Abstract
NRT1/PTR FAMILY (NPF) genes are characterized as nitrate and peptide transporters that played important roles in various substrates transport in plants. However, little is known about the NPF gene in tea plants. Here, a total of 109 CsNPF members were identified from the tea plant genome, and divided into 8 groups according to their sequence characteristics and phylogenetic relationship. Gene structure and conserved motif analysis supported the evolutionary conservation of CsNPFs. Many hormone and stress response cis-acting elements and transcription factor binding sites were found in CsNPF promoters. Syntenic analysis suggested that multiple duplication types contributed to the expansion of NPF gene family in tea plants. Selection pressure analysis showed that CsNPF genes experienced strong purifying selective during the evolution process. The distribution of NPF family genes revealed that 8 NPF subfamilies were formed before the divergence of eudicots and monocots. Transcriptome analysis showed that CsNPFs were expressed differently in different tissues of the tea plant. The expression of 20 CsNPF genes at different nitrate concentrations was analyzed, and most of those genes responded to nitrate resupply. Subcellular localization showed that both CsNPF2.3 and CsNPF6.1 were localized in the plasma membrane, which was consistent with the characteristics of transmembrane proteins involved in NO3- transport. This study provides a theoretical basis for further investigating the evolution and function of NPF genes.
1. Introduction
Nitrogen (N) is an essential nutrient for plant growth and development. Nitrate is a major N source for higher plants and also serves as a signal to regulate plant development [1,2,3]. The NO3− uptake system consists of low-affinity transport system (LATS) and high-affinity transport systems (HATS), which are mainly mediated by nitrate transporter 1/peptide transporter (NRT1/PTR) and nitrate transporter 2 (NRT2), respectively [4,5]. NRT1 belongs to the large PTR family, in which family members are also described as proton-coupled oligopeptide transporter (POT) or solute carrier 15 (SLC15) [5,6,7]. A study involving 31 fully sequenced plant genomes unified these proteins under the name NPF (NRT1/PTR FAMILY) [7].
The first functionally characterized NPF protein, AtNPF6.3/AtNRT1.1/CHL1, was identified in Arabidopsis thaliana as a nitrate transporter [8]. Since then, its homologs have been cloned and functionally characterized in various plants [9,10]. The NPF proteins well known for their roles in nitrate transport, are considered to be the main components of LATS of high NO3−. However, some NPF proteins, such as AtNPF6.3 in A. thaliana and MtNRT1.3 in Medicago truncatula, were dual-affinity transporters participating in both HATS and LATS [11,12]. These NPF proteins have a wide absorption range for both high and low nitrate concentrations. ZmNPF6.6 in Zea mays and MtNIP/LATD in M. truncatula have been shown to be high-affinity nitrate transporters [13,14]. To date, at least 21 Arabidopsis NPF members have been shown to transport nitrate, and many of them are also able to transport other substrates [15,16].
Tea plant, Camellia sinensis (L.) O. Kuntze, is an economically important crop and its leaves are the raw material for producing the non-alcoholic beverage “tea”. Tea is rich in tea polyphenols, theanine and other active substances beneficial to human health [17,18]. To improve the quality of tea, a large amount of nitrogen fertilizer was applied to increase the content of free amino acids in tea leaves. The N input in tea garden is about 2–5 times that of other crops [19]. Therefore, it is particularly important to understand the molecular mechanism of nitrogen uptake in tea plants. To date, NPF family genes have been identified in several species, such as Arabidopsis, rice [20], poplar [21], apple [22], sugarcane [23], wheat [16] and spinach [24]. However, research on NPF family gene in tea plants is still limited. Due to its important function, it is necessary to make further in-depth investigations of NPF genes in tea plants.
In this study, a total of 109 CsNPF genes were identified in the tea plant genome. The phylogenic distribution, chromosomal location, gene structure, conserved motifs, duplication pattern, and selection pressure of NPF genes in tea plant were systematically analyzed. Furthermore, we performed expression profile analysis of NPF genes in different tissues of tea plant. The expression of CsNPF genes under different nitrate concentrations was detected by reverse transcription-quantitative real-time PCR (RT-qPCR). This study provides a basis for studying the evolution and function of NPF genes in tea plant.
2. Results
2.1. Identification of Tea Plant CsNPFs
A total of 109 CsNPF protein sequences were identified from the tea plant genome (Tables S1 and S2). To better distinguish these genes, we named CsNPF genes as CsNPF1.X~CsNPF8.X according to the previously reported rules [7]. The lengths of the CsNPF proteins ranged from 72 (CsNPF5.17) to 957 (CsNPF2.5) amino acids, with molecular weight ranging from 7.8 kD to 105.81 kD and theoretical pI values ranging from 4.43 (CsNPF2.9) to 9.98 (CsNPF5.17). Subcellular localization prediction indicated that most CsNPFs were localized to the plasma membrane (Table S1).
2.2. Phylogenetic Analysis of CsNPFs
To investigate the evolutionary relationship of CsNPFs, multi-sequence alignment of the 53 AtNPFs, 85 PtNPFs, and 109 CsNPFs was conducted by using the clustalx1.83 software and the result was used for the construction of an unrooted phylogenetic tree. Notably, 36 CsNPFs were removed from the phylogenetic tree construction due to C-terminal or N-terminal deletions. According to the phylogenetic tree (Figure 1), the CsNPFs were divided into 8 subfamilies (NPF1–NPF8). Among the 8 subfamilies, the NPF5 subfamily (23) contained the most NPF genes, while the NPF6 subfamily (6) contained the least (Figure 2 and Table S2). All members of the NPF3, NPF6, and NPF7 subfamilies contained complete domains, while 70% of the members of the NPF4 subfamily contained incomplete domains.
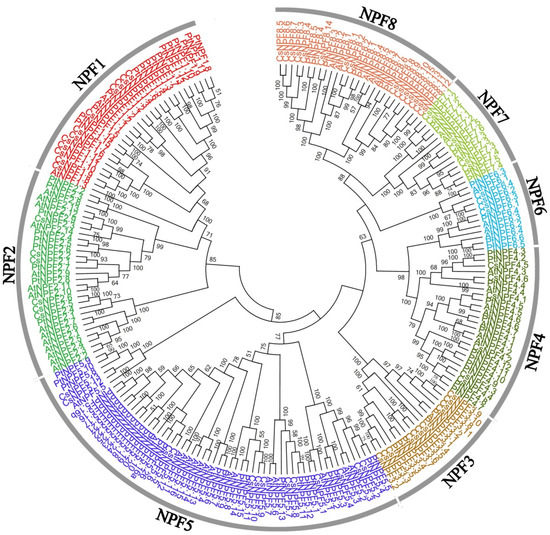
Figure 1.
Phylogenetic tree analysis of NPF proteins from C. sinensis, A. thaliana, and P. Trichocarpa.
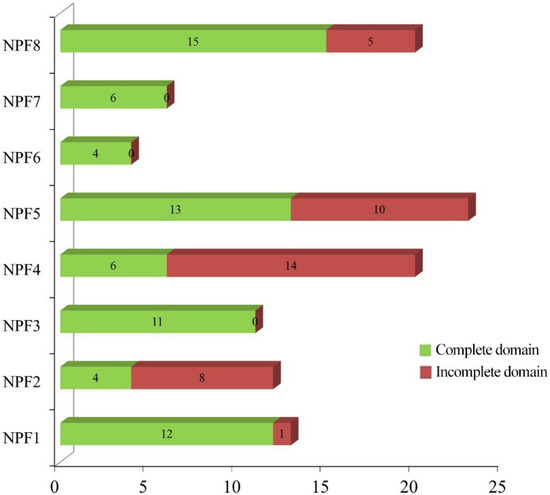
Figure 2.
Distribution of CsNPF genes in different subfamilies.
2.3. Distribution of NPF Members in Different Plant Species
To investigate the evolution of CsNPF in plant species, a comparison of NPF members in different plant species was conducted. Information on NPF family members in other species was extracted from previous studies [7,25]. A total of 36 plant species were analyzed, including 28 eudicots, 5 monocots, 1 other angiosperm (Amborella trichopoda), 1 spike moss (Selaginella moellendorffii), and 1 bryophyte (Physcomitrella patens). As shown in Figure 3, eight NPF subfamily members are present in almost all species, while NPF1 and NPF2 subfamily members are absent in lower plants (S. moellendorffii and P. patens). The NPF5 subfamily has the largest number of members of almost all species, especially in B. napus, with 63 members. The distribution of NPF members in eudicots and monocots was significantly different in different NPF subfamilies. The NPF1 and NPF2 subfamilies members were more abundant in eudicots than in monocots, while more NPF3, NPF7 and NPF8 subfamilies members were found in monocots. In the NPF6 subfamily, the number of NPF members in tea plant was less than the average of eudicots and monocots. However, in NPF1, NPF3, NPF4 and NPF8 subfamilies, the number of NPF members in tea plant was much more than the average number of eudicots, which was 1.8, 3.3, 1.8, and 2.9 times the number of of eudicots, respectively.
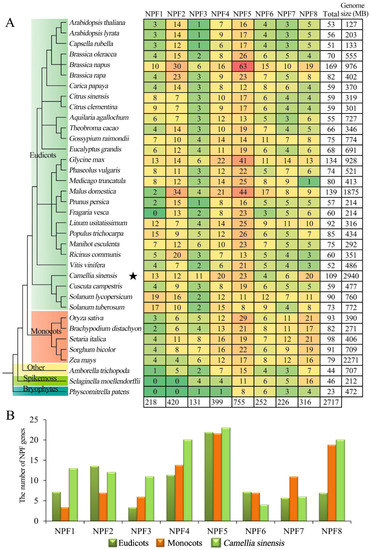
Figure 3.
Comparison of NPF genes in different species. (A) Classification of NPF genes from 36 plant species. (B) Distribution of NPF genes in tea plant compared with the average number in eudicots and monocots. The phylogenetic tree of the 36 plant species was downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi (accessed on 15 October 2021)) and reconstructed by MEGA5.
2.4. Gene Structure and Motif Analysis of CsNPFs
To characterize the protein sequences of the 73 CsNPF family members, 10 distinct motifs were predicted by the MEME program (Figure 4A). About 71% of CsNPF proteins contained all 10 conserved motifs, while other proteins appeared to be missing one or more of these motifs. Among them, all members of the NPF1 subfamily contain 10 motifs. Most closely related members share common motif compositions and arrangement orders. For example, motifs 1, 7, and 9 were lost in CsNPF2.5, and CsNPF2.6 of NPF2 subfamily. Motifs 3, 4 and 10 were absent in CsNPF5.9, CsNPF5.10 and CsNPF5.11 of NPF5 subfamily.
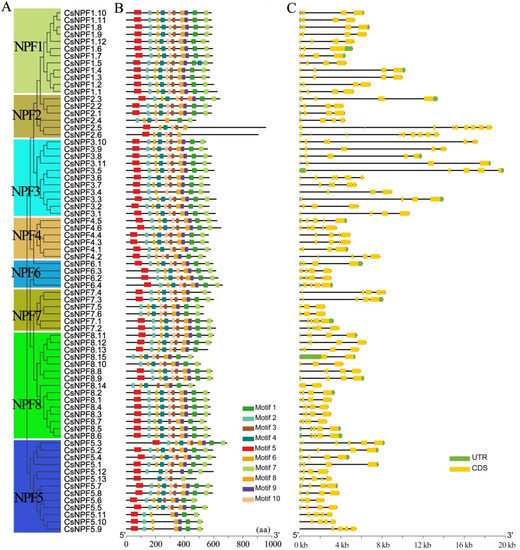
Figure 4.
Gene structure and conserved motif compositions of CsNPF genes. (A) Phylogenetic tree of CsNPFs. (B) Conserved motifs of CsNPF proteins. The motifs 1–10 are displayed in different colored boxes. (C) Gene structure of CsNPF genes. The green and yellow squares represent the untranslated region (UTR) and coding sequences (CDS), respectively.
Gene structure is an important parameter for gene evolution that further supports phylogenetic trees [26]. The coding sequences (CDS) of CsNPF genes were further analyzed to determine the structural diversity of these genes (Figure 4B). The numbers of introns of CsNPF genes ranged from 1 to 11. Of the 73 CsNPF genes, 53 (73%) had 3 or 4 introns. Fourteen CsNPF genes contained more than 4 introns, and 6 of them belonged to the NPF3 subfamily. CsNPF2.5 and CsNPF2.6 have the largest number of introns, with 10 and 11 introns respectively. However, both CsNPF8.14 and CsNPF8.15 contain only one intron. Overall, the most closely related CsNPF genes shared similar gene structures in terms of intron number and exon length.
2.5. Chromosome Distribution of CsNPFs
The chromosomal location of each CsNPF gene was analyzed based on tea plant genomic annotation information. As shown in Figure 5 and Table S1, 95 out of the 109 CsNPF were mapped on the 15 chromosomes, while the remaining 14 genes were located on Contig. The distribution of CsNPF on each chromosome was extremely uneven. For example, Chr10 possesses the most CsNPF genes (18), followed by Chr11 (15 genes), while Chr7 and Chr13 contain only one CsNPF gene. Notably, NPF3 subfamily genes have an obvious preference for Chr10, with 9 out of 10 CsNPF located on Chr10.
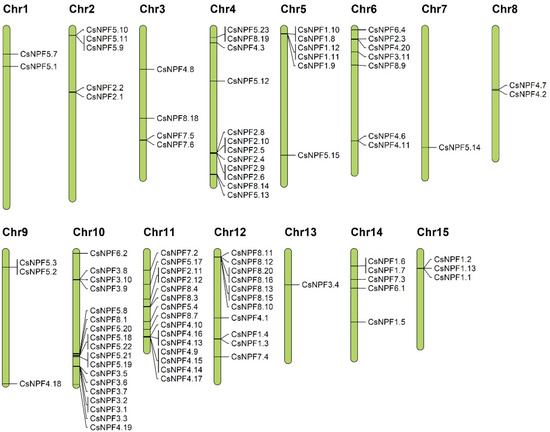
Figure 5.
Chromosomal locations of CsNPF genes in tea plant genome.
2.6. Expansion and Evolution of CsNPF Genes
To study the evolutionary processes of CsNPF family, the gene duplication pattern of CsNPF genes was analyzed (Figure 6 and Table S1). Thirty-three of the 109 CsNPF genes were found to be segmentally duplicated, 31 of which were located on chromosomes 1, 2, 4, 5, 6, 9, 10, 11, 12, 13, 14, and 15, and 2 on Contig (Figure 6A). There were 21 pairs of syntenic relationships in tea plant (Figure 6B). A total of 13 tandem repeats containing 20 CsNPF genes were identified. In addition, 25 and 31 CsNPFs were also found to be dispersed and proximal duplicates, respectively, which may also contribute to the expansion of the CsNPF family genes.
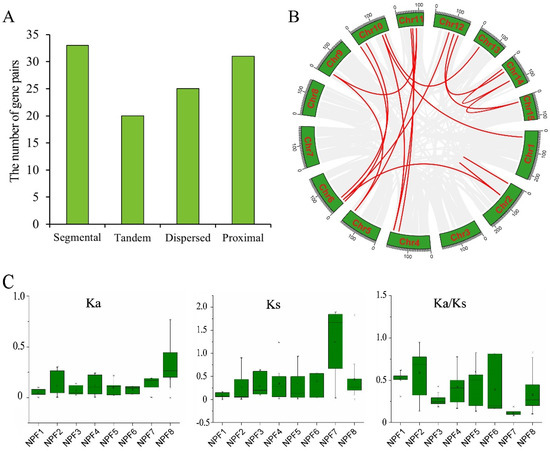
Figure 6.
The expansion and evolution analysis of NPF genes in tea plant. (A) The number of duplication types of CsNPF genes. (B) Syntenic relationships of CsNPF genes on tea plant chromosomes. (C) Values of Ka, Ks, and Ka/ Ks for CsNPF gene pairs of 8 subfamilies. Gray lines in the background indicate syntenic blocks in the whole tea plant genome, and red lines indicate segmental duplicates of CsNPF gene pairs.
To further study the selection pressure of tea plant NPF genes during evolution, the values of synonymous (Ks) and nonsynonymous (Ka) substitution rates, and Ka/Ks ratios for each duplicated pair from different subfamilies were calculated (Figure 6C and Table S3). The mean values of Ka of gene pairs in NPF1-NPF8 subfamilies were 0.05, 0.12, 0.07, 0.12, 0.10, 0.08, 0.14, and 0.09, respectively. The mean values of Ks were 0.09, 0.27, 0.29, 0.34, 0.35, 0.39, 1.24, and 0.44, respectively. The mean values of Ka/Ks were 0.52, 0.59, 0.27, 0.42, 0.6, 0.39, 0.11, and 0.33, respectively. Among the results, the Ks of the NPF7 subfamily was much higher than that of other NPF subfamilies, suggesting that they may have originated from more ancient duplication events. Except for CsNPF5.18/CsNPF5.19, the Ka/Ks ratio of almost all NPF duplicated pairs was less than 1, especially in the NPF7 subfamily. The results indicate that tea plant NPF genes might suffer from strong purifying selection throughout the long evolutionary events.
2.7. Synteny Analyses of NPF Genes between Tea Plant and Six Representative Species
To further explore the potential evolutionary mechanisms of the tea plant NPF family, we selected 6 representative species, including 4 eudicots (A. thaliana, P. trichocarpa, Vitis vinifera, and Solanum lycopersicu) and 2 monocots (O. sativa, and Sorghum bicolor), to construct comparative syntenic maps with tea plant (Figure 7 and Table S4). The results showed that the tea plant has more gene pairs with eudicots than with monocots. There were 75 pairs of syntenic pairs between tea plant and P. trichocarpa, followed by V. vinifera (65), S. lycopersicu (52), A. thaliana (45). However, there were only 7 and 6 pairs of syntenic relationships of tea plant with O. sativa, and S. bicolor, respectively. Among them, 3 NPF genes, CsNPF1.2, CsNPF2.2, and CsNPF7.4, were simultaneously identified between tea plant and the other six species, indicating that these genes may have vital roles in the evolution of the NPF gene family.
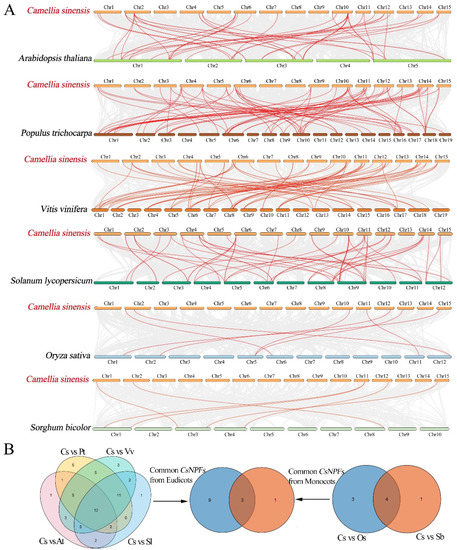
Figure 7.
Synteny analyses of the NPF genes between C. sinensis (Cs) and six representative plant species. (A) Synteny analyses of the NPF genes between C. sinensis and other six plant species. (B) Venn diagram analysis of collinear NPF genes between C. sinensis and other six plant species. Four eudicots include A. thaliana (At), P. trichocarpa (Pt), V. vinifera (Vv) and S. lycopersicum (Sl); 2 monocots include O. sativa (Os) and S. bicolor (Sb).
2.8. Regulatory Mechanism in the Promoter Regions of CsNPF Genes
To understand the potential functions of CsNPF genes, the cis-acting regulatory elements (CREs) in promoter regions of CsNPFs were predicted using PlantCARE. A total of 111 types of CREs were identified from 109 CsNPF gene promoters (Table S5). Each CsNPF contained 20 to 40 CREs in the promoter region, and some closely related genes showed similar CREs type. Several common CREs, such as CAAT-box and TATA-box and some light-responsive elements (G-box, Box 4, GT1-motif and TCT-motif) were obtained. Meanwhile, some CREs were shown to respond to stresses, such as drought-inducible MYB binding site elements (MBS, 50), low temperature-responsive elements (LTR, 67), wound-responsive elements (WUN-motif, 64), and anaerobic-inducible element (ARE, 100) (Figure 8A). Large numbers of CREs were involved in hormone responses, such as MeJA-responsive elements (TGACG-motif and CGTCA-motif), gibberellin-responsive elements (TATC-box, P-box and GARE-motif), Auxin (TGA-element and AuxRR-core), abscisic acid-responsive elements (ABREs), Ethylene (ERE) and salicylic acid reaction (TCA-elements) elements. In addition, many transcription factor (TF) binding sites were identified, such as WRKY binding sites (W box) and MYB binding sites (MRE, MYB, MBS, and MBSII).
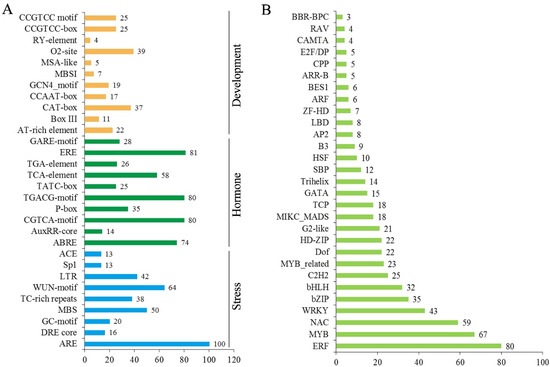
Figure 8.
Analysis of cis-acting regulatory elements and TF binding site in the CsNPF promoters. (A) Putative cis-acting regulatory elements of CsNPF genes. (B) The top 30 riched TF gene families with potential binding sites in the CsNPF promoter regions.
Transcription factors regulate the expression of target genes by binding to CREs in promoters. To further explore the transcriptional regulation of CsNPFs, we speculated the potential TFs that bind to the CsNPFs promoters (Figure 8B and Table S6). The results showed that 612 TF genes from 43 TF gene families had potential target binding sites in the CsNPFs promoter regions. Among the 612 TFs, MYB (90) gene family was the most abundant, followed by ERF (80), NAC (59), WRKY (43), bZIP (35), bHLH (32), and C2H2 (25) gene family.
2.9. Expression Patterns of CsNPFs in Different Tissues
To investigate the expression patterns of NPF genes in tea plant, we analyzed the expression levels of CsNPFs in 8 tissues (apical buds, young leaves, mature leaves, old leaves, immature stems, flowers, young fruits, and tender roots) (Figure 9 and Table S7). Of 109 CsNPF genes, 60 (55%) were ubiquitously expressed in all 8 tissues; 39 (36%) were low expressed with meanFPKM < 1. CsNPF genes showed different expression characteristics in different subfamilies. In NPF1, NPF6, and NPF8 subfamilies, most CsNPF genes were expressed at high levels in most tissues. In particular, all members of NPF6 subfamily genes were highly expressed in almost all tissues. Most members of the NPF4 and NPF7 subfamily genes showed lower expression levels. Some CsNPF genes displayed tissue-specific or preferential expression patterns. For example, in NPF3 subfamily, CsNPF2.6, CsNPF2.9 and CsNPF2.10 were preferentially expressed in young and mature leaves, while CsNPF2.7, CsNPF2.11 and CsNPF2.12 were preferentially expressed in root. In the NPF2 subfamily, CsNPF3.7 was preferentially expressed in root, while CsNPF3.10 was preferentially expressed in mature leaves. CsNPF5.13, CsNPF5.18, CsNPF5.21 and CsNPF5.22 in NPF4 subfamily were mainly expressed in root. In the NPF7 subfamily, CsNPF7.1, CsNPF7.3 and CsNPF7.6 had higher expression levels in flowers, while CsNPF7.3 and CsNPF7.4 were preferentially expressed in roots.
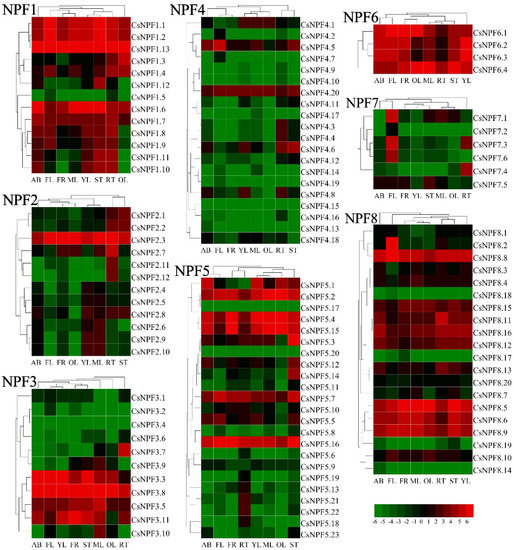
Figure 9.
Expression heatmap of CsNPF genes in 8 tissues of tea plant. The tissues are apical buds (AB), young leaves (YL), mature leaves (ML), old leaves (OL), young stems (ST), flowers (FL), young fruits (FR), and tender roots (RT). Gene expression level was evaluated log2 transformed RPKM value. Red represents high expression. Green indicates low expression.
2.10. Expression of CsNPFs under Different Nitrate Concentrations
Nitrate is the main substrate for NPF protein transport. Here, the expression profiles of 20 CsNPF genes were detected under different nitrate concentrations. The 20 CsNPF genes were selected from the 8 subfamilies, which were representative and can explain the expression profiles of genes from different subfamilies (Figure 10). CsNPF1.2, CsNPF1.8, CsNPF3.8, CsNPF5.4, and CsNPF5.7 were down-regulated with the increase of nitrate concentration. CsNPF2.2 was detected only at 0.1 mM nitrate concentration. CsNPF6.1 and CsNPF6.4 were decreased under 0.1 mM nitrate concentration. CsNPF2.12, CsNPF4.1 and CsNPF4.8 were induced under low nitrogen and then decreased with increasing nitrogen concentration. CsNPF7.3 and CsNPF7.4 showed an opposite trend, decreasing first and then increasing.
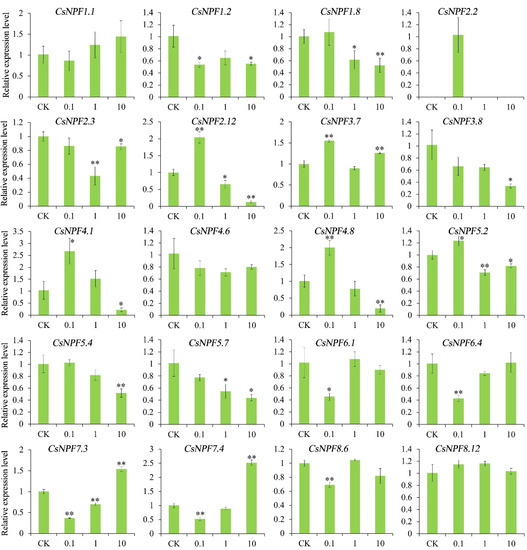
Figure 10.
Expression of selected CsNPF genes under different nitrate concentrations. Error bars represent standard deviation (SD). “*” and “**” represent the significance level of 0.05 and 0.01, respectively.
2.11. Subcellular Localization of CsNPF2.3 and CsNPF6.1
The CsNPF2.3-GFP fusion gene, CsNPF6.1-GFP fusion gene and empty vector containing GFP were transformed into tobacco epidermal cells. The results showed that the fusion proteins CsNPF2.3 and CsNPF6.1 were specifically located in the plasma membrane of tobacco leaves cells, and control GFP signals were observed in the nucleus, cytoplasm, and cell membrane (Figure 11). These results indicated that CsNPF2.3 and CsNPF6.1 were located in the plasma membrane, which was consistent with the results predicted by bioinformatics and consistent with the characteristics of transmembrane proteins involved in NO3− transport.
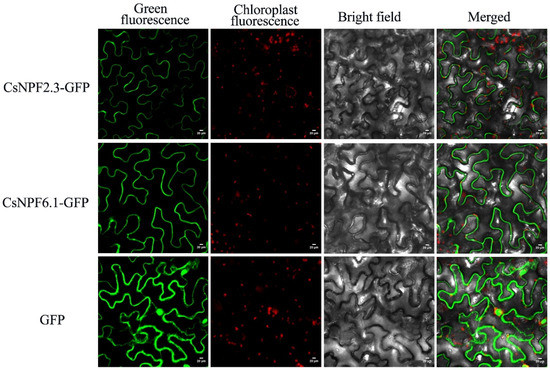
Figure 11.
Subcellular localization of CsNPF2.3 and CsNPF6.1 proteins.
3. Discussion
NPF genes comprise a large family of members widely distributed in eukaryotes. An increasing number of studies has confirmed that NPF genes are involved in the transport of nitrate/nitrite [8], di/tri-peptides [27], and various hormones [28,29,30]. As an important cash crop, tea plant is widely planted in China and all over the world because of its functional activity and important economic value. Tea plant is a plant that needs a lot of nitrogen fertilizer [19]. So far, functional studies of NPF gene have been carried out in Arabidopsis, rice, and other plants, but almost no reports on tea plant. In this study, 109 CsNPF genes were identified from the tea plant genome and divided into 8 subgroups based on the similarity of structure.
It is widely believed that the variation in gene structure is one of the representative traces of gene family evolution. According to the analysis of gene structure and conserved motifs, CsNPF was relatively conserved structurally. Most CsNPFs contain all 10 motifs, and no specific motifs belonging to a subfamily have been found. In the same subfamily, some closely related genes show the same motif deletion. Analysis of gene structure showed that most CsNPF genes (73%) contained 3 or 4 introns. Similar results have been reported in Arabidopsis, P. trichocarpa, soybean [31], and B. napus [25]. On the whole, CsNPFs within the same subfamily shared similar gene structures and motifs. Changes in gene structure and conserved motifs may be important reasons for the functional diversification of tea plant NPF family genes.
Gene duplication and loss events are the main drivers of species evolution, promoting the expansion of gene families and the generation of novel genes [32]. NPF genes are widely distributed in all subfamilies of 36 species, although NPF1 and NPF2 subfamilies are absent in lower plants S. Moellendorffii and P. Patens. In original angiosperms, all 8 subfamilies were present, suggesting that the duplication and diversification of NPF genes may have occurred before the differentiation of eudicots and monocots, and that NPF gene duplication in eudicots and monocots was an independent process. More NPF1 and NPF2 subfamily members were found in eudicots, while more NPF3, NPF7 and NPF8 subfamily members were found in monocots. The number of NPF1, NPF3, NPF4 and NPF8 subfamily members in tea plant was higher than that in eudicots, indicating these subfamilies seem to be more active in duplication and may have more function in the tea plant.
Gene family expansion mainly depends on tandem, segmental, or whole-genome duplications [33,34]. Recently completed genome sequencing of the tea plant indicated that tea plant has undergone two rounds of whole-genome duplications (WGD) [35,36,37]. Moreover, the large number of genes in the CsNPF family suggests that it evolved through many duplication events in tea plant. In B. napus, 137 (71%) of 193 BnaNPFs were found to be segmentally duplicated, and only 2 genes were identified as tandem duplication, suggesting that segmentation duplication was the main driver of NPF gene family expansion in B. napus [34]. Here, 33 (30%) and 20 (18%) of 109 CsNPFs were identified involved in segmental duplication and tandem events, respectively. Dispersed and proximal duplicates were also identified in the tea plant NPF gene family. These results indicated that the expansion forms of the NPF gene family in tea plant were more diversified. The number of NPF3 subfamily in tea plant was 3.3 times that in eudicots. Notably, there are two segmental and two tandem duplications in the NPF3 subfamily of tea plant, which may account for the higher number of NPF3 subfamily genes in tea plant than in other species. Few gene pairs were identified between tea plant and two monocots; while numerous colinearity gene pairs were found between tea plant and other 4 eudicots because of their close relationship. The results showed that the NPF gene differentiated rapidly in monocots and eudicots. In addition, three CsNPF genes, CsNPF1.2, CsNPF2.2, and CsNPF7.4, were colinear paired with all six species, suggesting that these colinear pairs already existed before the differentiation of monocots and eudicots ancestors.
The expression pattern of a gene is usually closely related to its CREs [38]. Many CREs related to development, stress, and phytohormone were detected in the CsNPF promoter regions, suggesting that CsNPFs may play different roles in biological processes. Numerous studies have demonstrated that NPF can transport hormone substrates. In Arabidopsis, at least 9 NPF members have ABA transport function and 18 NPF members have GA transport function [28]. In B. napus, 32.66% of BnaNPF expressions (65/199 gene) were regulated by at least one hormone [39]. In this study, most cis-elements involved in phytohormone responses, such as MeJA- (80/109 genes), Ethylene- (81/109), and ABA-responsive CRE (74/109 genes), were found in a series of CsNPF promoters, suggesting their potential hormone-inducing characteristics. The absorption and transport of nitrogen plays an important role in plant stress adaptation [40,41]. AtNPF6.3/AtNRT1.1 is involved in regulating drought tolerance of Arabidopsis [42]. AtNRT1.5 plays a negative role in salt tolerance and drought resistance in Arabidopsis, regulating the long-distance transport and spatial distribution of Na+, and affecting the expression of stress-responsive genes [43]. Many stress-related elements related to low temperature (LTR), drought (MBS), anaerobic (ARE) and wound (WUN-Motif) were also identified in the CsNPF promoters. Previous studies have shown the transcription factor AtERF59 binds directly to the AtNRT1.8 promoter and ethylene insensitive 3 (EIN3) binds to the AtNRT1.5 promoter [44]. Arabidopsis AtMYB59 positively regulates AtNRT1.5 expression to coordinate root-to-shoot K+/NO3– transport [45]. Many transcription factor binding sites were found in the CsNPF promoter regions. The results suggested that the expression of CsNPF genes might be regulated by multiple transcription factors.
Genes expressed in given tissues may have specific tissue functions, which provides crucial clues to understanding gene functions [46]. The CsNPF genes showed complex expression patterns, and closely related genes in the same subfamily showed similar expression patterns, reflecting the consistency of structure and function. Some CsNPF genes showed obvious tissue preference. For example, CsNPF2.6, CsNPF2.9, CsNPF2.10 were mainly expressed in leaf. CsNPF4.5, CsNPF7.3, CsNPF7.6, CsNPF8.2 showed preferred expression in flower. Most of the tissue-specific expression genes are preferentially expressed in root, such as CsNPF2.7, CsNPF2.11, CsNPF2.12, CsNPF3.7, CsNPF5.13, CsNPF5.21, CsNPF5.22, CsNPF7.3, CsNPF7.4. Similarly, most rice tissue-specific genes were preferentially expressed in root [20]. AtNPF6.3/AtNRT1.1 was a dual-affinity nitrate transporter mainly expressed in Arabidopsis root and involved in nascent organ development [47]. In T. aestivum, TaNPF6.2 was predominantly expressed in root; TaNPF6.1 was highly expressed in different tissues; while TaNPF6.4 was preferentially expressed in spike and node, but low in root and leaf [16]. All 4 CsNPF6 genes were highly expressed in tea plant except for the root and old leaf. The variation in expression patterns suggested the divergence of AtNPF6.3 orthologues in different species.
Nitrate is the main substrate for NPF protein transport [15,16]. In Arabidopsis, more than a third of the NPF genes have been shown to transport nitrates [16]. AtNPF4.6/AtNRT1.2 has been reported to be expressed mainly in the root of Arabidopsis and is involved in constitutive nitrate uptake [48]. In rice, OsNPF4.5 plays a key role in mycorrhizal NO3− acquisition [49]. Here, the transcripts of CsNPF4.1 and CsNPF4.8 in tea plant were significantly up-regulated under low nitrate conditions while down-regulated under high nitrate conditions. AtNPF7.3/AtNRT1.5 has been reported to be involved in nitrate loading in root xylem and transport nitrate from root to shoot [50]. OsNPF7.3 was mainly expressed in lateral root and stem of rice, and could improve nitrogen utilization efficiency in rice paddy [51]. Its homologous genes in tea plant were CsNPF7.3 and CsNPF7.4, which were preferentially expressed in root, and first decreased and then increased with the increase of nitrate concentration. In this study, the expression of several CsNPF genes changed significantly, indicating that these genes may be involved in nitrate uptake.
4. Materials and Methods
4.1. Plant Materials and Treatments
Fourteen-month-old cutting seedlings from tea plant, Zhongming 6 hao, were selected for this study. As previously [52], tea seedlings were cultured in full nitrogen hydroponic solution for 2 months. Then the seedlings were transferred to the nitrogen-deficient (without N) nutrient solution for 2 weeks of N-starvation treatment. Afterward, the seedlings were cultivated in the nutrient solution containing 0.1, 1, and 10 mM nitrate, respectively. Nitrogen starvation was used as the control (CK). The white roots were harvested after 2 h of treatment, rapidly frozen in liquid nitrogen, and then stored at −80 °C.
4.2. Identification of Tea Plant NPF Genes
Fifty-three NPF sequences of A. thaliana were obtained from the TARI website (http://www.arabidopsis.org/ (accessed on 15 July 2021)). The tea plant genome sequences were downloaded from the Tea Plant Information Archive (TPIA) website (http://tpdb.shengxin.ren/ (accessed on 15 July 2021)) [36]. The putative CsNPFs of tea plant were obtained via screening tea plant genome sequences by BlastP search, using AtNPFs as queries. The candidate sequences were then submitted to Pfam (http://pfam.xfam.org/ (accessed on 15 July 2021)) and NCBI CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 15 July 2021)) to confirm the presence of the MFS_1 or PTR2 (PF00854) domain. TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/ (accessed on 20 July 2021)) was applied for prediction of putative transmembrane (TM) regions of CsNPFs. Subcellular localization was predicted by the WoLF PSORT server (https://wolfpsort.hgc.jp/ (accessed on 20 July 2021)).
4.3. Phylogenetic Analysis and Structural Characterization
Sequence alignments of NPF proteins from tea plant, A. thaliana, and P. trichocarpa were performed clustalx1.83 software with default parameters. The unrooted phylogenetic tree was constructed using MEGA 5.0 software with the neighbor-joining (NJ) methods, bootstrapping with 1000 replicates. NPF sequences of A. thaliana and P. trichocarpa were obtained from previous reports (Table S8) [53]. The exon/intron structures of CsNPF genes were obtained from tea plant genome annotation [36]. Conserved motifs were analyzed using the Multiple Em for Motif Elicitation 5.4.1 (MEME, https://meme-suite.org/meme/ (accessed on 9 November 2021)) program.
4.4. Cis-Element Analysis
The sequence of 2000 bp upstream of the CsNPF gene transcription start site (ATG) was considered as promoter sequence. These promoter sequences were used to evaluate the putative cis-acting regulatory elements (CREs) using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 9 November 2021)). The transcription factor (TF) binding sites in CsNPF promoter sequences were predicted using the PlantTFDB database (http://planttfdb.gao-lab.org/ (accessed on 9 November 2021)).
4.5. Chromosomal Arrangement and Gene Duplication of CsNPF Genes
Physical positions of 109 CsNPF genes were extracted from the tea plant genome database. Mapchart software was used to visualize the chromosome localization [54]. Multiple Collinear Scan Toolkit (MCScanX) was used to determine the gene duplication events and collinearity relationships of tea plant NPF genes [55]. A chromosome region containing two or more homologous genes within a 200 kb range is defined as a tandem duplication event. The syntenic analysis maps of tea plant and other selected plants were constructed and visualized using the MCScanX and TBtools software [55,56]. The synonymous (Ks) and nonsynonymous (Ka) substitution rates, and Ka/Ks ratios for each duplicated pair (similarity of aligned regions > 70%) were calculated based on CDS alignments of the CsNPF genes using TBtools [56].
4.6. Transcript Abundance Analysis
For expression profiling of tea plant NPF genes, we utilized the transcriptome sequencing data of eight tissues of tea plant that were derived from the Tea Plant Information Archive (TPIA) (http://tpia.teaplant.org/ (accessed on 20 July 2021)) [35]. The eight tea plant tissues were apical buds (AB), young leaves (YL), mature leaves (ML), old leaves (OL), immature stems (ST), flowers (FL), young fruits (FR), and tender roots (RT). The expression cluster of CsNPF was analyzed and displayed by HemI 1.0 software.
4.7. RNA Isolation, Reverse Transcription, and RT-qPCR Detection
Total RNAs were extracted from samples using the EASYspin plant RNA extraction kit (Aidlab Biotechnologies, Beijing, China), and first-strand cDNA synthesis was performed using the PrimeScript RT reagent kit (TaKaRa, Dalian, China). RT-qPCR was performed using a LightCycler 480 machine (Roche Diagnostics) with SYBR Green reagents (Takara, Japan). RT-qPCR was performed in a 10 μL reaction mixture consisting of 5 µL of SYBR Premix Ex, 1 µL of diluted template cDNA, 0.3 µL of each primer, and 3.4 μL of ddH2O. The PCR amplification profile was as follows: denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, 58 °C for 10 s and 72 °C for 15 s. Three independent PCR reactions were performed on each gene, and the relative gene expression was calculated using the 2−∆∆CT method [57]. Tea plant GAPDH was selected as the reference gene [52]. All primers used are listed in Table S9.
4.8. Subcellular Localization
Subcellular localization analysis of tobacco epidermis was according to Zhang et al.’s method [58]. The CsNPF2.3 and CsNPF6.1 genes of the deletion termination codon were amplified using specific primers (Table S9) and inserted into the pBWA(V)HS-GFP vector. The constructed vector plasmid was transformed into Agrobacterium GV3101. The Agrobacterium cells were injected into the lower epidermis of one-month-old tobacco leaves and cultured under low light for 2 days. The GFP signals were observed and imaged under the confocal laser microscope (Zeiss, Germany).
5. Conclusions
This study performed a comprehensive analysis of NPF genes in tea plant. A total of 109 CsNPF genes were identified from the tea plant genome, and their gene structure, conserved motifs, chromosome location, cis-elements, gene duplication, evolutionary relationships, and expression patterns were systematically analyzed. Gene structure and motif analysis indicated that the CsNPF proteins were highly conserved in each subfamily. Syntenic analysis revealed that the expansion of tea plant NPF gene family was caused by multiple duplication types. The CsNPF family genes of tea plant experienced strong purifying selective pressure during evolution. Most of the selected CsNPF genes were induced by nitrate treatments. In addition, we proved that CsNPF2.3 and CsNPF6.1 were localized in the plasma membrane, which was consistent with the characteristics of transmembrane proteins involved in NO3− transport. These results provide a valuable basis for studying the evolution and function of CsNPF genes, and aid in improving N absorption in tea plant breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23126663/s1.
Author Contributions
Conceptualization, L.W. (Liyuan Wang) and H.C.; methodology, Y.W.; software, Y.W.; validation, Y.W.; formal analysis, Y.W.; investigation, Y.W., K.W., L.R., P.B., L.W. (Liyun Wu); resources, L.W. (Liyuan Wang) and H.C.; data curation, Y.W.; writing—original draft preparation, Y.W.; writing—review and editing, Y.W.; visualization, Y.W.; supervision, L.W. (Liyuan Wang); project administration, L.W. (Liyuan Wang) and H.C.; funding acquisition, L.W. (Liyuan Wang) and H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31570695), Earmarked Fund for China Agriculture Research System (CARS-19) and the Major Science and Technology Special Project of Variety Breeding of Zhejiang Province (2016C02053-6).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alboresi, A.; Gestin, C.; Leydecker, M.T.; Bedu, M.; Meyer, C.; Truong, H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Ann. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Fan, X.R.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Chiu, C.C.; Tsai, C.B.; Ho, C.H.; Hsu, P.K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [Green Version]
- Steiner, H.Y.; Naider, F.; Becker, J.M. The PTR family: A new group of peptide transporters. Mol. Microbiol. 1995, 16, 825–834. [Google Scholar] [CrossRef]
- Leran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Xia, X.D.; Fan, X.R.; Wei, J.; Feng, H.M.; Qu, H.Y.; Xie, D.; Miller, A.J.; Xu, G.H. Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 2015, 66, 317–331. [Google Scholar] [CrossRef]
- Jorgensen, M.E.; Xu, D.; Crocoll, C.; Ramirez, D.; Motawia, M.S.; Olsen, C.E.; Nour-Eldin, H.H.; Halkier, B.A. Origin and evolution of transporter substrate specificity within the NPF family. Elife 2017, 6, e19466. [Google Scholar] [CrossRef]
- Liu, K.H.; Tsay, Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003, 22, 1005–1013. [Google Scholar] [PubMed] [Green Version]
- Morere-Le Paven, M.C.; Viau, L.; Hamon, A.; Vandecasteele, C.; Pellizzaro, A.; Bourdin, C.; Laffont, C.; Lapied, B.; Lepetit, M.; Frugier, F.; et al. Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 2011, 62, 5595–5605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagchi, R.; Salehin, M.; Adeyemo, O.S.; Salazar, C.; Shulaev, V.; Sherrier, D.J.; Dickstein, R. Functional assessment of the Medicago truncatula NIP/LATD protein demonstrates that it is a high-affinity nitrate transporter. Plant Physiol. 2012, 160, 906–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.Y.; Tyerman, S.D.; Dechorgnat, J.; Ovchinnikova, E.; Dhugga, K.S.; Kaiser, B.N. Maize NPF6 proteins are homologs of Arabidopsis CHL1 that are selective for both nitrate and chloride. Plant Cell 2017, 29, 2581–2596. [Google Scholar] [PubMed] [Green Version]
- Corratge-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Wang, H.D.; Wan, Y.F.; Buchner, P.; King, R.; Ma, H.X.; Hawkesford, M.J. Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Triticum aestivum. J. Exp. Bot. 2020, 71, 4531–4546. [Google Scholar] [CrossRef]
- Mukhopadhyay, M.; Mondal, T.K.; Chand, P.K. Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): A review. Plant Cell Rep. 2016, 35, 255–287. [Google Scholar] [CrossRef]
- Weng, H.; Zeng, X.T.; Li, S.; Kwong, J.S.W.; Liu, T.Z.; Wang, X.H. Tea consumption and risk of bladder cancer: A dose-response meta-analysis. Front. Physiol. 2017, 7, 693. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yao, Z.S.; Pan, Z.L.; Wang, R.; Yan, G.X.; Liu, C.Y.; Su, Y.Y.; Zheng, X.H.; Butterbach-Bahl, K. Tea-planted soils as global hotspots for N2O emissions from croplands. Environ. Res. Lett. 2020, 15, 104018. [Google Scholar] [CrossRef]
- Zhao, X.B.; Huang, J.Y.; Yu, H.H.; Wang, L.; Xie, W.B. Genomic survey, characterization and expression profile analysis of the peptide transporter family in rice (Oryza sativa L.). BMC Plant Biol. 2010, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The Nitrate Transporter (NRT) gene family in poplar. PLoS ONE 2013, 8, e72126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, C.H.; Dong, Q.L.; Huang, D.; Li, C.Y.; Li, P.M.; Ma, F.W. Genome-wide identification and analysis of apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER Family (NPF) genes reveals MdNPF6.5 confers high capacity for nitrogen uptake under low-nitrogen conditions. Int. J. Mol. Sci. 2018, 19, 2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, Y.X.; Zhu, F.; Ming, R.; Chen, L.Q. Genome-wide analysis of nitrate transporter (NRT/NPF) family in sugarcane Saccharum spontaneum L. Trop. Plant Biol. 2019, 12, 133–149. [Google Scholar] [CrossRef]
- Wang, X.L.; Cai, X.F.; Xu, C.X.; Wang, Q.H. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2021, 158, 297–307. [Google Scholar] [CrossRef]
- Chao, H.B.; He, J.J.; Cai, Q.Q.; Zhao, W.G.; Fu, H.; Hua, Y.P.; Li, M.T.; Huang, J.Y. The expression characteristics of NPF genes and their response to vernalization and nitrogen deficiency in rapeseed. Int. J. Mol. Sci. 2021, 22, 4944. [Google Scholar] [CrossRef]
- Wang, Z.J.; Zhu, J.; Yuan, W.Y.; Wang, Y.; Hu, P.P.; Jiao, C.Y.; Xia, H.M.; Wang, D.D.; Cai, Q.W.; Li, J.; et al. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int. J. Biol. Macromol. 2021, 181, 1207–1223. [Google Scholar] [CrossRef]
- Hammes, U.Z.; Meier, S.; Dietrich, D.; Ward, J.M.; Rentsch, D. Functional properties of the Arabidopsis peptide transporters AtPTR1 and AtPTR5. J. Biol. Chem. 2010, 285, 39710–39717. [Google Scholar] [CrossRef] [Green Version]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar]
- Krouk, G.; Crawford, N.M.; Coruzzi, G.M.; Tsay, Y.F. Nitrate signaling: Adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010, 13, 266–273. [Google Scholar] [CrossRef]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubes, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zazimalova, E.; et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef]
- You, H.G.; Liu, Y.M.; Minh, T.N.; Lu, H.R.; Zhang, P.M.; Li, W.F.; Xiao, J.L.; Ding, X.D.; Li, Q. Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 2020, 61, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Xu, Z.S.; Tian, C.; Huang, Y.; Wang, F.; Xiong, A.S. Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 2016, 6, 23101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, S.; Shi, M.; Wang, S.; Shi, L.; Xu, F.; Ding, G. Genome-wide systematic characterization of the NPF family genes and their transcriptional responses to multiple nutrient stresses in allotetraploid rapeseed. Int. J. Mol. Sci. 2020, 21, 5947. [Google Scholar] [CrossRef]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [Green Version]
- Xia, E.H.; Tong, W.; Hou, Y.; An, Y.L.; Chen, L.B.; Wu, Q.; Liu, Y.L.; Yu, J.; Li, F.D.; Li, R.P.; et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Wang, X.C.; Feng, H.; Chang, Y.X.; Ma, C.L.; Wang, L.Y.; Hao, X.Y.; Li, A.L.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Bilas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss. Org. 2016, 127, 269–287. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Li, P.F.; Ran, F.; Guo, P.C.; Zhu, J.T.; Yang, J.; Zhang, L.L.; Chen, P.; Li, J.N.; Du, H. Genome-wide characterization, expression analyses, and functional prediction of the NPF family in Brassica napus. BMC Genom. 2020, 21, 871. [Google Scholar] [CrossRef]
- Dodd, I.C.; Tan, L.P.; He, J. Do increases in xylem sap pH and/or ABA concentration mediate stomatal closure following nitrate deprivation? J. Exp. Bot. 2003, 54, 1281–1288. [Google Scholar] [CrossRef]
- Banedjschafie, S.; Bastani, S.; Widmoser, P.; Mengel, K. Improvement of water use and N fertilizer efficiency by subsoil irrigation of winter wheat. Eur. J. Agron. 2008, 28, 1–7. [Google Scholar] [CrossRef]
- Guo, F.O.; Young, J.; Crawford, N.M. The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 2003, 15, 107–117. [Google Scholar] [PubMed] [Green Version]
- Chen, C.Z.; Lv, X.F.; Li, J.Y.; Yi, H.Y.; Gong, J.M. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 2012, 159, 1582–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.B.; Yi, H.Y.; Gong, J.M. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 2014, 26, 3984–3998. [Google Scholar] [CrossRef] [Green Version]
- Du, X.Q.; Wang, F.L.; Li, H.; Jing, S.; Yu, M.; Li, J.; Wu, W.H.; Kudla, J.; Wang, Y. The Transcription Factor MYB59 Regulates K+/NO3− translocation in the Arabidopsis response to low K+ Stress. Plant Cell 2019, 31, 699–714. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.Q.; Wang, R.C.; Chen, M.S.; Crawford, N.M. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1. (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 2001, 13, 1761–1777. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.C.; Liu, K.H.; Lo, H.J.; Tsay, Y.F. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 1999, 11, 1381–1392. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Chen, A.; Xie, K.; Yang, X.; Luo, Z.; Chen, J.; Zeng, D.; Ren, Y.; Yang, C.; Wang, L.; et al. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 16649–16659. [Google Scholar] [CrossRef]
- Lin, S.H.; Kuo, H.F.; Canivenc, G.; Lin, C.S.; Lepetit, M.; Hsu, P.K.; Tillard, P.; Lin, H.L.; Wang, Y.Y.; Tsai, C.B.; et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar]
- Fang, Z.; Bai, G.; Huang, W.; Wang, Z.; Wang, X.; Zhang, M. The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front. Plant Sci. 2017, 8, 1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Wang, L.Y.; Bai, P.X.; Wei, K.; Zhang, Y.Z.; Ruan, L.; Wu, L.Y.; Cheng, H. Identification of regulatory networks and hub genes controlling nitrogen uptake in tea plants [Camellia sinensis (L.) O. Kuntze]. J. Agric. Food Chem. 2020, 68, 2445–2456. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rodriguez, V.; Canas, R.A.; de la Torre, F.N.; Pascual, M.B.; Avila, C.; Canovas, F.M. Molecular fundamentals of nitrogen uptake and transport in trees. J. Exp. Bot. 2017, 68, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological Data. Molec. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, F.; He, W.; Yuan, Q.; Wei, K.; Ruan, L.; Wang, L.; Cheng, H. Transcriptome analysis identifies CsNRT genes involved in nitrogen uptake in tea plants, with a major role of CsNRT2.4. Plant Physiol. Biochem. 2021, 167, 970–979. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).