Single-Step Fast Tissue Clearing of Thick Mouse Brain Tissue for Multi-Dimensional High-Resolution Imaging
Abstract
:1. Introduction
2. Results
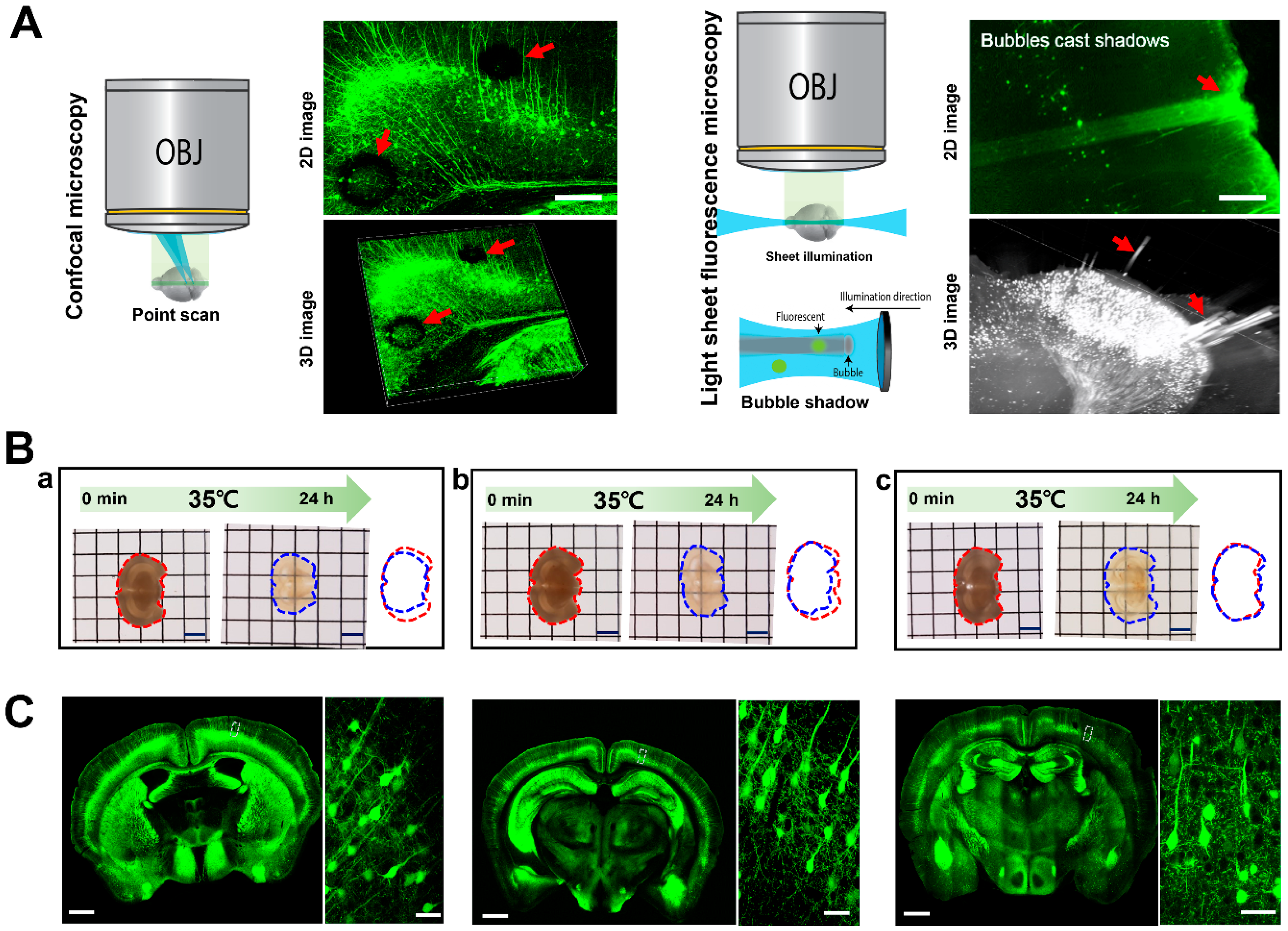
2.1. Limitations in Three-Dimensional Imaging for Cleared Tissue
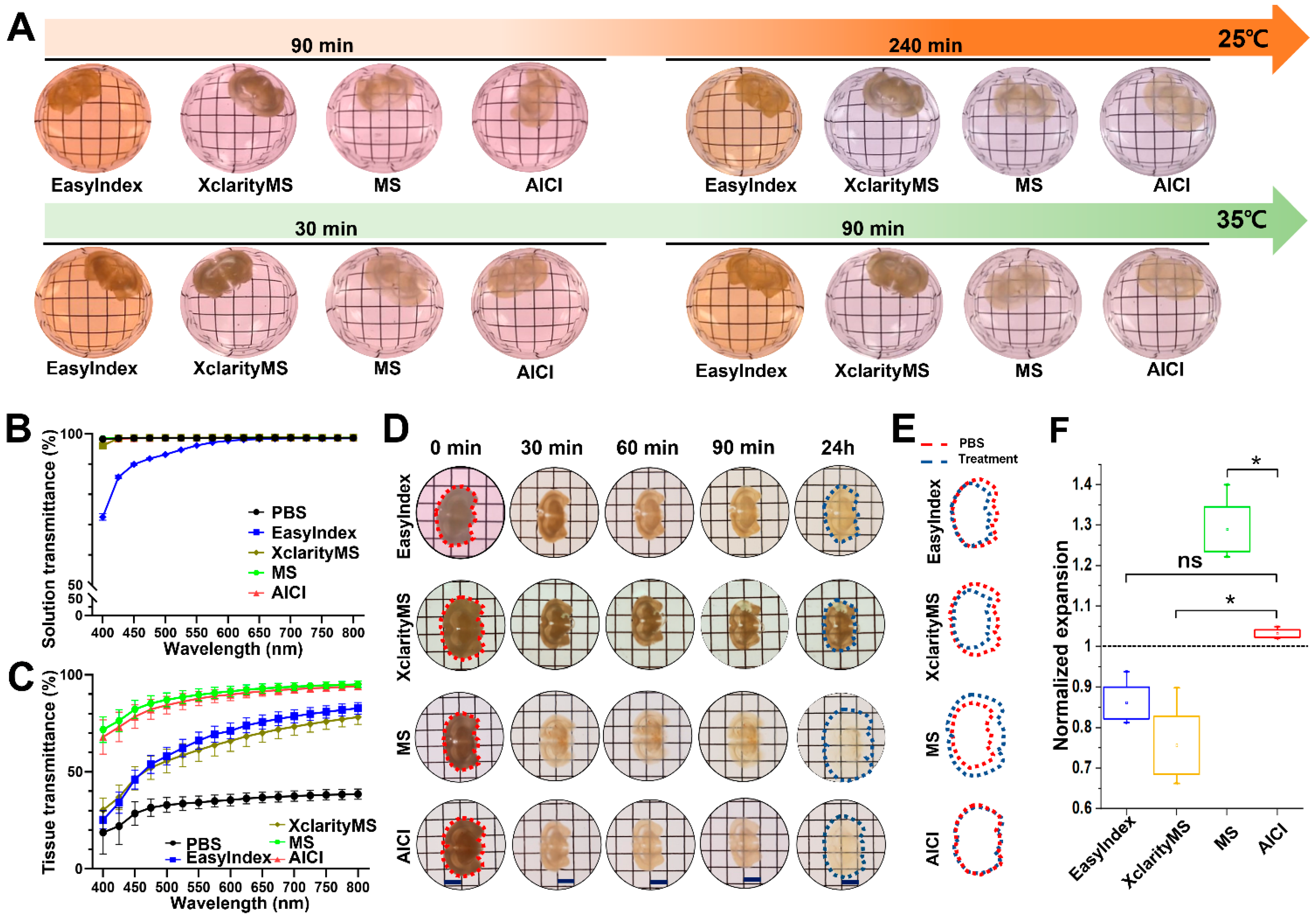
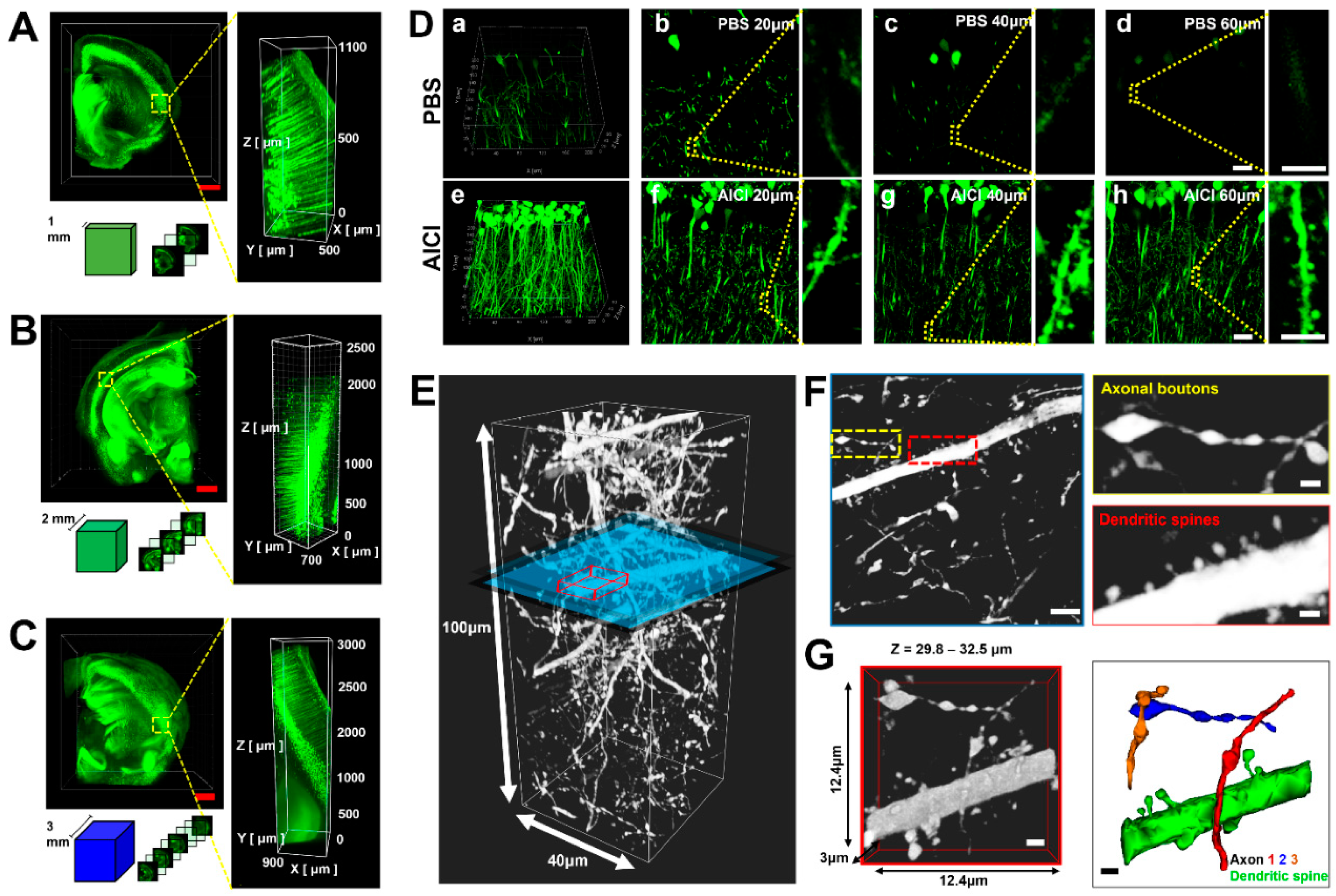
2.2. AICI Enables Rapid Tissue Clearing and Low Sample Distortion
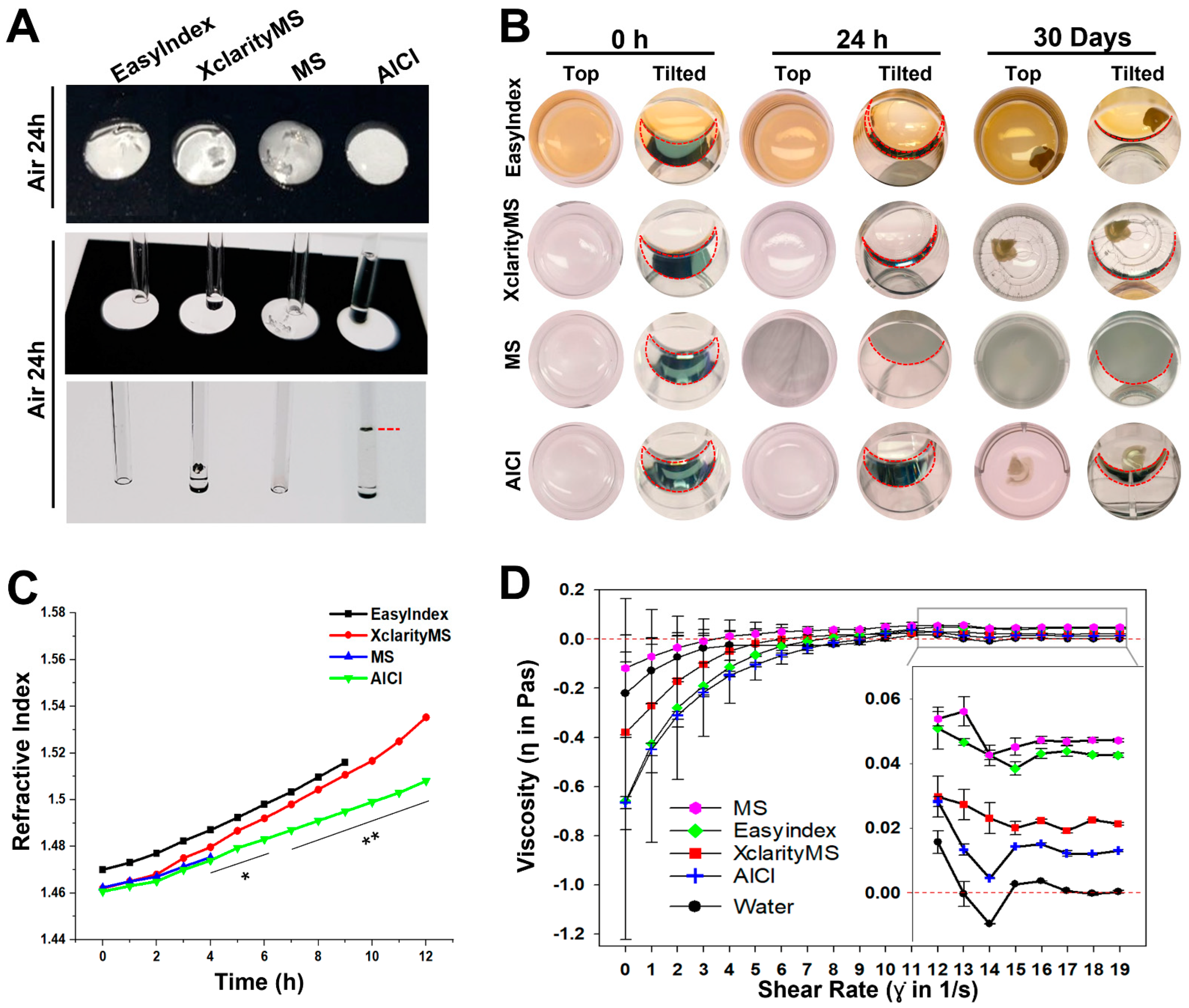
2.3. AICI Is an Aqueous, High-Refractive-Index Solution with Low Viscosity
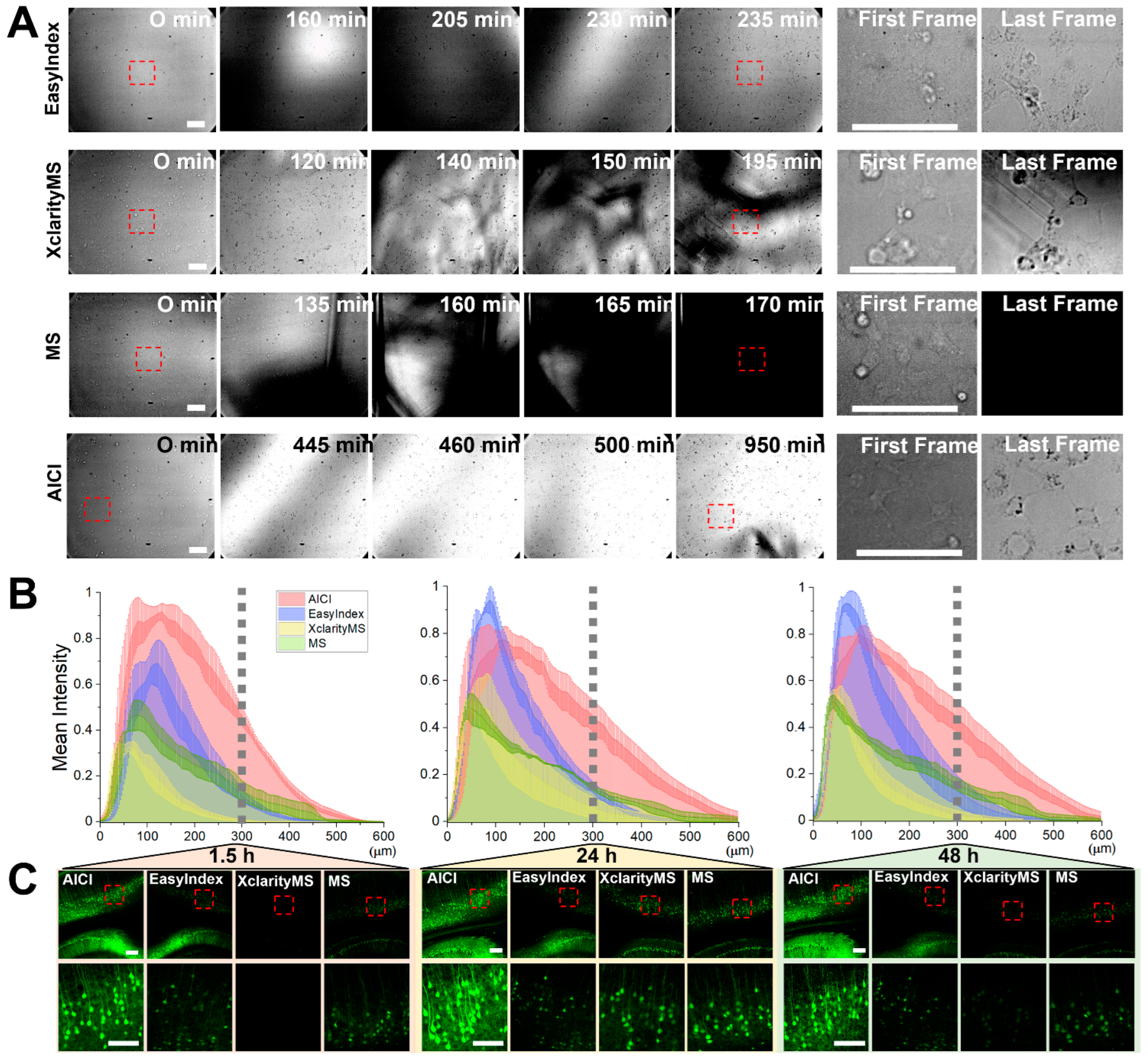
2.4. AICI Minimizes Reduction of Fluorescence Intensity and Distortion of Neuronal Structure
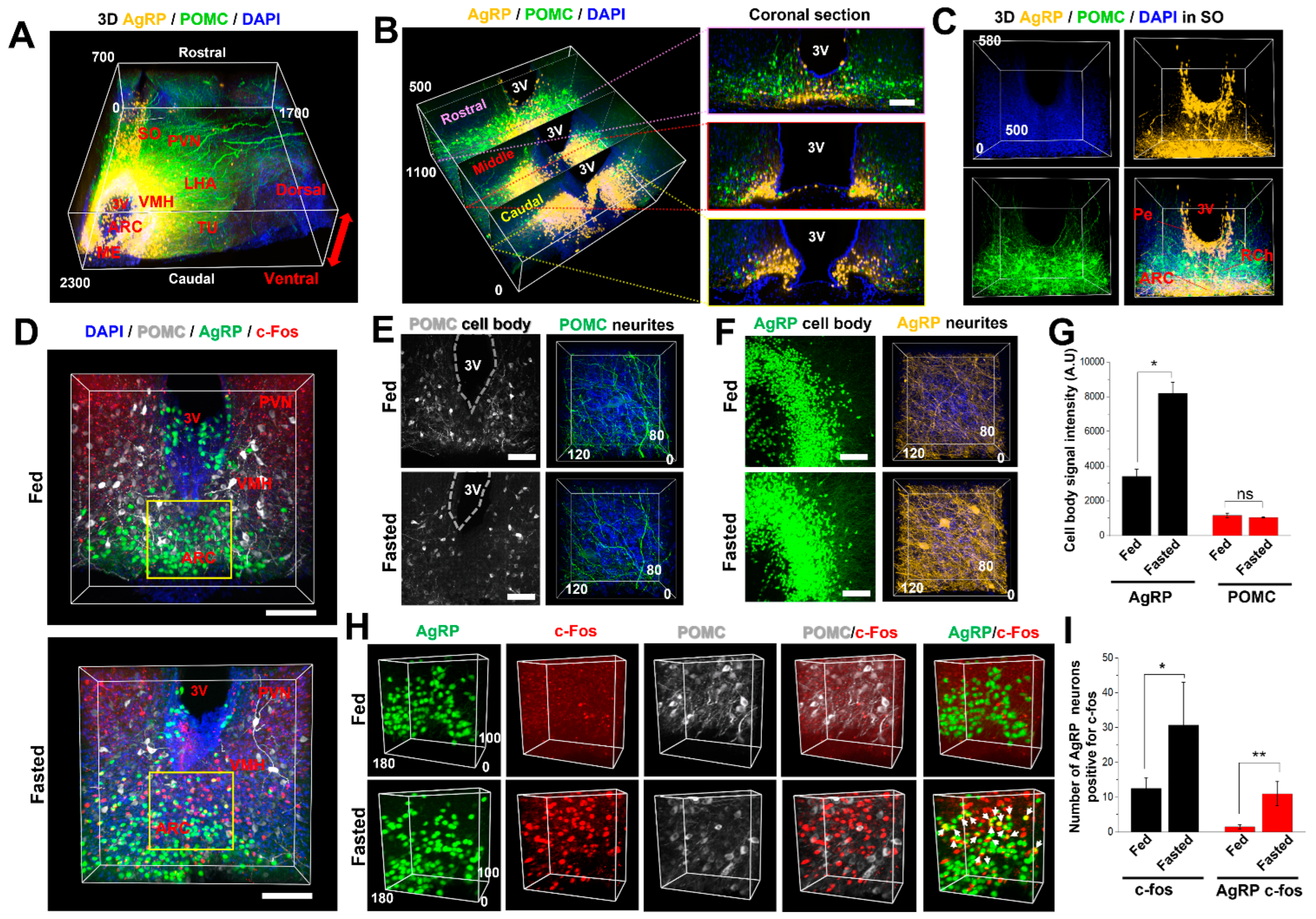
2.5. AICI Is Suitable for Three-Dimensional and Multifunctional Imaging Using Multicolor Fluorescence
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. AICI Reagent Preparation
4.3. Transparency and Linear Expansion of Brain Slice
4.4. Transmission Measurements
4.5. Hardening and Viscosity Measurement
4.6. Durability of Reagents for Imaging
4.7. 3D-Imaging of Brain Slices Treated with AICI
4.8. Immunostaining for Multifunctional Imaging by AICI
4.9. Stitching and 3D-Rendering of the Stack Images
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RI | Refractive index |
| WD | Working distance |
| PFS | Perfect focus system |
| ARC | Hypothalamic arcuate nucleus |
| AgRP | Agouti-related peptide |
| POMC | Pro-opiomelanocortin |
| ARH | Arcuate hypothalamic nucleus |
| VMH | Ventromedial hypothalamic nucleus |
| MPO | Medial preoptic nucleus |
| LHA | Lateral hypothalamic area |
| ME | Median eminence |
| TU | Tuberal nucleus |
| OCA | Optical clear agent |
| Pe | Periventricular hypothalamic nucleus |
| RCh | Retrochiasmatic area |
| SO | Supraoptic nucleus |
| PVN | Paraventricular nucleus |
| GABA | Gamma-Aminobutyric acid |
| FOCM | Ultrafast optical clearing method |
References
- Richardson, D.S.; Lichtman, J.W. Clarifying Tissue Clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, N.; Sato, T.R.; Betzig, E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Natl. Acad. Sci. USA 2012, 109, 22–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, P.S.; Kaufhold, J.P.; Blinder, P.; Friedman, B.; Drew, P.J.; Karten, H.J.; Lyden, P.D.; Kleinfeld, D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 2009, 29, 14553–14570. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.M.; Judkewitz, B.; Dimarzio, C.A.; Yang, C. Deep-tissue focal fluorescence imaging with digitally time-reversed ultrasound-encoded light. Nat. Commun. 2012, 3, 928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, K.; Fiolka, R.; Cui, M. Fluorescence imaging beyond the ballistic regime by ultrasound pulse guided digital phase conjugation. Nat. Photonics 2012, 6, 657–661. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Sun, W.; Richie, C.T.; Harvey, B.K.; Betzig, E.; Ji, N. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat. Commun. 2015, 6, 7276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Kong, L.; Zhou, Y.; Cui, M. Large-field-of-view imaging by multi-pupil adaptive optics. Nat. Methods 2017, 14, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Dodt, H.U.; Leischner, U.; Schierloh, A.; Jahrling, N.; Mauch, C.P.; Deininger, K.; Deussing, J.M.; Eder, M.; Zieglgansberger, W.; Becker, K. Ultramicroscopy: Three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 2007, 4, 331–336. [Google Scholar] [CrossRef]
- Erturk, A.; Becker, K.; Jahrling, N.; Mauch, C.P.; Hojer, C.D.; Egen, J.G.; Hellal, F.; Bradke, F.; Sheng, M.; Dodt, H.U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012, 7, 1983–1995. [Google Scholar] [CrossRef]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, J.; Wan, P.; Yu, T.; Zhu, D. Optimization of GFP Fluorescence Preservation by a Modified uDISCO Clearing Protocol. Front. Neuroanat. 2018, 12, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, C.; Cai, R.; Quacquarelli, F.P.; Ghasemigharagoz, A.; Lourbopoulos, A.; Matryba, P.; Plesnila, N.; Dichgans, M.; Hellal, F.; Erturk, A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 2016, 13, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Yu, T.; Xu, J.; Wan, P.; Ma, Y.; Zhu, J.; Li, Y.; Gong, H.; Luo, Q.; Zhu, D. FDISCO: Advanced solvent-based clearing method for imaging whole organs. Sci. Adv. 2019, 5, eaau8355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, C.; Becker, K.; Saghafi, S.; Pende, M.; Avdibasic, A.; Foroughipour, M.; Heinz, D.E.; Wotjak, C.T.; Dodt, H.U. High-resolution imaging of fluorescent whole mouse brains using stabilised organic media (sDISCO). J. Biophotonics 2019, 12, e201800368. [Google Scholar] [CrossRef] [Green Version]
- Hama, H.; Kurokawa, H.; Kawano, H.; Ando, R.; Shimogori, T.; Noda, H.; Fukami, K.; Sakaue-Sawano, A.; Miyawaki, A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011, 14, 1481–1488. [Google Scholar] [CrossRef]
- Susaki, E.A.; Tainaka, K.; Perrin, D.; Kishino, F.; Tawara, T.; Watanabe, T.M.; Yokoyama, C.; Onoe, H.; Eguchi, M.; Yamaguchi, S.; et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014, 157, 726–739. [Google Scholar] [CrossRef] [Green Version]
- Tainaka, K.; Kubota, S.I.; Suyama, T.Q.; Susaki, E.A.; Perrin, D.; Ukai-Tadenuma, M.; Ukai, H.; Ueda, H.R. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 2014, 159, 911–924. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.C.; Mano, T.; Saikawa, S.; Horiguchi, S.A.; Shigeta, D.; Baba, K.; Sekiya, H.; Shimizu, Y.; Tanaka, K.F.; Kiyonari, H.; et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 2018, 21, 625–637. [Google Scholar] [CrossRef]
- Hama, H.; Hioki, H.; Namiki, K.; Hoshida, T.; Kurokawa, H.; Ishidate, F.; Kaneko, T.; Akagi, T.; Saito, T.; Saido, T.; et al. ScaleS: An optical clearing palette for biological imaging. Nat. Neurosci. 2015, 18, 1518–1529. [Google Scholar] [CrossRef]
- Chung, K.; Wallace, J.; Kim, S.Y.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mirzabekov, J.J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomer, R.; Ye, L.; Hsueh, B.; Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014, 9, 1682–1697. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.T.; Fujimoto, S.; Imai, T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013, 16, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Vartak, M.; Rahman, S.; Madden, S.; Parameswaran, A.; Polyzotis, N. SeeDB: Efficient Data-Driven Visualization Recommendations to Support Visual Analytics. Proc. VLDB Endow. 2015, 8, 2182–2193. [Google Scholar] [CrossRef]
- Costantini, I.; Ghobril, J.P.; Di Giovanna, A.P.; Allegra Mascaro, A.L.; Silvestri, L.; Mullenbroich, M.C.; Onofri, L.; Conti, V.; Vanzi, F.; Sacconi, L.; et al. A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 2015, 5, 9808. [Google Scholar] [CrossRef] [Green Version]
- Aoyagi, Y.; Kawakami, R.; Osanai, H.; Hibi, T.; Nemoto, T. A rapid optical clearing protocol using 2,2′-thiodiethanol for microscopic observation of fixed mouse brain. PLoS ONE 2015, 10, e0116280. [Google Scholar] [CrossRef] [Green Version]
- Kuwajima, T.; Sitko, A.A.; Bhansali, P.; Jurgens, C.; Guido, W.; Mason, C. ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013, 140, 1364–1368. [Google Scholar] [CrossRef] [Green Version]
- Hou, B.; Zhang, D.; Zhao, S.; Wei, M.; Yang, Z.; Wang, S.; Wang, J.; Zhang, X.; Liu, B.; Fan, L.; et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 2015, 9, 19. [Google Scholar] [CrossRef]
- Staudt, T.; Lang, M.C.; Medda, R.; Engelhardt, J.; Hell, S.W. 2,2′-thiodiethanol: A new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 2007, 70, 1–9. [Google Scholar] [CrossRef]
- Meglinski, I.V.; Bashkatov, A.N.; Genina, E.A.; Churmakov, D.Y.; Tuchin, V.V. The enhancement of confocal images of tissues at bulk optical immersion. Laser Phys. 2003, 13, 65–69. [Google Scholar]
- Chiang, A.S.; Lin, W.Y.; Liu, H.P.; Pszczolkowski, M.A.; Fu, T.F.; Chiu, S.L.; Holbrook, G.L. Insect NMDA receptors mediate juvenile hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 37–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Cho, J.H.; Murray, E.; Bakh, N.; Choi, H.; Ohn, K.; Ruelas, L.; Hubbert, A.; McCue, M.; Vassallo, S.L.; et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. USA 2015, 112, E6274–E6283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamur, M.C.; Oliver, C. Permeabilization of cell membranes. Methods Mol. Biol. 2010, 588, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.; Lee, M.; Seo, J.M.; Park, H.S.; Cho, Y.E. Optimization of the optical transparency of rodent tissues by modified PACT-based passive clearing. Exp. Mol. Med. 2016, 48, e274. [Google Scholar] [CrossRef] [Green Version]
- Loren, M.; Crouzet, C.; Bahani, A.; Vasilevko, V.; Choi, B. Optical clearing potential of immersion-based agents applied to thick mouse brain sections. PLoS ONE 2019, 14, e0216064. [Google Scholar] [CrossRef]
- Feng, G.; Mellor, R.H.; Bernstein, M.; Keller-Peck, C.; Nguyen, Q.T.; Wallace, M.; Nerbonne, J.M.; Lichtman, J.W.; Sanes, J.R. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 2000, 28, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Betley, J.N.; Cao, Z.F.; Ritola, K.D.; Sternson, S.M. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell 2013, 155, 1337–1350. [Google Scholar] [CrossRef] [Green Version]
- Uner, A.G.; Kecik, O.; Quaresma, P.G.F.; De Araujo, T.M.; Lee, H.; Li, W.; Kim, H.J.; Chung, M.; Bjorbaek, C.; Kim, Y.B. Role of POMC and AgRP neuronal activities on glycaemia in mice. Sci. Rep. 2019, 9, 13068. [Google Scholar] [CrossRef]
- Timper, K.; Bruning, J.C. Hypothalamic circuits regulating appetite and energy homeostasis: Pathways to obesity. Dis. Model. Mech. 2017, 10, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Lemus, M.B.; Stark, R.; Bayliss, J.A.; Reichenbach, A.; Lockie, S.H.; Andrews, Z.B. The temporal pattern of cfos activation in hypothalamic, cortical, and brainstem nuclei in response to fasting and refeeding in male mice. Endocrinology 2014, 155, 840–853. [Google Scholar] [CrossRef] [Green Version]
- Cabral, A.; Fernandez, G.; Tolosa, M.J.; Rey Moggia, A.; Calfa, G.; De Francesco, P.N.; Perello, M. Fasting induces remodeling of the orexigenic projections from the arcuate nucleus to the hypothalamic paraventricular nucleus, in a growth hormone secretagogue receptor-dependent manner. Mol. Metab. 2020, 32, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Henry, F.E.; Sugino, K.; Tozer, A.; Branco, T.; Sternson, S.M. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife 2015, 4, e09800. [Google Scholar] [CrossRef] [PubMed]
- Oliet, S.H. Functional consequences of morphological neuroglial changes in the magnocellular nuclei of the hypothalamus. J. Neuroendocrinol. 2002, 14, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Prevot, V. Glial-neuronal-endothelial interactions are involved in the control of GnRH secretion. J. Neuroendocrinol. 2002, 14, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.W. Network of hypothalamic neurons that control appetite. BMB Rep. 2015, 48, 229–233. [Google Scholar] [CrossRef]
- Briggs, D.I.; Lemus, M.B.; Kua, E.; Andrews, Z.B. Diet-induced obesity attenuates fasting-induced hyperphagia. J. Neuroendocrinol. 2011, 23, 620–626. [Google Scholar] [CrossRef]
- Fekete, C.; Zseli, G.; Singru, P.S.; Kadar, A.; Wittmann, G.; Fuzesi, T.; El-Bermani, W.; Lechan, R.M. Activation of anorexigenic pro-opiomelanocortin neurones during refeeding is independent of vagal and brainstem inputs. J. Neuroendocrinol. 2012, 24, 1423–1431. [Google Scholar] [CrossRef]
- Pinto, S.; Roseberry, A.G.; Liu, H.; Diano, S.; Shanabrough, M.; Cai, X.; Friedman, J.M.; Horvath, T.L. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science 2004, 304, 110–115. [Google Scholar] [CrossRef]
- Atasoy, D.; Betley, J.N.; Su, H.H.; Sternson, S.M. Deconstruction of a neural circuit for hunger. Nature 2012, 488, 172–177. [Google Scholar] [CrossRef]
- Wu, Q.; Clark, M.S.; Palmiter, R.D. Deciphering a neuronal circuit that mediates appetite. Nature 2012, 483, 594–597. [Google Scholar] [CrossRef]
- Krashes, M.J.; Shah, B.P.; Koda, S.; Lowell, B.B. Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab. 2013, 18, 588–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.Y.; Han, Y.; Zhang, W.; Wang, S.R.; Wei, Y.C.; Li, S.S.; Lin, J.K.; Yan, J.J.; Chen, A.X.; Zhang, X.; et al. AGRP Neurons Project to the Medial Preoptic Area and Modulate Maternal Nest-Building. J. Neurosci. 2019, 39, 456–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Huang, L.; Zheng, Y.; Song, Y.; Xu, Q.; Wang, J.; Si, K.; Duan, S.; Gong, W. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. USA 2019, 116, 11480–11489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, Y.; Kim, Y.; Lim, H.R.; Kim, H.-J.; Park, B.S.; Kim, J.G.; Park, S.-J.; Ha, C.M. Single-Step Fast Tissue Clearing of Thick Mouse Brain Tissue for Multi-Dimensional High-Resolution Imaging. Int. J. Mol. Sci. 2022, 23, 6826. https://doi.org/10.3390/ijms23126826
Ryu Y, Kim Y, Lim HR, Kim H-J, Park BS, Kim JG, Park S-J, Ha CM. Single-Step Fast Tissue Clearing of Thick Mouse Brain Tissue for Multi-Dimensional High-Resolution Imaging. International Journal of Molecular Sciences. 2022; 23(12):6826. https://doi.org/10.3390/ijms23126826
Chicago/Turabian StyleRyu, Youngjae, Yoonju Kim, Hye Ryeong Lim, Hyung-Joon Kim, Byong Seo Park, Jae Geun Kim, Sang-Joon Park, and Chang Man Ha. 2022. "Single-Step Fast Tissue Clearing of Thick Mouse Brain Tissue for Multi-Dimensional High-Resolution Imaging" International Journal of Molecular Sciences 23, no. 12: 6826. https://doi.org/10.3390/ijms23126826
APA StyleRyu, Y., Kim, Y., Lim, H. R., Kim, H.-J., Park, B. S., Kim, J. G., Park, S.-J., & Ha, C. M. (2022). Single-Step Fast Tissue Clearing of Thick Mouse Brain Tissue for Multi-Dimensional High-Resolution Imaging. International Journal of Molecular Sciences, 23(12), 6826. https://doi.org/10.3390/ijms23126826





