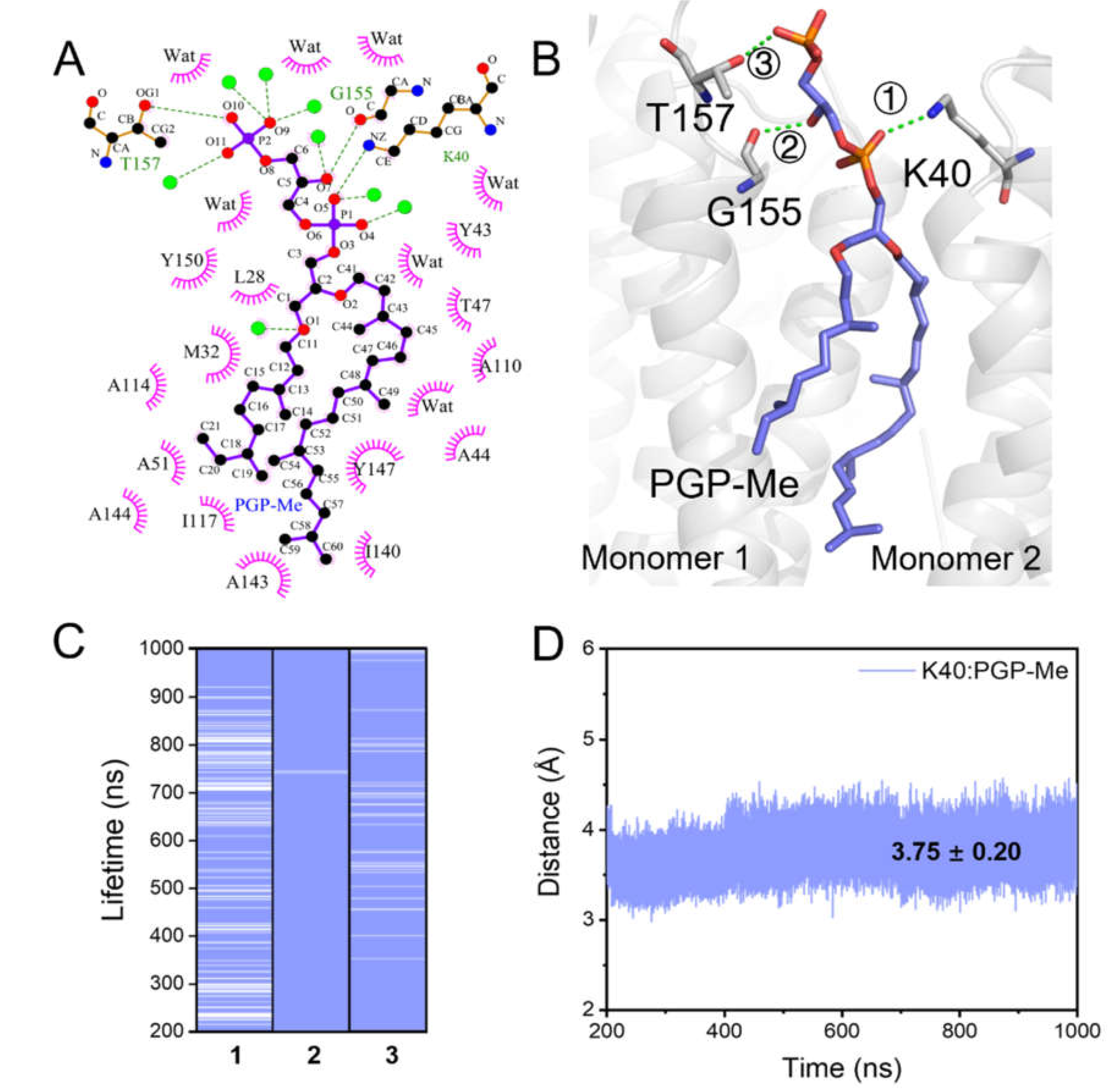
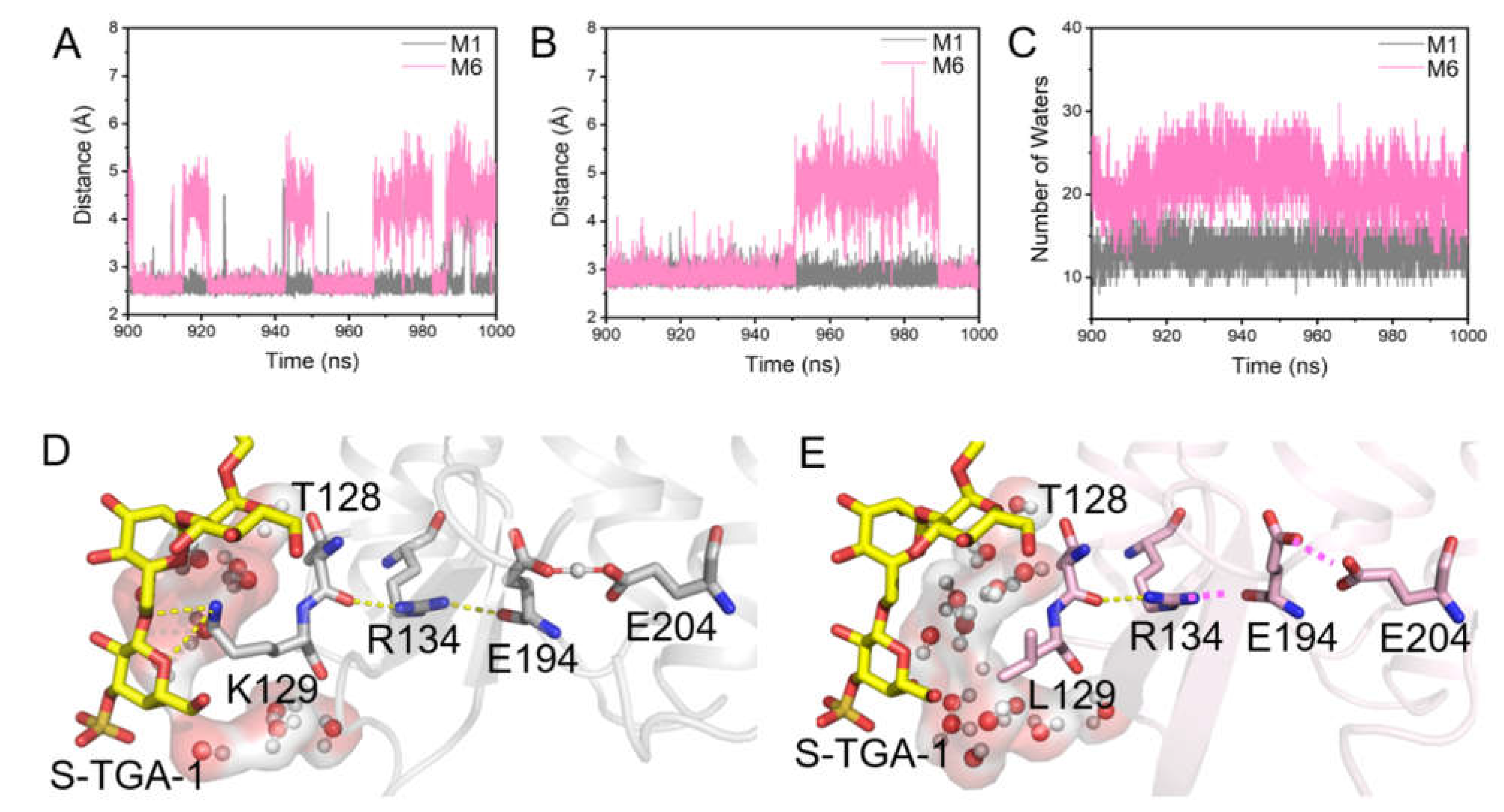
2.3. Coupling of PGP-Me with the Cytoplasmic Surface of bR for Proton Uptake
3. Conclusions
4. Materials and Methods
4.1. Molecular Models and Parameters
4.2. Molecular Dynamics Simulation
4.3. Analysis of the Simulations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| M1 | Membrane containing bR trimer, POPC, S-TGA-1, and PGP-Me lipids |
| M2 | Membrane containing bR trimer and POPC lipids |
| M3 | Membrane containing bR trimer, POPC, and S-TGA-1 lipids |
| M4 | Membrane containing bR trimer, POPC, and PGP-Me lipids |
| M5 | Membrane containing W80A-bR trimer, POPC, S-TGA-1, and PGP-Me lipids |
| M6 | Membrane containing K129L-bR trimer, POPC, S-TGA-1, and PGP-Me lipids |
| M7 | Membrane containing W80A-bR trimer and POPC lipids |
| M8 | Membrane containing K129L-bR trimer and POPC lipids |
| M9 | Membrane containing K40L-bR trimer, POPC, S-TGA-1, and PGP-Me lipids |
| M10 | Membrane containing K40L-bR trimer and POPC lipids |
References
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Flynn, A.D. Drugging Membrane Protein Interactions. Annu. Rev. Biomed. Eng. 2016, 18, 51–76. [Google Scholar] [CrossRef]
- Hancock, J.F. Lipid rafts: Contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006, 7, 456–462. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Nishimura, T.; Tooze, S.A. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020, 6, 32. [Google Scholar] [CrossRef]
- Janmey, P.A.; Kinnunen, P.K.J. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 2006, 16, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Reading, E.; Allison, T.M.; Ulmschneider, M.B.; Degiacomi, M.T.; Baldwin, A.J.; Robinson, C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510, 172–175. [Google Scholar] [CrossRef]
- Yen, H.-Y.; Hoi, K.K.; Liko, I.; Hedger, G.; Horrell, M.R.; Song, W.; Wu, D.; Heine, P.; Warne, T.; Lee, Y.; et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 2018, 559, 423–427. [Google Scholar] [CrossRef]
- Lee, A.G. Lipid-protein interactions in biological membranes: A structural perspective. Biochim. Biophys. Acta 2003, 1612, 1–40. [Google Scholar] [CrossRef]
- Lee, A.G. Lipid–protein interactions. Biochem. Soc. Trans. 2011, 39, 761–766. [Google Scholar] [CrossRef]
- Weingarth, M.; Prokofyev, A.; van der Cruijsen, E.A.W.; Nand, D.; Bonvin, A.M.J.J.; Pongs, O.; Baldus, M. Structural Determinants of Specific Lipid Binding to Potassium Channels. J. Am. Chem. Soc. 2013, 135, 3983–3988. [Google Scholar] [CrossRef]
- Schmidt, D.; Jiang, Q.-X.; MacKinnon, R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 2006, 444, 775–779. [Google Scholar] [CrossRef]
- Domene, C.; Bond, P.J.; Deol, S.S.; Sansom, M.S.P. Lipid/Protein Interactions and the Membrane/Water Interfacial Region. J. Am. Chem. Soc. 2003, 125, 14966–14967. [Google Scholar] [CrossRef]
- Sansom, M.S.; Bond, P.J.; Deol, S.S.; Grottesi, A.; Haider, S.; Sands, Z.A. Molecular simulations and lipid-protein interactions: Potassium channels and other membrane proteins. Biochem. Soc. Trans. 2005, 33, 916–920. [Google Scholar] [CrossRef]
- Duncan, A.L.; Song, W.; Sansom, M.S.P. Lipid-Dependent Regulation of Ion Channels and G Protein–Coupled Receptors: Insights from Structures and Simulations. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 31–50. [Google Scholar] [CrossRef]
- Zhou, Y.; Morais-Cabral, J.H.; Kaufman, A.; MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 2001, 414, 43–48. [Google Scholar] [CrossRef]
- Belrhali, H.; Nollert, P.; Royant, A.; Menzel, C.; Rosenbusch, J.; Landau, E.; Pebay-Peyroula, E. Protein, lipid and water organization in bacteriorhodopsin crystals: A molecular view of the purple membrane at 1.9 Å resolution. Structure 1999, 7, 909–917. [Google Scholar] [CrossRef]
- Cartailler, J.-P.; Luecke, H. X-Ray Crystallographic Analysis of Lipid-Protein Interactions in the Bacteriorhodopsin Purple Membrane. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 285–310. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like Protein from the Purple Membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef]
- Ernst, O.; Lodowski, D.; Elstner, M.; Hegemann, P.; Brown, L.; Kandori, H. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 2013, 114, 126–163. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. USA 1973, 70, 2853–2857. [Google Scholar] [CrossRef] [PubMed]
- Stoeckenius, W.; Lozier, R.H.; Bogomolni, R.A. Bacteriorhodopsin and the purple membrane of halobacteria. Biochim. Biophys. Acta 1979, 505, 215–278. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Bacteriorhodopsin—The movie. Nature 2000, 406, 569–570. [Google Scholar] [CrossRef] [PubMed]
- Lanyi, J.K. Bacteriorhodopsin. Annu. Rev. Physiol. 2004, 66, 665–688. [Google Scholar] [CrossRef]
- Lanyi, J.K. Mechanism of proton transport from crystallographic structures of the nine states of the bacteriorhodopsin photocycle. Biochim. Biophys. Acta 2004, 1658, 78. [Google Scholar]
- Wickstrand, C.; Nogly, P.; Nango, E.; Iwata, S.; Standfuss, J.; Neutze, R. Bacteriorhodopsin: Structural Insights Revealed Using X-Ray Lasers and Synchrotron Radiation. Annu. Rev. Biochem. 2019, 88, 59–83. [Google Scholar] [CrossRef]
- Lanyi, J.K.; Schobert, B. Mechanism of Proton Transport in Bacteriorhodopsin from Crystallographic Structures of the K, L, M1, M2, and M2’ Intermediates of the Photocycle. J. Mol. Biol. 2003, 328, 439–450. [Google Scholar] [CrossRef]
- Corcelli, A.; Colella, M.; Mascolo, G.; Fanizzi, F.P.; Kates, M. A Novel Glycolipid and Phospholipid in the Purple Membrane. Biochemistry 2000, 39, 3318–3326. [Google Scholar] [CrossRef]
- Corcelli, A.; Lattanzio, V.M.T.; Mascolo, G.; Papadia, P.; Fanizzi, F. Lipid-protein stoichiometries in a crystalline biological membrane: NMR quantitative analysis of the lipid extract of the purple membrane. J. Lipid Res. 2002, 43, 132–140. [Google Scholar] [CrossRef]
- Renner, C.; Kessler, B.; Oesterhelt, D. Lipid composition of integral purple membrane by 1H and 31P NMR. J. Lipid Res. 2005, 46, 1755–1764. [Google Scholar] [CrossRef]
- Kushwaha, S.C.; Kates, M.; Martin, W.G. Characterization and composition of the purple and red membrane from Halobacterium cutirubrum. Can. J. Biochem. 1975, 53, 284–292. [Google Scholar] [CrossRef]
- Essen, L.; Siegert, R.; Lehmann, W.D.; Oesterhelt, D. Lipid patches in membrane protein oligomers: Crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. USA 1998, 95, 11673–11678. [Google Scholar] [CrossRef]
- Dracheva, S.; Bose, S.; Hendler, R.W. Chemical and functional studies on the importance of purple membrane lipids in bacteriorhodopsin photocycle behavior. FEBS Lett. 1996, 382, 209–212. [Google Scholar] [CrossRef]
- Watts, A. Bacteriorhodopsin: The mechanism of 2D-array formation and the structure of retinal in the protein. Biophys. Chem. 1995, 55, 137–151. [Google Scholar] [CrossRef]
- Mitsuoka, K.; Hirai, T.; Murata, K.; Miyazawa, A.; Kidera, A.; Kimura, Y.; Fujiyoshi, Y. The structure of bacteriorhodopsin at 3.0 Å resolution based on electron crystallography: Implication of the charge distribution. J. Mol. Biol. 1999, 286, 861–882. [Google Scholar] [CrossRef]
- Sternberg, B.; L’Hostis, C.; Whiteway, C.A.; Watts, A. The essential role of specific Halobacterium halobium polar lipids in 2D-array formation of bacteriorhodopsin. Biochim. Biophys. Acta 1992, 1108, 21–30. [Google Scholar] [CrossRef]
- Bryl, K.; Yoshihara, K. The role of retinal in the long-range protein-lipid interactions in bacteriorhodopsin-phosphatidylcholine vesicles. Eur Biophys. J. Biophy 2001, 29, 628–640. [Google Scholar] [CrossRef]
- Cui, J.; Kawatake, S.; Umegawa, Y.; Lethu, S.; Yamagami, M.; Matsuoka, S.; Sato, F.; Matsumori, N.; Murata, M. Stereoselective synthesis of the head group of archaeal phospholipid PGP-Me to investigate bacteriorhodopsin–lipid interactions. Org. Biomol. Chem. 2015, 13, 10279–10284. [Google Scholar] [CrossRef]
- Hendler, R.W.; Barnett, S.M.; Dracheva, S.; Bose, S.; Levin, I.W. Purple membrane lipid control of bacteriorhodopsin conformational flexibility and photocycle activity. Eur. J. Biochem. 2003, 270, 1920–1925. [Google Scholar] [CrossRef]
- Hendler, R.W.; Dracheva, S. Importance of Lipids for Bacteriorhodopsin Structure, Photocycle, and Function. Biochemistry 2001, 66, 1311–1314. [Google Scholar] [CrossRef]
- Mukhopadhay, A.K.; Bose, S.; Hendler, R.W. Membrane-Mediated Control of the Bacteriorhodopsin Photocycle. Biochemistry 1994, 33, 10889–10895. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.K.; Dracheva, S.; Bose, S.; Hendler, R.W. Control of the Integral Membrane Proton Pump, Bacteriorhodopsin, by Purple Membrane Lipids of Halobacterium halobium. Biochemistry 1996, 35, 9245–9252. [Google Scholar] [CrossRef] [PubMed]
- Höjeberg, B.; Lind, C.; Khorana, H.G. Reconstitution of bacteriorhodopsin vesicles with Halobacterium halobium lipids. Effects of variations in lipid composition. J. Biol. Chem. 1982, 257, 1690–1694. [Google Scholar] [CrossRef]
- Joshi, M.K.; Dracheva, S.; Mukhopadhyay, A.K.; Bose, S.; Hendler, R.W. Importance of Specific Native Lipids in Controlling the Photocycle of Bacteriorhodopsin. Biochemistry 1998, 37, 14463–14470. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Manas, J.M.; Virto, M.D.; Gurtubay, J.I.; Goni, F.M. The interaction of Triton X-100 with purple membranes. Detergent binding, spectral changes and membrane solubilization. Eur. J. Biochem. 1990, 188, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Manas, J.M.; Goni, F.M.; Tribout, M.; Paredes, S. Kinetics of purple membrane dark-adaptation in the presence of Triton X-100. Arch. Biochem. Biophys. 1990, 282, 239–243. [Google Scholar] [CrossRef]
- Hu, K.; Sun, Y.; Chen, D.; Zhang, Y. The effect of lipid environment in purple membrane on bacteriorhodopsin. J. Photochem. Photobiol. B Biol. 2000, 58, 163–169. [Google Scholar] [CrossRef]
- Lopez, F.; Lobasso, S.; Colella, M.; Agostiano, A.; Corcelli, A. Light-dependent and Biochemical Properties of Two Different Bands of Bacteriorhodopsin Isolated on Phenyl-Sepharose CL-4B. Photochem. Photobiol. 1999, 69, 599–604. [Google Scholar] [CrossRef]
- Weik, M.; Patzelt, H.; Zaccai, G.; Oesterhelt, D. Localization of Glycolipids in Membranes by In Vivo Labeling and Neutron Diffraction. Mol. Cell 1998, 1, 411–419. [Google Scholar] [CrossRef]
- Barnett, S.M.; Dracheva, S.; Hendler, R.W.; Levin, I.W. Lipid-Induced Conformational Changes of an Integral Membrane Protein: An Infrared Spectroscopic Study of the Effects of Triton X-100 Treatment on the Purple Membrane of Halobacterium halobium ET1001. Biochemistry 1996, 35, 4558–4567. [Google Scholar] [CrossRef]
- Milder, S.J.; Thorgeirsson, T.E.; Miercke, L.J.W.; Stroud, R.M.; Kliger, D.S. Effects of detergent environments on the photocycle of purified monomeric bacteriorhodopsin. Biochemistry 1991, 30, 1751–1761. [Google Scholar] [CrossRef]
- Inada, M.; Kinoshita, M.; Matsumori, N. Archaeal Glycolipid S-TGA-1 Is Crucial for Trimer Formation and Photocycle Activity of Bacteriorhodopsin. ACS Chem. Biol. 2020, 15, 197–204. [Google Scholar] [CrossRef]
- Inada, M.; Kinoshita, M.; Sumino, A.; Oiki, S.; Matsumori, N. A concise method for quantitative analysis of interactions between lipids and membrane proteins. Anal. Chim. Acta 2019, 1059, 103–112. [Google Scholar] [CrossRef]
- Matsui, Y.; Sakai, K.; Murakami, M.; Shiro, Y.; Adachi, S.-I.; Okumura, H.; Kouyama, T. Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin. J. Mol. Biol. 2002, 324, 469–481. [Google Scholar] [CrossRef]
- Sato, H.; Takeda, K.; Tani, K.; Hino, T.; Okada, T.; Nakasako, M.; Kamiya, N.; Kouyama, T. Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 1251–1256. [Google Scholar] [CrossRef]
- Muller, M.P.; Jiang, T.; Sun, C.; Lihan, M.; Pant, S.; Mahinthichaichan, P.; Trifan, A.; Tajkhorshid, E. Characterization of Lipid-Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation. Chem. Rev. 2019, 119, 6086–6161. [Google Scholar] [CrossRef]
- Gu, R.-X.; de Groot, B.L. Lipid-protein interactions modulate the conformational equilibrium of a potassium channel. Nat. Commun 2020, 11, 2162. [Google Scholar] [CrossRef]
- Yang, J.; Aslimovska, L.; Glaubitz, C. Molecular Dynamics of Proteorhodopsin in Lipid Bilayers by Solid-State NMR. J. Am. Chem. Soc. 2011, 133, 4874–4881. [Google Scholar] [CrossRef]
- Agasid, M.T.; Robinson, C.V. Probing membrane protein–lipid interactions. Curr. Opin. Struct. Biol. 2021, 69, 78–85. [Google Scholar] [CrossRef]
- Corradi, V.; Sejdiu, B.I.; Mesa-Galloso, H.; Abdizadeh, H.; Noskov, S.Y.; Marrink, S.J.; Tieleman, D.P. Emerging Diversity in Lipid–Protein Interactions. Chem. Rev. 2019, 119, 5775–5848. [Google Scholar] [CrossRef]
- Duncan, A.; Corey, R.; Sansom, M. Defining how multiple lipid species interact with inward rectifier potassium (Kir2) channels. Proc. Natl. Acad. Sci. USA 2020, 117, 7803–7813. [Google Scholar] [CrossRef] [PubMed]
- Amos, S.T.A.; Kalli, A.C.; Shi, J.; Sansom, M.S.P. Membrane Recognition and Binding by the Phosphatidylinositol Phosphate Kinase PIP5K1A: A Multiscale Simulation Study. Structure 2019, 27, 1336–1346. [Google Scholar] [CrossRef] [PubMed]
- Chavent, M.; Duncan, A.L.; Sansom, M.S.P. Molecular dynamics simulations of membrane proteins and their interactions: From nanoscale to mesoscale. Curr. Opin. Struct. Biol. 2016, 40, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Crozier, P.S.; Stevens, M.J.; Woolf, T.B. How Environment Supports a State: Molecular Dynamics Simulations of Two States in Bacteriorhodopsin Suggest Lipid and Water Compensation. Biophys. J. 2004, 87, 129–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kandt, C.; Gerwert, K.; Schlitter, J. Water dynamics simulation as a tool for probing proton transfer pathways in a heptahelical membrane protein. Proteins Struct. Funct. Bioinform. 2005, 58, 528–537. [Google Scholar] [CrossRef]
- Tieleman, D.; Berendsen, H.; Sansom, M. An Alamethicin Channel in a Lipid Bilayer: Molecular Dynamics Simulations. Biophys. J. 1999, 76, 1757–1769. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.; Cerutti, D.; Cheatham, T.E.; Darden, T.; Duke, R.; Giese, T.J.; Gohlke, H.; Götz, A.; Homeyer, N. Amber 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuserial, G.E.; Robb, M.A. Gaussian 16; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Mayer, K.L.; Earley, M.R.; Gupta, S.; Pichumani, K.; Regan, L.; Stone, M.J. Covariation of backbone motion throughout a small protein domain. Nat. Struct. Biol. 2003, 10, 962–965. [Google Scholar] [CrossRef]
- Lange, O.F.; Grubmüller, H.; de Groot, B.L. Molecular Dynamics Simulations of Protein G Challenge NMR-Derived Correlated Backbone Motions. Angew. Chem. Int. Ed. 2005, 44, 3394–3399. [Google Scholar] [CrossRef]
- Ghosh, A.; Vishveshwara, S. A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 15711–15716. [Google Scholar] [CrossRef]
- Kumari, M.; Singh, R.; Subbarao, N. Exploring the interaction mechanism between potential inhibitor and multi-target Mur enzymes of mycobacterium tuberculosis using molecular docking, molecular dynamics simulation, principal component analysis, free energy landscape, dynamic cross-correlation matrices, vector movements, and binding free energy calculation. J. Biomol. Struct. Dyn. 2021, 1–30. [Google Scholar] [CrossRef]
- Luecke, H.; Schobert, B.; Richter, H.-T.; Cartailler, J.-P.; Lanyi, J.K. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 1999, 291, 899–911. [Google Scholar] [CrossRef]
- Wolf, S.; Freier, E.; Gerwert, K. A delocalized proton-binding site within a membrane protein. Biophys. J. 2014, 107, 174–184. [Google Scholar] [CrossRef]
- Goyal, P.; Ghosh, N.; Phatak, P.; Clemens, M.; Gaus, M.; Elstner, M.; Cui, Q. Proton storage site in bacteriorhodopsin: New insights from quantum mechanics/molecular mechanics simulations of microscopic pKa and infrared spectra. J. Am. Chem. Soc. 2011, 133, 14981–14997. [Google Scholar] [CrossRef]
- Phatak, P.; Ghosh, N.; Yu, H.; Cui, Q.; Elstner, M. Amino acids with an intermolecular proton bond as proton storage site in bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 2008, 105, 19672–19677. [Google Scholar] [CrossRef]
- Tripathi, R.; Forbert, H.; Marx, D. Settling the Long-Standing Debate on the Proton Storage Site of the Prototype Light-Driven Proton Pump Bacteriorhodopsin. J. Phys. Chem. B 2019, 123, 9598–9608. [Google Scholar] [CrossRef]
- Nakai, H.; Takemura, T.; Ono, J.; Nishimura, Y. Quantum-Mechanical Molecular Dynamics Simulations on Secondary Proton Transfer in Bacteriorhodopsin Using Realistic Models. J. Phys. Chem. B 2021, 125, 10947–10963. [Google Scholar] [CrossRef]
- Clemens, M.; Phatak, P.; Cui, Q.; Bondar, A.-N.; Elstner, M. Role of Arg82 in the Early Steps of the Bacteriorhodopsin Proton-Pumping Cycle. J. Phys. Chem. B 2011, 115, 7129–7135. [Google Scholar] [CrossRef][Green Version]
- Govindjee, R.; Imasheva, E.S.; Misra, S.; Balashov, S.P.; Ebrey, T.G.; Chen, N.; Menick, D.R.; Crouch, R.K. Mutation of a Surface Residue, Lysine-129, Reverses the Order of Proton Release and Uptake in Bacteriorhodopsin; Guanidine Hydrochloride Restores It. Biophys. J. 1997, 72, 886–898. [Google Scholar] [CrossRef][Green Version]
- Lórenz-Fonfría, V.A.; Furutani, Y.; Kandori, H. Active Internal Waters in the Bacteriorhodopsin Photocycle. A Comparative Study of the L and M Intermediates at Room and Cryogenic Temperatures by Infrared Spectroscopy. Biochemistry 2008, 47, 4071–4081. [Google Scholar] [CrossRef]
- Dioumaev, A.K.; Brown, L.S.; Needleman, R.; Lanyi, J.K. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry 1998, 37, 9889–9893. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.; Skopintsev, P.; James, D.; Dworkowski, F.; Panepucci, E.; Kekilli, D.; Furrer, A.; Brünle, S.; Mous, S.; Ozerov, D.; et al. Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography. Science 2019, 365, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Freier, E.; Wolf, S.; Gerwert, K. Proton transfer via a transient linear water-molecule chain in a membrane protein. Proc. Natl. Acad. Sci. USA 2011, 108, 11435–11439. [Google Scholar] [CrossRef] [PubMed]
- Otomo, J.; Tomioka, H.; Sasabe, H. Properties and the primary structure of a new halorhodopsin from halobacterial strain mex. Biochim. Biophys. Acta 1992, 1112, 7–13. [Google Scholar] [CrossRef]
- Chen, D.; Lanyi, J.K. Structural changes in the N and N’ states of the bacteriorhodopsin photocycle. Biophys. J. 2009, 96, 2779–2788. [Google Scholar] [CrossRef]
- Shibata, M.; Yamashita, H.; Uchihashi, T.; Kandori, H.; Ando, T. High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat. Nanotechnol. 2010, 5, 208–212. [Google Scholar] [CrossRef]
- Luecke, H.; Schobert, B.; Richter, H.T.; Cartailler, J.P.; Lanyi, J.K. Structural Changes in Bacteriorhodopsin During Ion Transport at 2 Angstrom Resolution. Science 1999, 286, 255–260. [Google Scholar] [CrossRef]
- Kato, H.E.; Kamiya, M.; Sugo, S.; Ito, J.; Taniguchi, R.; Orito, A.; Hirata, K.; Inutsuka, A.; Yamanaka, A.; Maturana, A.D.; et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat. Commun. 2015, 6, 7177. [Google Scholar] [CrossRef]
- Inoue, K. Diversity, Mechanism, and Optogenetic Application of Light-Driven Ion Pump Rhodopsins; Springer: Singapore, 2021; pp. 89–126. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2007, 2, e880. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Kandt, C.; Schlitter, J.; Gerwert, K. Dynamics of Water Molecules in the Bacteriorhodopsin Trimer in Explicit Lipid/Water Environment. Biophys. J. 2004, 86, 705–717. [Google Scholar] [CrossRef][Green Version]
- Grudinin, S.; Büldt, G.; Gordeliy, V.; Baumgaertner, A. Water Molecules and Hydrogen-Bonded Networks in Bacteriorhodopsin—Molecular Dynamics Simulations of the Ground State and the M-Intermediate. Biophys. J. 2005, 88, 3252–3261. [Google Scholar] [CrossRef]
- del Val, C.; Bondar, L.; Bondar, A.-N. Coupling between inter-helical hydrogen bonding and water dynamics in a proton transporter. J. Struct. Biol. 2014, 186, 95–111. [Google Scholar] [CrossRef]
- Feller, S.E.; MacKerell, A.D. An Improved Empirical Potential Energy Function for Molecular Simulations of Phospholipids. J. Phys. Chem. B 2000, 104, 7510–7515. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Schrödinger, LLC. Schrödinger Suites; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Schauperl, M.; Nerenberg, P.S.; Jang, H.; Wang, L.-P.; Bayly, C.I.; Mobley, D.L.; Gilson, M.K. Non-bonded force field model with advanced restrained electrostatic potential charges (RESP2). Commun. Chem. 2020, 3, 44. [Google Scholar] [CrossRef]
- Dickson, C.J.; Madej, B.D.; Skjevik, Å.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; DeBolt, S.; Ferguson, D.; Seibel, G.; Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Cerutti, D.S.; Duke, R.; Freddolino, P.L.; Fan, H.; Lybrand, T.P. A Vulnerability in Popular Molecular Dynamics Packages Concerning Langevin and Andersen Dynamics. J. Chem. Theory Comput. 2008, 4, 1669–1680. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- McDonald, I.K.; Thornton, J.M. Satisfying Hydrogen Bonding Potential in Proteins. J. Mol. Biol. 1994, 238, 777–793. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Sun, X.; Zhao, X. Investigating the Impact of Asp181 Point Mutations on Interactions between PTP1B and Phosphotyrosine Substrate. Sci. Rep. 2014, 4, 5095. [Google Scholar] [CrossRef]
- Cao, Z.; Ding, X.; Peng, B.; Zhao, Y.; Ding, J.; Watts, A.; Zhao, X. Novel expression and characterization of a light-driven proton pump archaerhodopsin 4 in a Halobacterium salinarum strain. Biochim. Biophys. Acta. 2015, 1847, 390–398. [Google Scholar] [CrossRef]
- Krebs, M.P.; Mollaaghababa, R.; Khorana, H.G. Gene replacement in Halobacterium halobium and expression of bacteriorhodopsin mutants. Proc. Natl. Acad. Sci. USA 1993, 90, 1987–1991. [Google Scholar] [CrossRef]
- Krebs, M.P.; Hauss, T.; Heyn, M.P.; RajBhandary, U.L.; Khorana, H.G. Expression of the bacterioopsin gene in Halobacterium halobium using a multicopy plasmid. Proc. Natl. Acad. Sci. USA 1991, 88, 859–863. [Google Scholar] [CrossRef]
- Cline, S.W.; Lam, W.L.; Charlebois, R.L.; Schalkwyk, L.C.; Doolittle, W.F. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 1989, 35, 148–152. [Google Scholar] [CrossRef]
- Ming, M.; Lu, M.; Balashov, S.P.; Ebrey, T.G.; Li, Q.; Ding, J. pH dependence of light-driven proton pumping by an archaerhodopsin from Tibet: Comparison with bacteriorhodopsin. Biophys. J. 2006, 90, 3322–3332. [Google Scholar] [CrossRef]
- Luecke, H.; Schobert, B.; Cartailler, J.-P.; Richter, H.-T.; Rosengarth, A.; Needleman, R.; Lanyi, J.K. Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin. J. Mol. Biol. 2000, 300, 1237–1255. [Google Scholar] [CrossRef] [PubMed]
- Lanyi, J.K.; Schobert, B. Crystallographic Structure of the Retinal and the Protein after Deprotonation of the Schiff Base: The Switch in the Bacteriorhodopsin Photocycle. J. Mol. Biol. 2002, 321, 727–737. [Google Scholar] [CrossRef]
- Wang, T.; Sessions, A.O.; Lunde, C.S.; Rouhani, S.; Glaeser, R.M.; Duan, Y.; Facciotti, M.T. Deprotonation of D96 in Bacteriorhodopsin Opens the Proton Uptake Pathway. Structure 2013, 21, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Schobert, B.; Brown, L.S.; Lanyi, J.K. Crystallographic Structures of the M and N Intermediates of Bacteriorhodopsin: Assembly of a Hydrogen-bonded Chain of Water Molecules Between Asp-96 and the Retinal Schiff Base. J. Mol. Biol. 2003, 330, 553–570. [Google Scholar] [CrossRef]
- Nango, E.; Royant, A.; Kubo, M.; Nakane, T.; Wickstrand, C.; Kimura, T.; Tanaka, T.; Tono, K.; Song, C.; Tanaka, R.; et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 2016, 354, 1552–1557. [Google Scholar] [CrossRef]
- Neutze, R.; Pebay-Peyroula, E.; Edman, K.; Royant, A.; Navarro, J.; Landau, E.M. Bacteriorhodopsin: A high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta. 2002, 1565, 144–167. [Google Scholar] [CrossRef]
- Wickstrand, C.; Dods, R.; Royant, A.; Neutze, R. Bacteriorhodopsin: Would the real structural intermediates please stand up? Biochim. Biophys. Acta. 2015, 1850, 536–553. [Google Scholar] [CrossRef]
- Lanyi, J.K. Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta. 2006, 1757, 1012–1018. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Ding, X.; Sun, C.; Wang, F.; He, X.; Watts, A.; Zhao, X. Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake. Int. J. Mol. Sci. 2022, 23, 6913. https://doi.org/10.3390/ijms23136913
Chen S, Ding X, Sun C, Wang F, He X, Watts A, Zhao X. Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake. International Journal of Molecular Sciences. 2022; 23(13):6913. https://doi.org/10.3390/ijms23136913
Chicago/Turabian StyleChen, Sijin, Xiaoyan Ding, Chao Sun, Fei Wang, Xiao He, Anthony Watts, and Xin Zhao. 2022. "Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake" International Journal of Molecular Sciences 23, no. 13: 6913. https://doi.org/10.3390/ijms23136913
APA StyleChen, S., Ding, X., Sun, C., Wang, F., He, X., Watts, A., & Zhao, X. (2022). Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake. International Journal of Molecular Sciences, 23(13), 6913. https://doi.org/10.3390/ijms23136913





