Abstract
Fungal basic leucine zipper (bZIP) proteins play a vital role in biological processes such as growth, biotic/abiotic stress responses, nutrient utilization, and invasion. In this study, genome-wide identification of bZIP genes in the fungus Fusarium fujikuroi, the pathogen of bakanae disease, was carried out. Forty-four genes encoding bZIP transcription factors (TFs) from the genome of F. fujikuroi (FfbZIP) were identified and functionally characterized. Structures, domains, and phylogenetic relationships of the sequences were analyzed by bioinformatic approaches. Based on the phylogenetic relationships with the FfbZIP proteins of eight other fungi, the bZIP genes can be divided into six groups (A–F). The additional conserved motifs have been identified and their possible functions were predicted. To analyze functions of the bZIP genes, 11 FfbZIPs were selected according to different motifs they contained and were knocked out by genetic recombination. Results of the characteristic studies revealed that these FfbZIPs were involved in oxygen stress, osmotic stress, cell wall selection pressure, cellulose utilization, cell wall penetration, and pathogenicity. In conclusion, this study enhanced understandings of the evolution and regulatory mechanism of the FfbZIPs in fungal growth, abiotic/biotic stress resistance, and pathogenicity, which could be the reference for other fungal bZIP studies.
1. Introduction
Rice is one of the most important food crops in the world and more than 50% of the world population depends on rice as their staple food. The bakanae disease of rice what caused by Fusarium fujikuroi was one of a seed born disease and wide distribution in rice growing area in the world. In terms of the rice plant, bakanae disease causes slenderly growth, seedling and plant death, and substantial yield reduction, bringing about enormous economic loss and even threatening food security. According to the statistics, bakanae disease could cause a 0–10% yield reduction in a mild case, but up to 30–95% yield loss in a serious case [1,2,3,4].
Transcription factor (TF), also known as trans-acting factor, is a protein that regulates gene expression in eukaryotes. It participates in specifically binding to cis-acting elements in the upstream promoter sequence of structural genes and takes an important role in activating or inhibiting transcription [5]. TFs activate the activity of transcription complexes containing RNA polymerase II, thereby regulating the coordinated transcription and expression of multiple target structural genes and playing a crucial role in the life process of organisms [6]. So far, more than 90 types of TFs have been reported in eukaryotes [7]. In the whole genome of more than 200 fungi, over 80 typical TF types were annotated, in addition to basic region/leucine zipper motif (bZIP), including TFs common in fungi such as Zn(II)2Cys6 zinc cluster protein (C6 zinc fingers) [8], Cys2His2 zinc finger protein (C2H2 zinc fingers), basic Helix-Loop-Helix (bHLH), GATA protein, MADS-box protein, MYB-like protein [9,10], and and homeodomain protein.
bZIP TFs are one of the most widely distributed and conserved TFs of eukaryotes. bZIP proteins contain two domains—a highly conserved DNA-binding basic region and a diverse leucine zipper [11]. In the form of homodimers or heterodimers, bZIP TFs regulate downstream-related gene expression such as ERF5, COR78, CYP707A3, and ADS by binding to cis-acting elements G-box, C-box, ABRE, LTRE, and Box II [9]. bZIP TFs are located in the nucleus and participate in various biological processes [12], for example, YAP-type bZIP TFs respond to oxidative stress [13,14], Atf-type bZIP TFs respond to osmotic stress [15,16,17], and Hac-type bZIP TFs are associated with non-folded proteins [18,19].
Scholars have carried out a lot of research on bakanae disease pathogens, but the interaction mechanism between bakanae disease pathogens and rice is still unclear, the same as the related molecular regulatory mechanism. Lack of knowledge of the molecular mechanism of bakanae disease pathogens is not conducive to understanding the regulation of the pathogenicity of bakanae disease pathogens, as well as the screening and utilization of related resistance genes. In this study, we identified 44 bZIP TFs from F. fujikuroi, analyzed genome-wide systematic characterization, and performed phylogenetic analysis with eight other common fungal bZIP TFs to understand their phylogenetic relationships with each other. In order to elucidate the biological function of FfbZIP genes, 11 FfbZIP genes were selected according to different domains, and deletion mutants were obtained by gene knockout. Results of functional experiments indicated that FfbZIP TFs not only played a regulatory role in the process of fungal growth and development, nutrient absorption and utilization, and stress response, but also regulated the invasion process of F. fujikuroi. The genome-wide and functional analysis of FfbZIP TFs in this study contributed to the research on the molecular mechanism of pathogenicity and expanded the research field of fungal bZIP TFs.
2. Results
2.1. Identification of the FfbZIP Transcription Factor Family from F. fusarium
In the Pfam database, there are three conserved bZIP domain sequences with domain IDs PF00170 (bZIP_1 domain), PF07716 (bZIP_2), and PF03131 (bZIP_Maf). Based on this, an alignment search was performed on the F. fujikuroi bZIP TF family proteins using HMM, and 37 matches were obtained. 186 bZIP protein sequnces of F. graminearum, F. oxysporum, F. verticillioides, F. solani, S. cerevisiae, U. virens, N. crassa, and M. oryzae were used as templates to search through the F. fujikuroi genome with TBLASTN, and 57 matches were obtained. Repeated sequences were deleted, and the obtained protein sequences were screened by CD-Search. Some protein sequences without bZIP domain were eliminated. Finally, a total of 44 predicted F. fujikuroi bZIP TF protein sequences were obtained from the F. fujikuroi genome. These genes were designated as FfbZIP1 to FfbZIP44 according to their locus number ordering. The gene name, locus number, gene symbol, gene ID, chromosome position, genomic location, protein size and ORF size on supercontig of all 44 FfbZIPs are indicated in Table 1.

Table 1.
List of bZIP TF family identified in the F. fujikuroi genome.
2.2. Conserved Domain and Structure of FfbZIP Protein
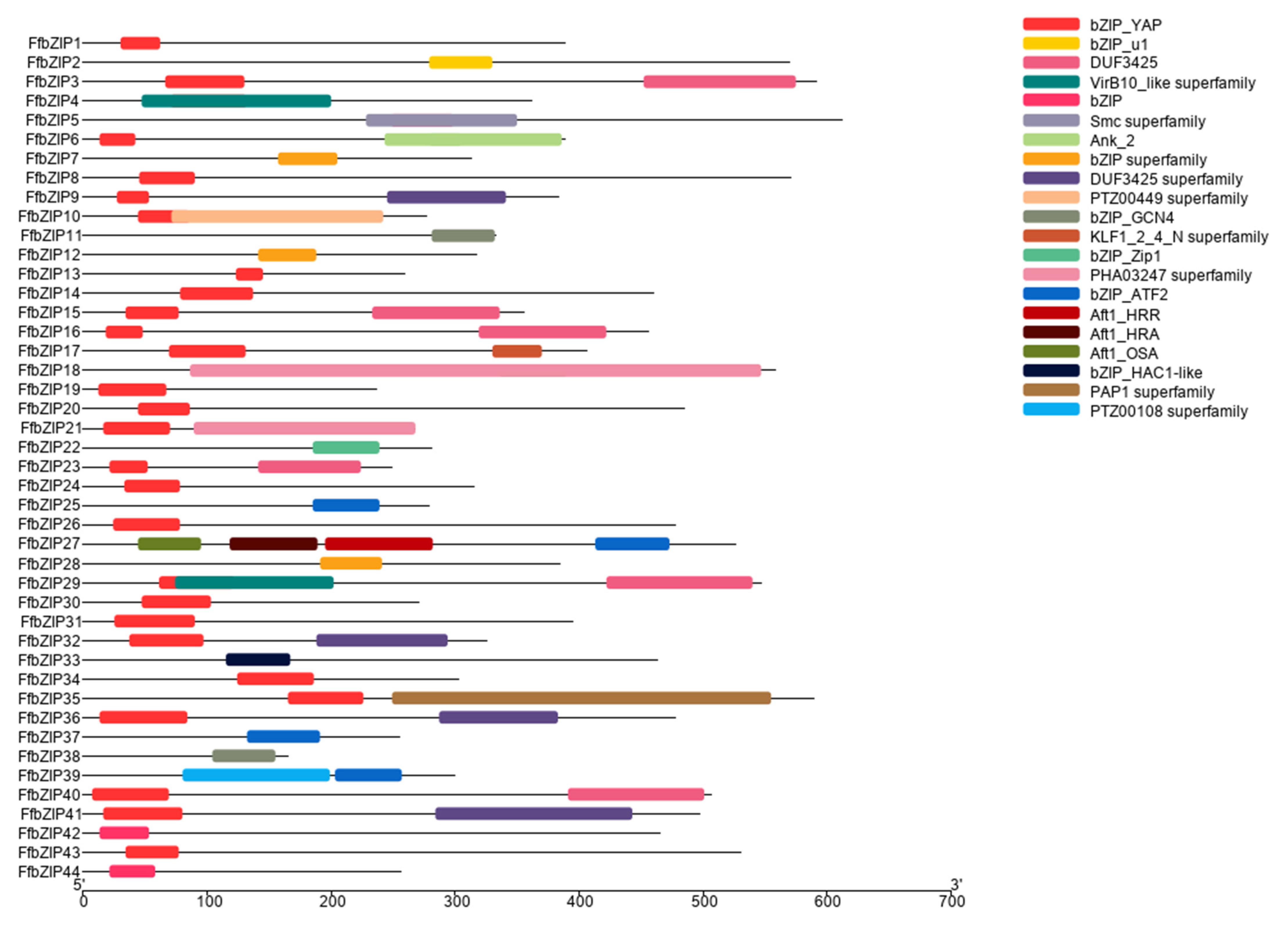
In the predicted FfbZIP TF protein sequences, in addition to 19 FfbZIP TFs with only one domain, the bZIP domain, 12 of these protein sequences also contained other predicted domains. FfbZIP3 contained VirB10_like domain, and FfbZIP4 contained Smc superfamily domain, and both domains overlapped with the bZIP domain. FfbZIP7 with PTZ00449 superfamily domain at the N-terminal of the protein, and the domain partial overlapped with the C-terminus of the bZIP domain. FfbZIP13 contained KLF1_2_4_N superfamily domain, FfbZIP22 contained PAP1 superfamily domain and FfbZIP24 contained PTZ00108 superfamily domain, all domains do not show overlap with bZIP domain. FfbZIP17 contained four kinds of Atf domains, including Atf1_HRR, Atf1_HRA, Atf1_OSA and bZIP_ATF2. In addition, FfbZIP11, FfbZIP12, FfbZIP23, FfbZIP27, and FfbZIP28 all contained DUF domains, which are DUF3425 and DUF3425 superfamily (Figure 1).
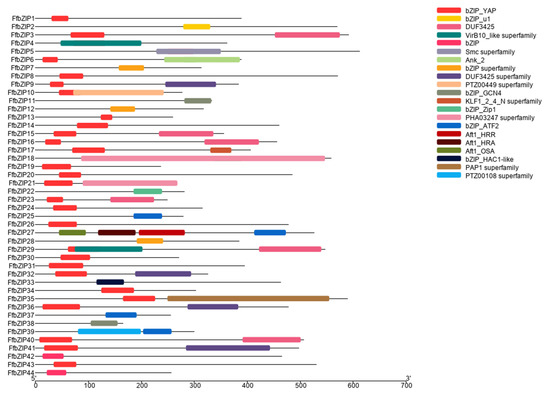
Figure 1.
Schematic diagram of the conserved domains of FfbZIP proteins.
The N-terminal basic amino acid region of the bZIP domain was highly conserved, containing a nuclear localization signal (NLS) [20] and an N-x7-R/K domain consisting of 16–20 amino acid residues, among which the last amino acid is arginine (R) or lysine (K) [9]. The leucine zipper region was not conserved and consists of several leucine repeat heptapeptides or hydrophobic amino acid residues (such as Ile, Val, Phe or Met). The leucine zipper region involved in oligomerization was connected to the basic region. Leucine is at position seventh of every seven amino acids, and sometimes replaced by isoleucine, valine, phenylalanine, or methionine [21]. In order to study the characteristics of FfbZIP domains, the amino acid sequences were compared and analyzed. All basic regions of FfbZIP TFs contained an N-x7-R domain but not an N-x7-K domain, suggesting that arginine was more conserved than lysine in FfbZIP proteins. In the leucine zipper region, the leucine in the first repeat heptapeptide was highly conserved. Two leucines (L) were replaced by valine (V) or arginine (R). In the latter two repeat heptapeptides, there were a lot of variation (Figure 2).

Figure 2.
Sequence alignment of FfbZIP domains.
2.3. Intron Distribution Analysis of FfbZIP Gene
To gain insights into the structural evolution of the FfbZIP genes, the intron distribution in FfbZIP genes was subjected to alignment analysis using GSDS. As shown in Figure 3, all FfbZIP genes contained 1–4 introns except FfbZIP12, FfbZIP18, FfbZIP25, FfbZIP28, FfbZIP38 and FfbZIP39. The FfbZIP genes were classified into five classes depending on their different intron numbers. Most FfbZIP genes contained two introns (34.09%, 15) and single-intron FfbZIP genes (34.09%, 15), followed by three introns (13.63%, 6), intronless (13.63%, 6), and four introns (4.56%, 2). There was no obvious relationship between exon length, intron length and intron number, neither the relationship between exon length and intron length (Supplementary Figure S1).

Figure 3.
Intron and exon distribution patterns in the coding sequence of FfbZIP genes.
2.4. Phylogenetic Analysis of FfbZIP Proteins
To investigate the phylogenetic relationships of the FfbZIP genes and relationships with other fungal bZIP proteins, 230 bZIP proteins from F. fujikuroi, F. graminearum, F. oxysporum, F. verticillioides, F. solani, S. cerevisiae, U. virens, N. crassa, and M. oryzae were analyzed (Supplementary Figure S2). The bZIP proteins were divided into six clades, and designated as A to F, indicating that the evolution of these bZIP genes were conserved between the nine fungi. In clades B and C there were bZIP proteins from nine fungi, while clade A was missing the bZIP protein of S. cerevisiae, clade D and E were missing the bZIP protein of N. crassa, and clade F was missing the bZIP protein of F. graminearum, F. verticillioides and N. crassa. Evolutionary analysis also revealed a closer homology relationship between F. fujikuroi and F. oxysporum (nine bZIP proteins with high homology), five with F. graminearum, four with F. verticillioides, three with U. virens, two with F. solani and M. oryzae, and one with S. cerevisiae.
2.5. Construction and Identification of FfbZIP Deletion Mutants
Generally, each domain had a unique spatial conformation and undertook different biological functions. To explore the effect of different domains on the function of FfbZIP TFs, the deletion mutants of FfbZIP genes with different domains were constructed. FfbZIP TFs contained bZIP_u1, bZIP_YAP, VirB10, bZIP, Smc, PTZ00449, bZIP_GCN4, DUF3425, KLF1_2_4_N, bZIP_Zip1, PAP1 domains, and the corresponding genes were FfbZIP2, FfbZIP4, FfbZIP5, FfbZIP8, FfbZIP10, FfbZIP11, FfbZIP16, FfbZIP17, FfbZIP22, FfbZIP35, and FfbZIP44. To characterize the functional roles of FfbZIP genes on biological functions and pathogenicity, targeted deletion of these genes was achieved by homologous recombination.
F/R primers were used to detect the presence or absence of HygR or target gene at a specific position at the same time. Due to the difference in the length of the target gene and the hygromycin resistance gene, if the wild-type and transformant amplified fragments have different lengths and no identical bands, then it shows that the hygromycin resistance gene has replaced the target gene, that is, the target gene has been knocked out. Taking the construction of the ΔFfbZIP10 mutant as an example, the hygromycin resistance gene fragment was inserted into the upstream and downstream 2000 bp gene sequences of the FfbZIP10 gene using SnapGene software, and the NCBI Primer-BLAST website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome/, accessed on 5 June 2022) found two upstream primers within the upstream 500 bp range, named UbZIP10F1 and UbZIP10F2, respectively, and similarly found two downstream primers in the downstream sequence and named them as DbZIP10R2, DbZIP10R1, at the upstream junction with the hygromycin resistance gene, took 20 bp of gene sequence totaling 40 bp, set as primer UbZIP10R, and similarly obtained primer DbZIP10F at the junction between the downstream gene and the resistance gene. Two primers, H2 and H3, were obtained at the position of about three equal parts of the hygromycin resistance gene, the knockout fragment was obtained by PCR technology, and the transformant was obtained by PEG mediation. The inserted gene was stably inherited after three generations of continuous culture, and a total of 21 transformants were obtained. The transformants were detected by PCR using DbZIP10F/UbZIP10R primers. The amplification product of FfbZIP10 is shorter than the amplification product of the resistance gene, and the band of the wild-type PCR amplification product was lower than that of the transformant, that is, T1, T2, T4 and T6 were positive transformants, and T3 and T5 indicated that the hygromycin resistance gene was inserted but not at the location of the FfbZIP10 gene, but was inserted randomly, so the PCR amplification result was two bands (Supplementary Figure S3).
2.6. Phenotypic Observation of FfbZIP Deletion Mutants
2.6.1. Fungi Morphology Observation
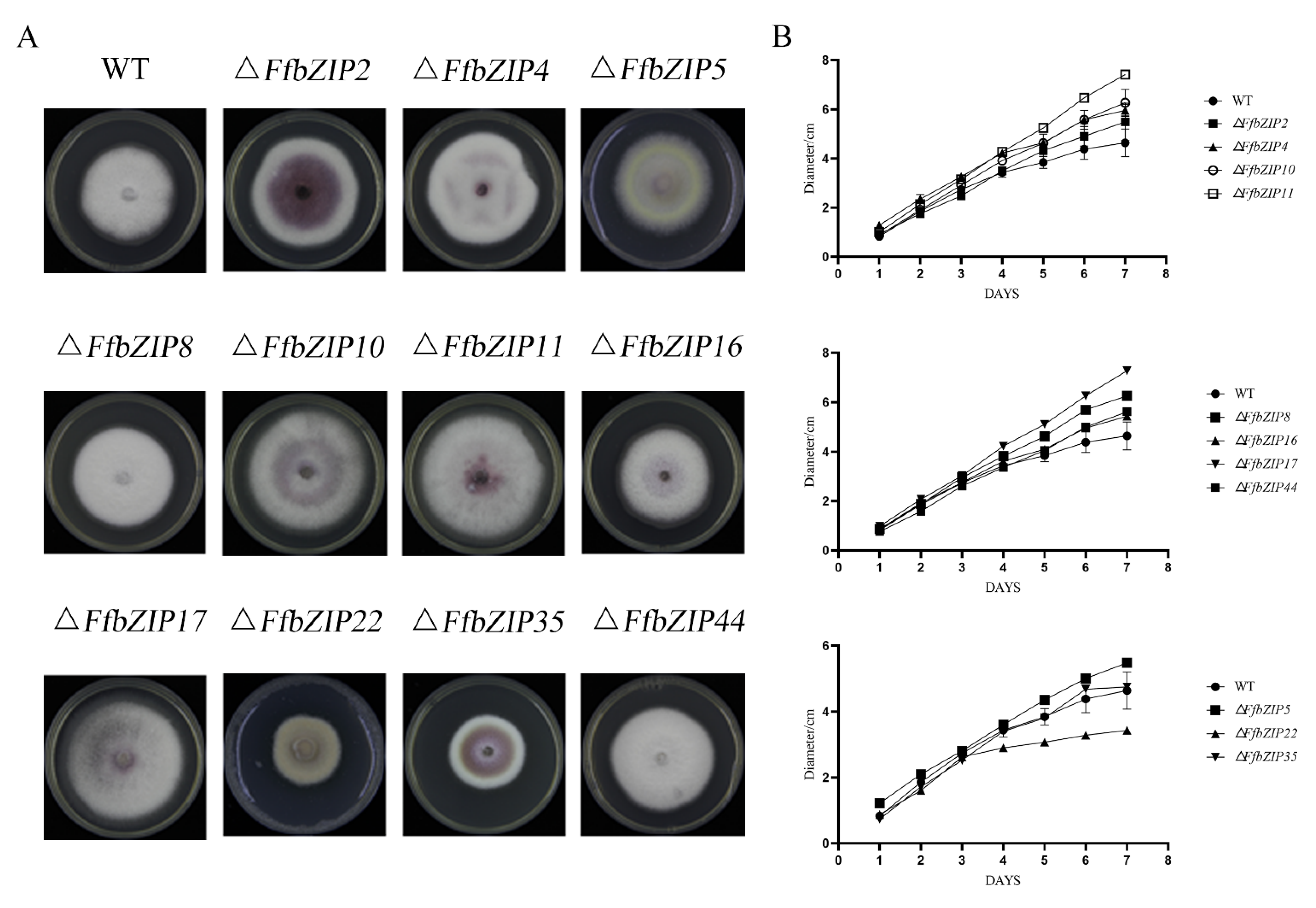
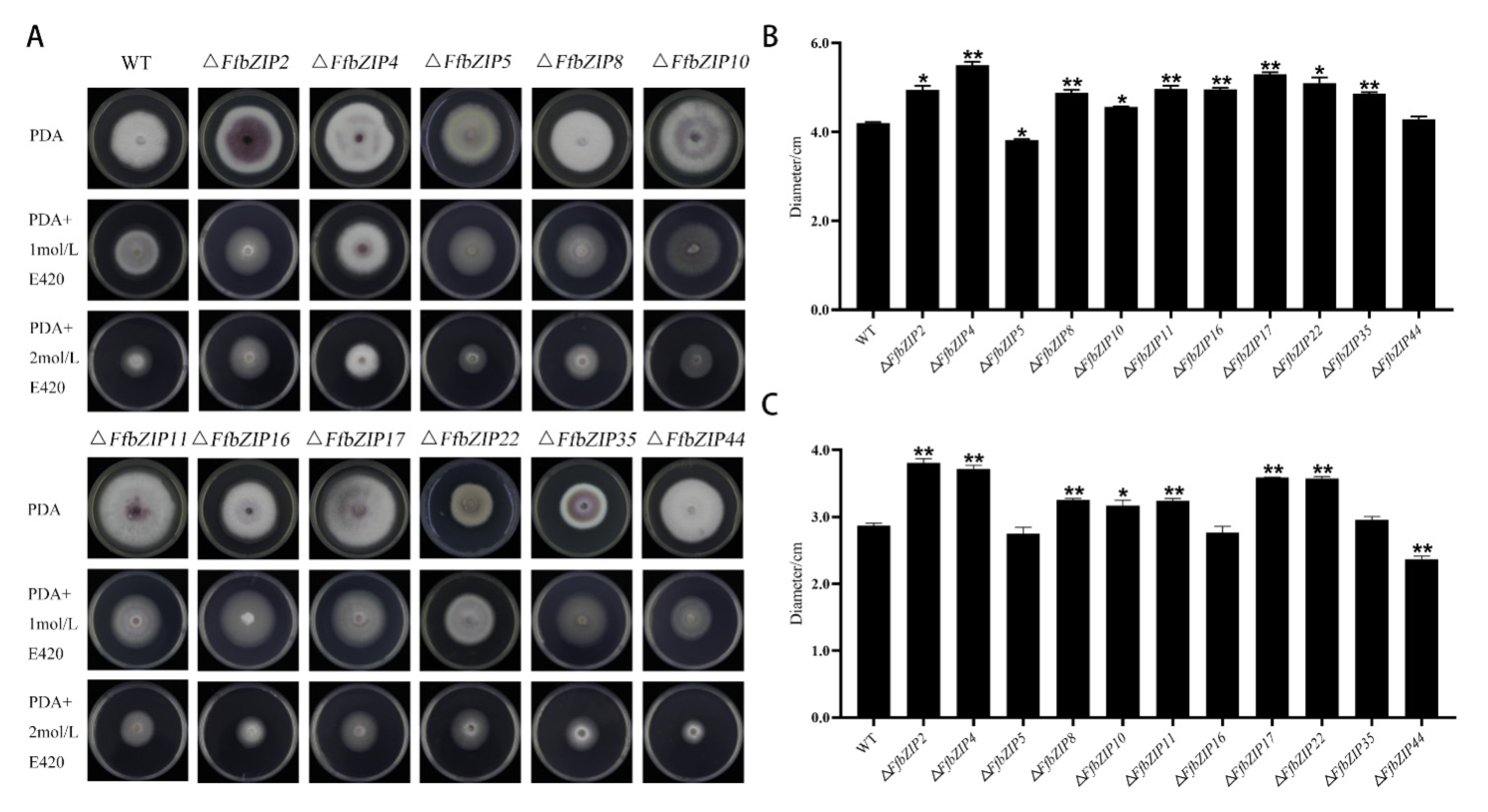
The comparison of the strain morphology between F. fujikuroi wild-type and FfbZIP deletion mutants on PDA medium in Figure 4 showed that ΔFfbZIP2, ΔFfbZIP4, ΔFfbZIP5, ΔFfbZIP16, and ΔFfbZIP35 were not significantly different from wild type at 7 days post inoculation (DPI). ΔFfbZIP35 was smaller than wild type, and the growth rate was slower than wild type after 3 days’ culturing by observation. The strains of other deletion mutants were larger than the wild type, and the growth rates were faster after 2–4 days’ culturing. The results showed that FfbZIP2, FfbZIP4, FfbZIP5, FfbZIP16, and FfbZIP35 had nothing to do with strain growth, FfbZIP22 promoted strain growth, and the other FfbZIP genes inhibited strain growth of F. fujikuroi.
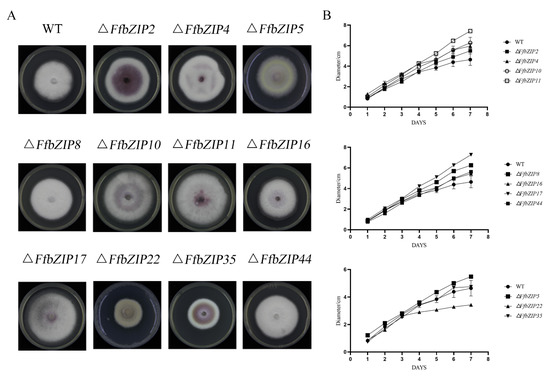
Figure 4.
Fungi morphology observation. (A) Colony morphology of FfbZIP knockout mutant strain. (B) Growth rate of FfbZIP knockout mutants versus wild type. Error bars represent means ± SEs. Some error bars were not plotted because the error bar would be shorter than the size of the symbol.
F. fujikuroi wild-type strains were white, and only a small amount of purple pigment could be observed at the bottom. ΔFfbZIP2 and ΔFfbZIP35 produced a large amount of purple pigment, while ΔFfbZIP4, ΔFfbZIP10, ΔFfbZIP11 and ΔFfbZIP17 produced slightly more purple pigment than the wild type, and the rest of the deletion mutants were no different from the wild type. The coloration phenotypes were maintained in the whole population of positive deletion mutants. The results revealed that FfbZIP2 and FfbZIP35 inhibited the synthesis of purple pigment, while FfbZIP4, FfbZIP10, FfbZIP11, and FfbZIP17 inhibited to a certain extent, and other FfbZIP genes did not participate in the synthesis of purple pigment.
We discovered by observation that wild type produced a large number of aerial hyphae, while ΔFfbZIP5 and ΔFfbZIP22 were without it. The aerial hyphae of ΔFfbZIP10 and ΔFfbZIP17 were slightly less than those of the wild type, and the other deletion mutants were consistent with the wild type. The results indicated that FfbZIP5 and FfbZIP22 promoted the formation of aerial hyphae, FfbZIP10 and FfbZIP17 had positive regulatory effects on the formation of aerial hyphae, and the other FfbZIP genes did not affect the formation of aerial hyphae.
2.6.2. Oxidative Stress Experiment
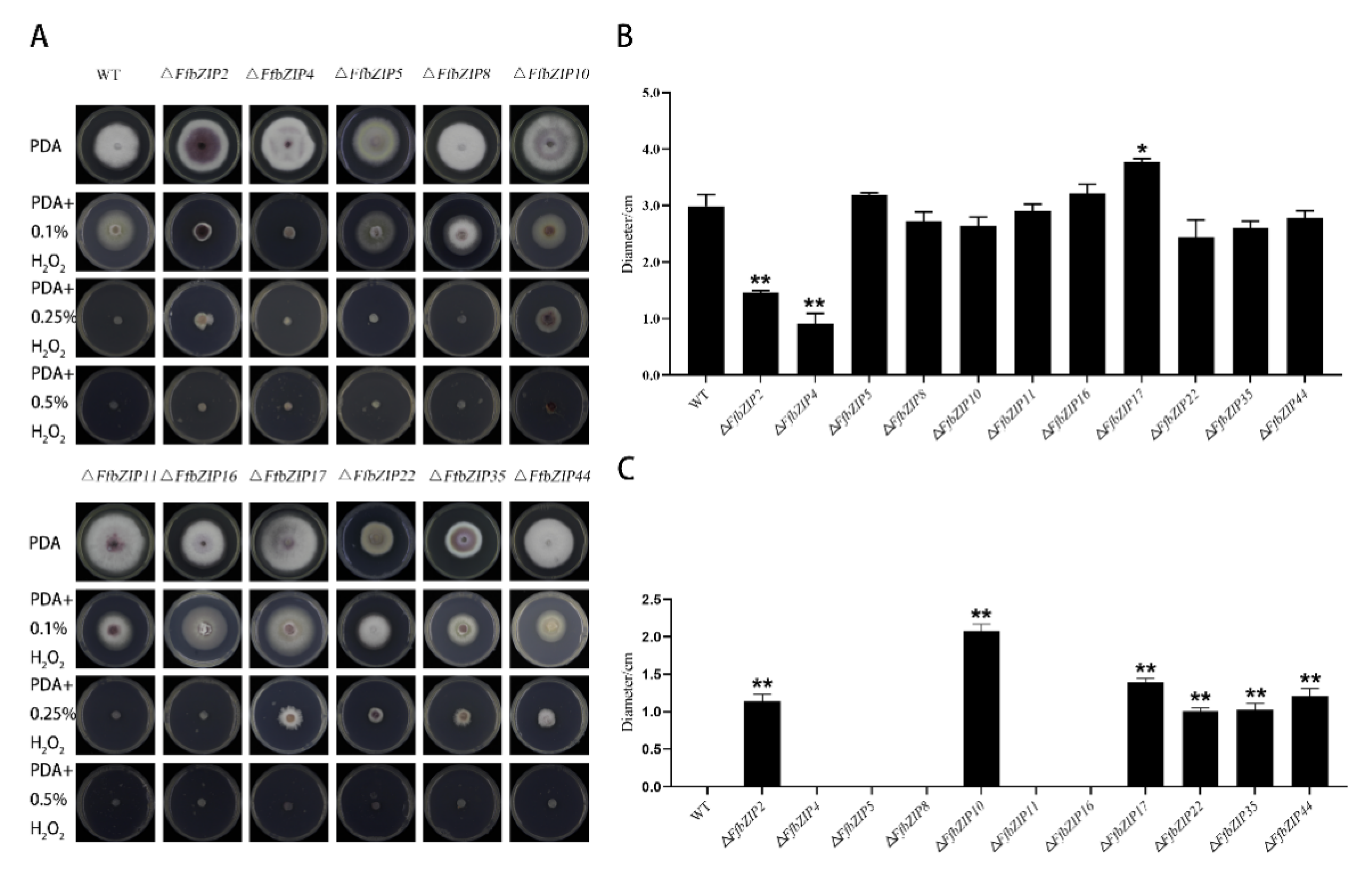
ΔFfbZIP4 could not grow under 0.1% H2O2, while ΔFfbZIP2 grew much smaller than the wild type (Figure 5). ΔFfbZIP17 was significantly larger than wild type at p = 0.05 level, but not at p = 0.01 level. The other deletion mutants were not significantly different from the wild type. Under 0.25% H2O2, ΔFfbZIP5, ΔFfbZIP8, ΔFfbZIP11, and ΔFfbZIP16 lost the growth ability, like the wild type. ΔFfbZIP2, ΔFfbZIP10, ΔFfbZIP17, ΔFfbZIP22, ΔFfbZIP35 and ΔFfbZIP44 grew to be tiny. Neither wild type nor deletion mutants could grow under 0.5% H2O2. The results showed that FfbZIP2 and FfbZIP4 positively regulated the anti-oxidative stress of F. fujikuroi under low oxidative stress conditions, while FfbZIP17 inhibited the ability of F. fujikuroi to resist oxidative stress, and other FfbZIP genes were not involved in the relevant regulation. FfbZIP2, FfbZIP10, FfbZIP17, FfbZIP22, FfbZIP35, and FfbZIP44 inhibited the oxidative stress tolerance of F. fujikuroi under high oxidative stress, and other FfbZIP genes were not involved.
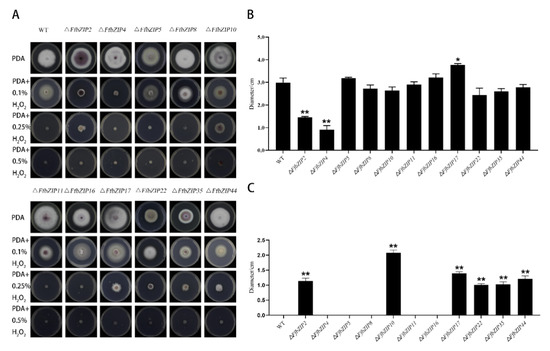
Figure 5.
Oxidative stress experiment. (A) Growth of wild type and FfbZIP knockout mutants under exogenous oxygen stress. (B) Histogram of strain diameter under 0.1% H2O2 oxidative stress. Error bars represent means ± SEs. (C) Histogram of strain diameter under 0.25% H2O2 oxidative stress. Error bars represent means ± SEs. * p < 0.05, ** p < 0.01.
2.6.3. Osmotic Stress Experiment
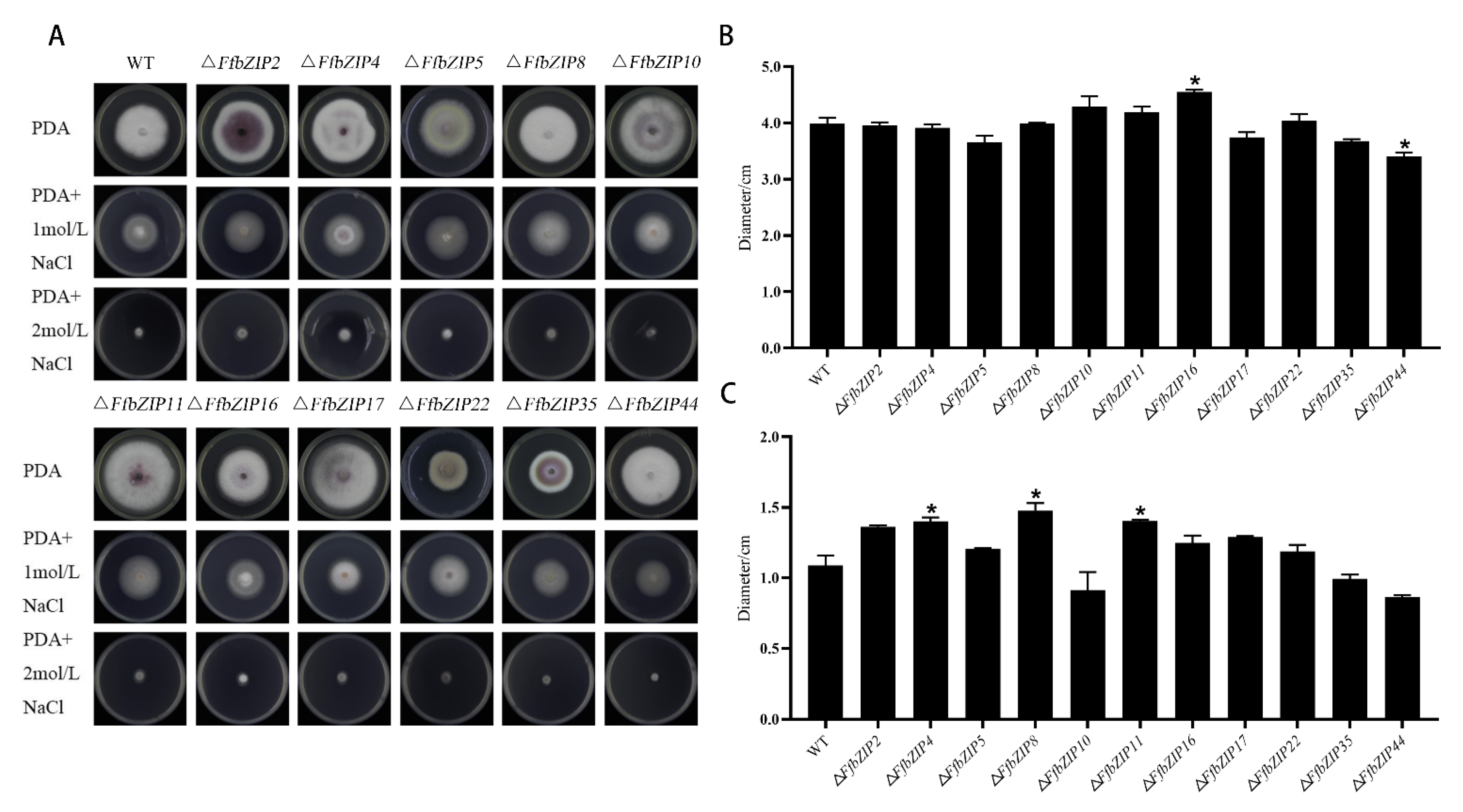
The osmotic stress experiment used NaCl to simulate a high osmotic pressure environment. Under 1 mol/L NaCl, only ΔFfbZIP44 deletion mutant was smaller than the wild type, and it was significantly different from the wild type (p = 0.05) (Figure 6). ΔFfbZIP16 was significantly larger than the wild type (p = 0.05), and the other deletion mutants had similar inhibitory effects to the wild type. Under 2 mol/L NaCl, ΔFfbZIP4, ΔFfbZIP8 and ΔFfbZIP11 were larger than the wild type, and there was a significant difference between the deletion mutants and the wild type at p = 0.05 level, but no difference between the wild type and all the deletion mutants at p = 0.01 level. The results indicated that FfbZIP44 could enhance the resistance to osmotic stress of F. fusarium, under 1 mol/L NaCl, while other FfbZIP genes could not. Under 2 mol/L NaCl, FfbZIP4, FfbZIP8 and FfbZIP11 had a certain inhibitory effect on the osmotic tolerance of F. fusarium, while the other FfbZIP genes were without it.
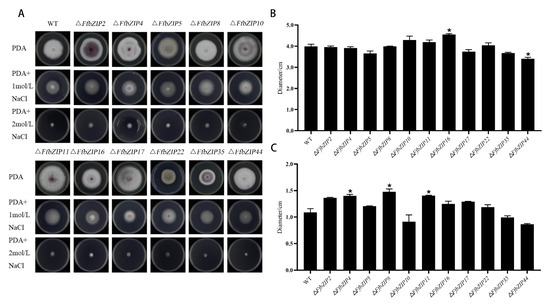
Figure 6.
Osmotic stress experiment. (A) Growth of wild type and FfbZIP knockout mutants under NaCl osmotic stress. (B) Histogram of strain diameter under 1 mol/L NaCl osmotic pressure. Error bars represent means ± SEs. (C) Histogram of Histogram of strain diameter under 2 mol/L NaCl osmotic pressure. Error bars represent means ± SEs. * p < 0.05.
2.6.4. Cell Wall Selection Pressure Experiment
Cell wall selective stress experiment used sorbitol to simulate a cell wall stress environment. Under 1 mol/L sorbitol (E420), there was no significant difference between ΔFfbZIP44 and the wild type, ΔFfbZIP5 was significantly smaller than wild type (p = 0.05), and other mutants were significantly larger than wild type (Figure 7). Under 2 mol/L E420, the growth of ΔFfbZIP5, ΔFfbZIP16, and ΔFfbZIP35 was similar to the wild type, and ΔFfbZIP10 was significantly larger than the wild type (p = 0.05). ΔFfbZIP44 was significantly smaller (p = 0.01) than the wild type, and the other deletion mutants were significantly larger than the wild type (p = 0.01). The results revealed that under 1 mol/L sorbitol (E420), FfbZIP44 had no effect on the strain growth of F. fujikuroi, and the other FfbZIP genes were involved in the reverse regulation of the cell wall selective stress tolerance.
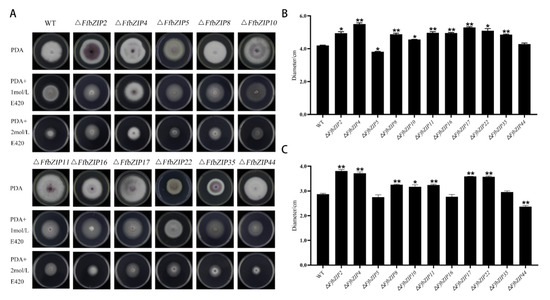
Figure 7.
Cell wall selection pressure experiment. (A) Growth of wild type and FfbZIP knockout mutants under cell wall selection pressure. (B) Histogram of strain diameter under 1 mol/L E420 cell wall selection pressure. Error bars represent means ± SEs. (C) Histogram of strain diameter under 2 mol/L E420 cell wall selection pressure. Error bars represent means ± SEs, * p < 0.05, ** p < 0.01.
2.6.5. Cellulose Utilization Experiment
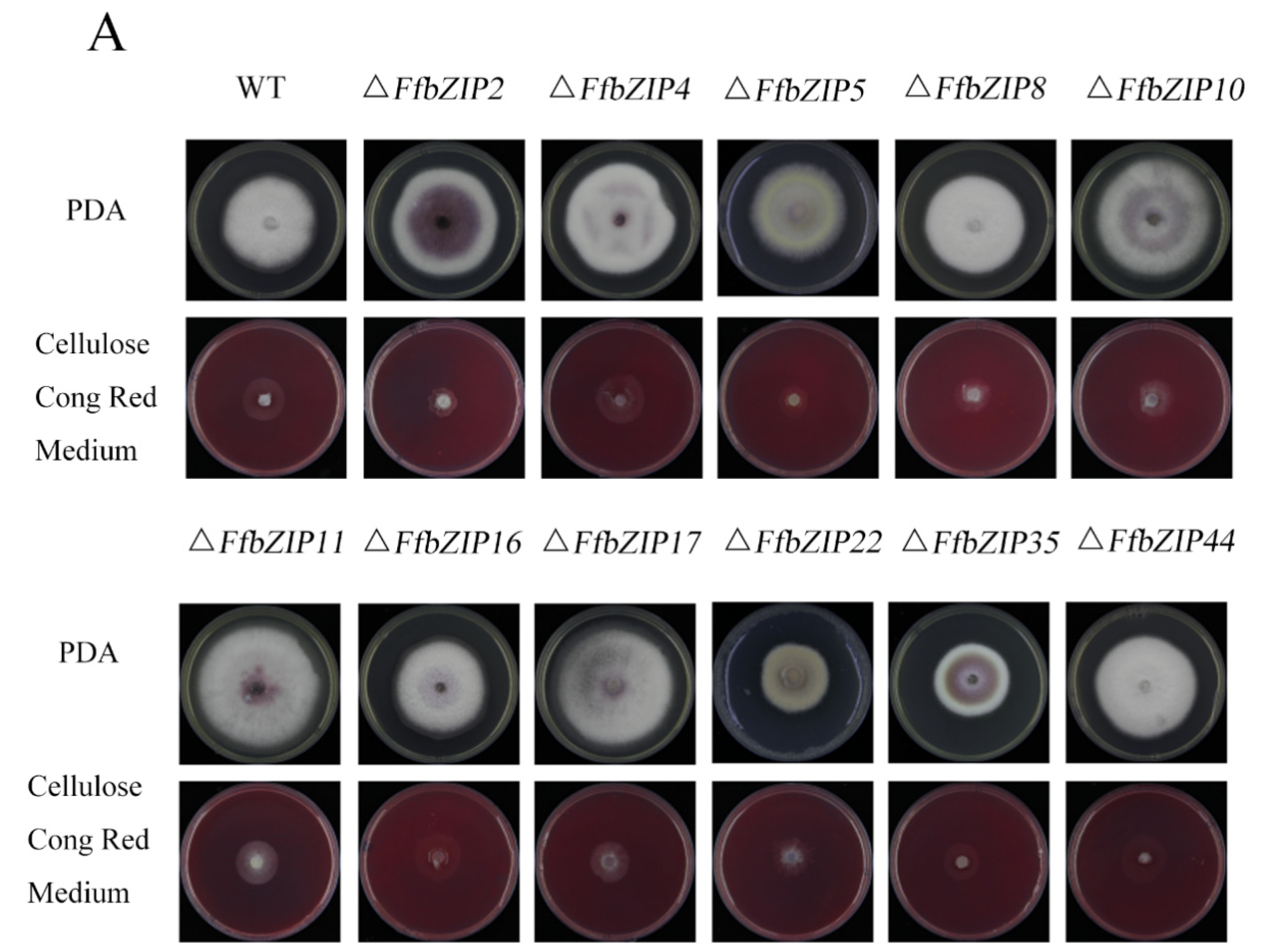
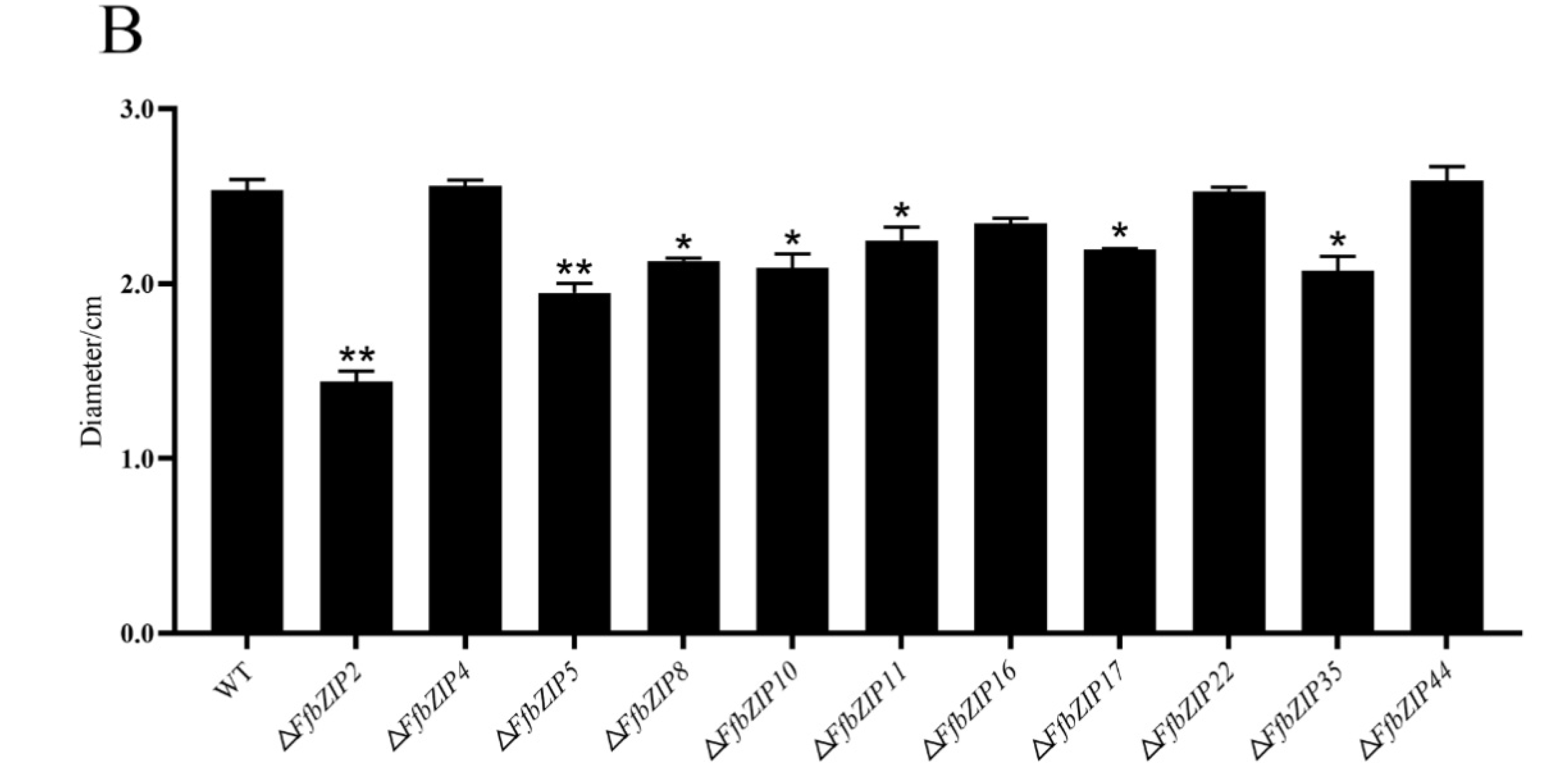
ΔFfbZIP2 and ΔFfbZIP5 cultured on cellulose Congo red medium were much smaller than the wild type (p = 0.01), and ΔFfbZIP8, ΔFfbZIP10, ΔFfbZIP11, ΔFfbZIP17 and ΔFfbZIP35 were significantly smaller than the wild type (p = 0.05), while the rest were not (Figure 8). The results showed that FfbZIP2, FfbZIP5, FfbZIP8, FfbZIP10, FfbZIP11, FfbZIP16, FfbZIP17 and FfbZIP35 were involved in the absorption and utilization of cellulose, and the other FfbZIP genes had no effect.
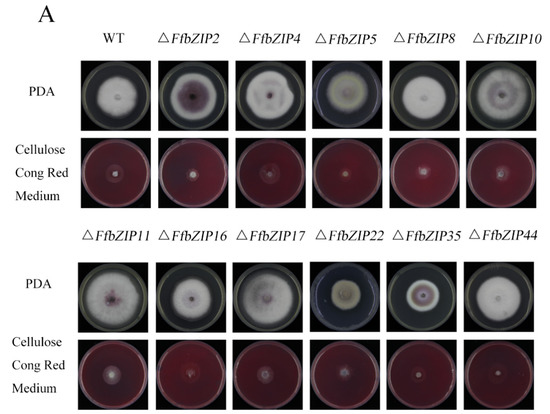
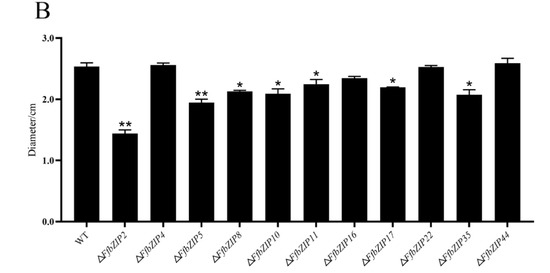
Figure 8.
Cellulose utilization experiment. (A) Growth of wild type and FfbZIP knockout mutants in Cellulose Congo red medium. (B) Histogram of strain diameter under cellulose Congo red culture conditions. Error bars represent means ± SEs. * p < 0.05, ** p < 0.01.
2.6.6. Cellophane Penetration Experiment
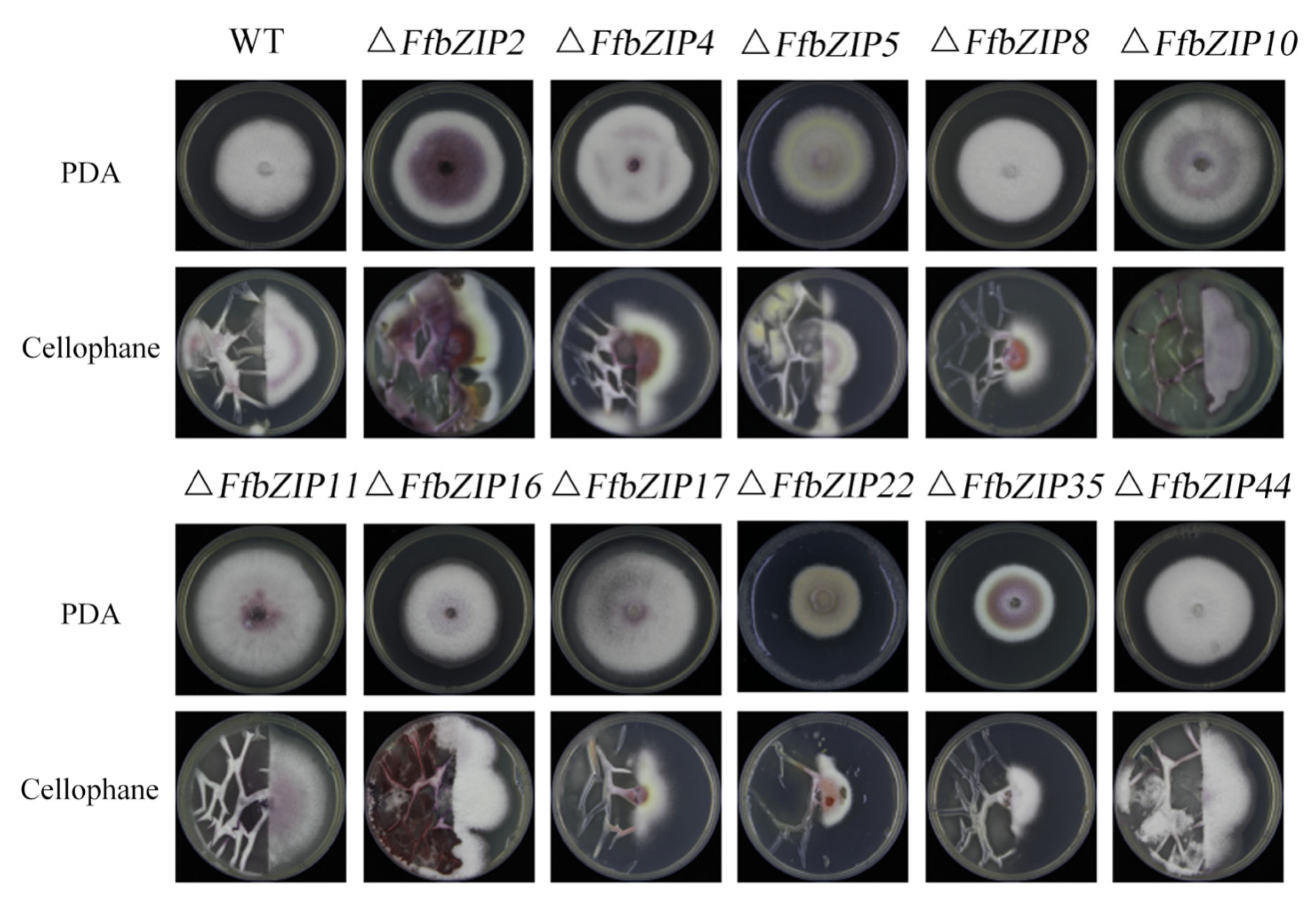
To clarify the ability of the deletion mutants to penetrate the plant cell wall, cellophane was used to simulate the plant cell wall. ΔFfbZIP2, ΔFfbZIP4, ΔFfbZIP5, ΔFfbZIP16, ΔFfbZIP44 and wild type grew through cellophane, indicating their ability to penetrate plant cell walls, while ΔFfbZIP8, ΔFfbZIP10, ΔFfbZIP11, ΔFfbZIP17, ΔFfbZIP22 and ΔFfbZIP35 could not (Figure 9). The results revealed that FfbZIP8, FfbZIP10, FfbZIP11, FfbZIP17, FfbZIP22, and FfbZIP35 played a decisive role in the penetration of F. fujikuroi infection into plant cell walls, while FfbZIP2, FfbZIP4, FfbZIP5, FfbZIP16, and FfbZIP44 did not.
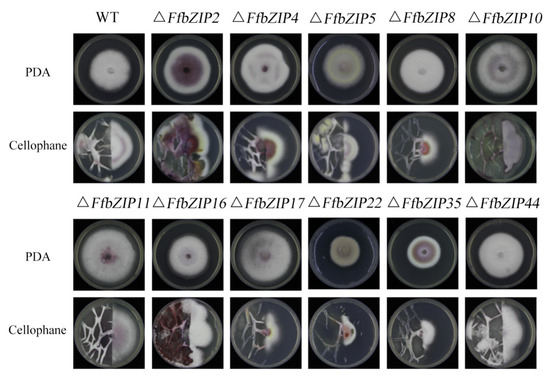
Figure 9.
Determination of cellophane penetration ability of wild type and FfbZIP knockout mutants.
2.7. Pathogenicity Assay of FfbZIP Deletion Mutants
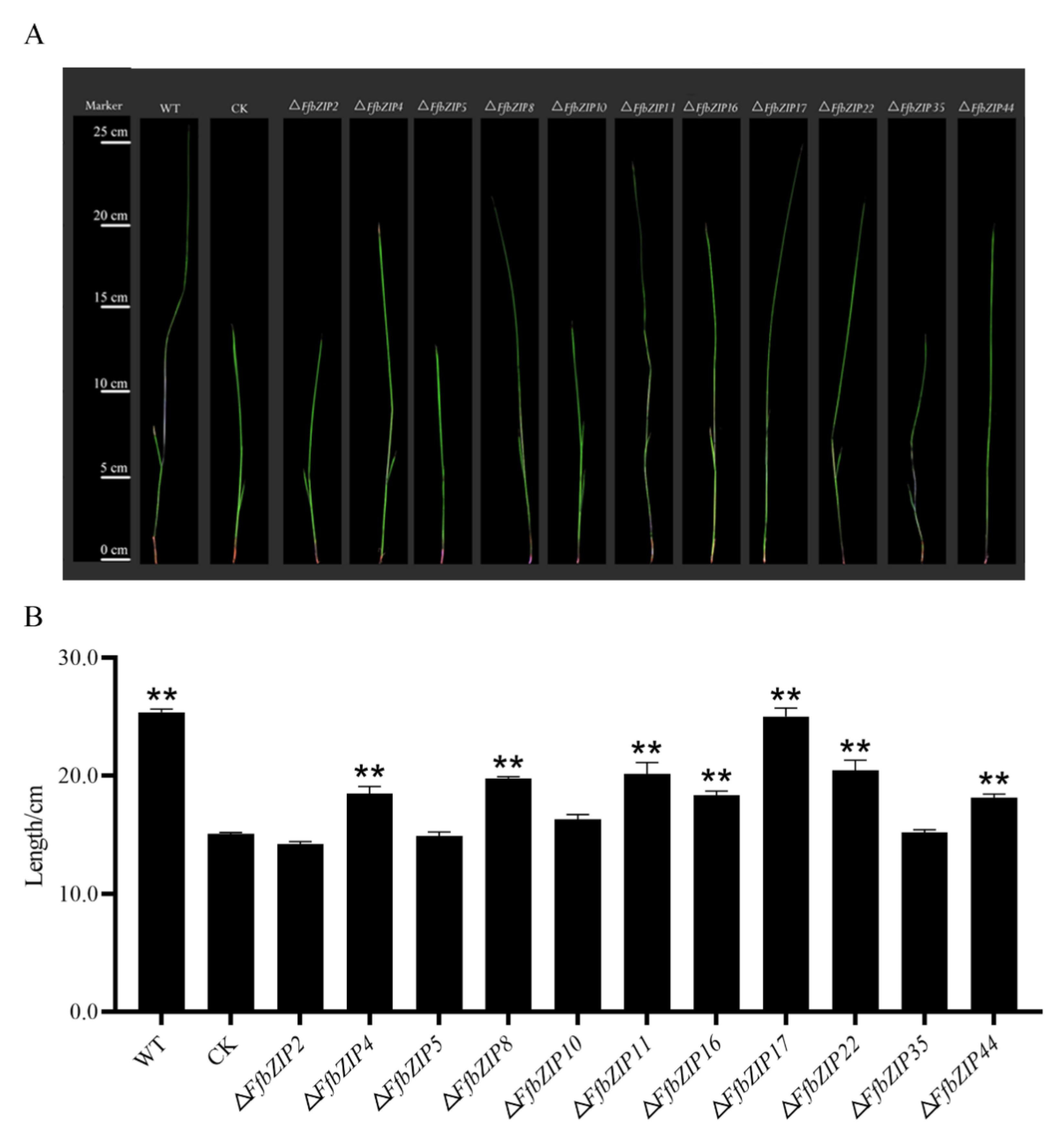
The pathogenicity assays used bakanae disease-susceptible rice varieties Zhongzao 39 and FfbZIP deletion mutants as test materials. Cultivated for 21 days after artificial inoculation, the growth of rice seedlings varies greatly (Figure 10A). The shoot length of rice seedlings was measured, and there was no significant difference in the growth of rice seedlings inoculated with ΔFfbZIP2, ΔFfbZIP5, and ΔFfbZIP10 and uninoculated rice seedlings. Although the shoot length of rice seedlings inoculated with ΔFfbZIP35 was not significantly different from that of uninoculated rice seedlings, the plants were thin, pale green and slightly whitish, showing typical symptoms of bakanae disease. The rice seedlings inoculated with ΔFfbZIP4, ΔFfbZIP8, ΔFfbZIP11, ΔFfbZIP16, ΔFfbZIP22 and ΔFfbZIP44 all became diseased, and the shoot length was significantly higher than that of the uninoculated rice seedlings, but significantly smaller than that of the wild-type seedlings. The difference was that the shoot length of rice seedlings inoculated with ΔFfbZIP17 was not significantly different from that inoculated with wild type. The results showed that FfbZIP2, FfbZIP5 and FfbZIP10 were indispensable in the pathogenesis of F. fujikuroi. FfbZIP4, FfbZIP8, FfbZIP11, FfbZIP16, FfbZIP22, FfbZIP35 and FfbZIP44 had positive regulatory effects on the pathogenesis of F. fujikuroi, while FfbZIP17 did not participate at all.
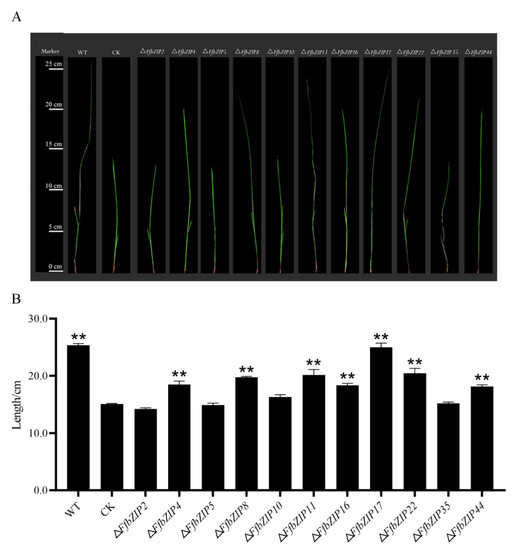
Figure 10.
Pathogenicity observation of inoculated rice seedlings. (A) Pathogenicity alignment of FfbZIP knockout mutants and wild type. (B) Shoot length determination of rice seedlings inoculated with wild type and FfbZIP deletion mutants. CK stood for the shoot length of uninoculated rice seedlings, ** p < 0.01.
3. Discussion
In recent years, with the continuous improvement of genome databases, more and more TF families have been identified, such as bZIP [22], MYB [23], GATA [24], and PacC [25]. bZIP TFs are one of the most widely distributed and conserved proteins of eukaryotes. At present, a variety of fungal bZIP TF families have been identified. According to the published literature, a large number of bZIP TFs have been found in the genomes of many fungi such as Alternaria sp. [26], Phytophthora sojae [27], Sclerotinia sclerotiorum [28], Aspergillus niger [29], F. graminearum [30,31] and F. pseudograminearum [32]. Only a few bZIP TFs in fungi have been systematically identified, such as those in M. oryzae [9] and F. graminearum [33].
Currently, only one FfbZIP protein, MeaB, has been identified in F. fujikuroi. This protein cooperates with AreA to mediate nitrogen metabolite inhibition in F. fujikuroi [34]. This study focused on the bZIP TF family in F. fujikuroi and identified 44 FfbZIP proteins genome-wide for the first time. The number of bZIP TFs in F. oxysporum was 56. U. virens, M. oryzae, F. graminearum, F. verticillium, F. solani, N. crassa and S. cerevisiae, respectively, contained 28, 22, 21, 19, 17, 13 and 12 bZIP transcription factors, all less than F. fujikuroi. The results of phylogenetic analysis indicated that bZIP TFs in several common fungi were conserved in regulating the biological functions.
In the bZIP protein sequences of F. fujikuroi, in addition to the bZIP domain, there were some other domains (Table 2). This phenomenon was very common not only in plant species, but also in fungi. The function could be predicted through the domains FfbZIP TFs contained.

Table 2.
FfbZIP transcription factor domains and their function.
General features and species specificity are reflected in the exon-intron structures of Arabidopsis, Rice, Nematode, and Homo sapiens genes, whose average exon length all decreased with increasing intron number [55], while the exon length in the FfbZIP genes had no significant relationship with intron length and intron number. The features and species specificity among intron number, exon length and intron length of Arabidopsis, Rice, Nematode, and Homo sapiens genes were not available for FfbZIP genes.
Phylogenetic analysis of the total bZIP protein sequences of nine fungi including F. fujikuroi were divided into six clades according to the phylogenetic tree. Fungal bZIP TFs have diverse functions. By aligning bZIP TFs with model fungi, we predicted the function of FfbZIP proteins. Eight FfbZIP TFs were relatively specific in F. fujikuroi, indicating that these TFs may have different functions in F. fujikuroi relative to others.
Eleven FfbZIP deletion mutants were obtained by gene knockout. FfbZIP4, FfbZIP8, FfbZIP10, FfbZIP16, FfbZIP17, FfbZIP35, and FfbZIP44 containing bZIP_YAP domain took part in stress response and osmotic stress response, and FfbZIP35 containing PAP1 domain played a role in response to H2O2. The results of phenotype and pathogenicity analysis showed that the above FfbZIP genes were involved in the growth, stress response and invasion pathogenic process of F. fujikuroi, which was consistent with the previous prediction of domain functions (Table 3).

Table 3.
The biological function and pathogenicity of FfbZIP genes.
Though fungi have significant species diversity, due to their complex life cycle, unknown growth environment and other factors, researchers continue to pose new questions. Due to the large number of bZIP TFs, the signaling mechanism of each subfamily and the cross-function between subfamilies need further study. It is expected that researchers will isolate more bZIP TFs, so as to further explore their roles in hormone signaling, anti-stress pathways, pathogenicity, as well as the regulatory mechanism of energy metabolism pathways, and further improve the functional research of bZIP TFs.
In view of the diversity and importance of the biological functions of bZIP TFs, gene cloning technology can be combined with transcriptomics, proteomics, and other technical means to expand the research field of bZIP TFs, strengthen the study of bZIP in a wider range of species, and explore more theoretical and applied values. In addition, more attention needs to be paid to the response regulation pathway of bZIP TFs and explore the dynamic process of the bZIP network regulation mechanism using fungal tracer technology. By applying system biology methods and exploring the regulatory network of the upstream and downstream genes of bZIP TFs, the network regulatory mechanism of bZIP in response to environmental stress will be revealed. In general, a comprehensive understanding of the distribution, classification, structure, and function of bZIP TFs, and the realization of their global and dynamic research will be the future research direction.
4. Materials and Methods
4.1. Identification of bZIP Transcription Factors in F. fujikuroi
The protein data of F. fujikuroi (IMI 58289) was downloaded from the National Center for Biological Information (http://www.ncbi.com, accessed on 8 June 2022), and the gene sequences with PF00170 (bZIP_1), PF07716 (bZIP_2) and PF03131 (bZIP_Maf) were searched and aligned by the HMMER software using the Hidden Markov Model (HMM) profile of Pfam [56,57]. The E value threshold was set to be ≤500, and the HMM search operation was performed. The amino acid sequences of the bZIP transcription factor of F. graminearum, F. oxysporum, Saccharomyces cerevisiae, Ustilaginoidea virens, Neurospora crassa, Magnaporthe oryzae, F. verticillioides and F. solani were downloaded from the Fungal Transcription Factor Database (http://ftfd.snu.ac.kr/, accessed on 1 June 2022) as the reference sequences. 21, 56, 12, 27, 13, 22, 19 and 17 bZIP protein sequences were obtained, respectively [58,59]. These sequences were used as queries for TBLASTN searches (E-value cutoff less than 1 × 10−5) against the F. fujikuroi genome. We removed repeated sequences and kept similar sequences. The presence of typical bZIP domains in predicted protein sequences were verified through CD-Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/, accessed on 5 June 2022) and Multiple Em for Motif Elicitation (MEME) (https://meme-suite.org/meme/tools/meme/, accessed on 5 June 2022), with an E-value threshold of 0.01 [9]. After deleting protein sequences that did not contain bZIP domains, the remaining sequences were considered as bZIP protein candidates in F. fujikuroi for further analysis.
4.2. Structural Analysis of bZIP Transcription Factors
The intron and exon structure information of FfbZIP genes was annotated in the genome.gff file (GCF_900079805.1_Fusarium_fujikuroi_IMI58289_V2_genomic.gff) downloading from the National Center for Biological Information (http://www.ncbi.com/, accessed on 9 June 2022). The Gene Structure Display Server (GSDS) 2.0 (http://gsds.gao-lab.org/, accessed on 2 June 2022) was used to annotate the intron and exon structure information [60]. The protein sequences of annotated DNA-binding domains and other functional domains from CD-Search screening were graphically displayed on full-length genes using TBtools [61]. Multiple sequence alignments of the amino acid sequences of bZIP were aligned using the DNAMAN [62].
4.3. Phylogenetic Analysis of bZIP Transcription Factors
Phylogenetic analysis of F. fujikuroi with F. graminearum, F. oxysporum, Saccharomyces cerevisiae, Ustilaginoidea virens, Neurospora crassa, Magnaporthe oryzae, F. verticillioides and F. solani was performed using iqTREE [63], and the optimal model calculated by Model Finder was used to complete the verification of 1000 bootstraps [64]. Phylogenetic analysis results were visualized by iTOL software [65].
4.4. Generation of FfbZIP Gene Deletion Mutants
The principle of homologous replacement was used to knock out the FfbZIP genes, and the FfbZIP genes were replaced with the hygromycin resistance gene. PCR technology was used for DNA fragment amplification and ligation. The principle was shown in Supplementary Figure S4. The upstream/downstream fragment of the target gene with the hygromycin resistance gene linkers were amplified by F1 + R/R1 + F primers, and the hygF/hygR hygmycin resistance genes were amplified by universal hygmycin resistance gene primers. The three fragments were directly connected by PCR technology, and then the PCR products were directly amplified by F2 + H3/R2 + H2 primers to obtain upstream and downstream fragments of the target gene with partially overlapping hygromycin resistance genes respectively.
Using F. fujikuroi IMI 58289 genomic DNA as a template, 44 primers (Supplementary Table S1) were used to amplify the upstream and downstream sequences of 11 genes, including FfbZIP2, FfbZIP4, FfbZIP5, FfbZIP6, FfbZIP10, FfbZIP11, FfbZIP16, FfbZIP17, FfbZIP22, FfbZIP35 and FfbZIP44, about 2 kb, for gene knockout. Using the LiGFP plasmid as a template, the hygromycin resistance gene HygR was amplified. 50 μL PCR reaction system: 25 μL Mix enzyme, 2 μL F. fujikuroi DNA template, 3 μL upstream/downstream primers (F1 + R/F + R1), 17 μL H2O. PCR reaction program: pre-denaturation at 94 °C for 2 min; 30 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 2 min; 72 °C for 10 min; 16 °C for 10 min. The PCR products were electrophoresed on agarose gels and recovered by cutting the gel using the TIANgel Midi Purification Kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China).
The upstream and downstream of the target gene and the hygromycin resistance gene three fragments were connected by PCR reaction. 25 μL PCR reaction system: 12.5 μL Mix enzyme, 1 μL upstream and downstream fragments of target gene, 3 μL hygromycin resistance gene fragment, 7.5 μL H2O. PCR reaction program: pre-denaturation at 94 °C for 2 min; 94 °C for 30 s, 60 °C for 10 min, 72 °C for 5 min, 9 cycles of these conditions; 72 °C for 10 min; 16 °C for 10 min. We used the above PCR products as the DNA template to directly repeat the PCR operation in the previous steps, changed the primers to F2 + H3/H2 + R2, and kept other conditions unchanged. The PCR products were electrophoresed on agarose gels and recovered by cutting the gel using the TIANgel Midi Purification Kit.
Fungal protoplasts of the wild-type strain FF43 were directly transformed by the standard PEG-mediated method [66]. The fungus grown on the PDA (containing 200 μg/mL hygromycin) plate was transferred to a new PDA (containing 200 μg/mL hygromycin) plate, cultured at 28 °C for 5 days, using the KAPA3G Plant PCR Kit (Merck & Co. Inc., Kenilworth, NJ, USA) PCR detection. In addition, we set primers on the upstream and downstream of the target gene to amplify the target fragment by f/r PCR. If the wild-type and transformant-amplified fragments had different lengths and no identical bands, it meant that the hygromycin resistance gene had replaced the target gene, that is, the target gene had been knocked out.
4.5. Phenotype Assays
- (a)
- Strain growth rate observation. The fungi block with 7 mm diameter was cut from the outer periphery of the wild-type strain and deletion mutants cultured for 5 days. We cultured these fungi blocks in PDA (Biological Technology Co., Shanghai, China Zhaorui) medium with natural light at 28 °C, measured the diameter of the strains every 24 h for 7 days and observed the morphological characteristics of the strains (color, shape, growth of aerial hyphae, etc.). Analysis of variance (ANOVA) was used to determine the growth between wild-type and mutant strains (Statistical Product Service Solutions (SPSS)). Each strain was inoculated in three replicates [67].
- (b)
- Stress sensitivity test. Oxidative stress experiment: the volume percentages concentration of 0.1%, 0.25%, and 0.5% of H2O2 (Aladdin Biochemical Technology Co., Ltd., Shanghai, China) were added to the PDA medium to prepare H2O2 PDA mediums with different concentrations; osmotic stress experiment: different concentrations of NaCl (National Pharmaceutical Group Chemical Reagent Co., Ltd., Beijing, China) were added to the PDA medium to prepare PDA mediums containing NaCl concentrations of 1 mol·L−1 and 2 mol·L−1; cell wall selection pressure experiment: sorbitol PDA mediums with 1 mol·L−1 and 2 mol·L−1 concentrations were prepared by adding sorbitol; cellulose utilization experiment: cellulose Congo red mediums (Zhaorui Biological Technology Co., Shanghai, China) were used. The 7 mm diameter fungi blocks of F. fujikuroi cultured for 5 days were transferred in the center of the each medium. They were cultured in a 28 °C incubator for 5 days, we observed and measured the strain size and took pictures to record. Analysis of variance was used to determine the growth between wild-type and mutant strains (Data Processing System (DPS)). The above experiments were repeated three times for each treatment [68,69].
- (c)
- Cellophane penetration experiment: tested the penetration ability of F. fujikuroi with cellophane (Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China) equivalent to plant cell wall. Took 5 mm fungi blocks of mutants and wild type strains of F. fujikuroi and placed the mycelium side down in the center of a 9 mm PDA medium. We sterilized a semicircular cellophane with a radius of 4 cm, and stock it on the medium with 3/4 fungi block covered. Each strain was inoculated in three replicates. Analysis of variance was used to determine the growth between wild-type and mutant strains (DPS). Strains were cultured in an incubator at 28 °C for 5 days and photographed for recording [70].
4.6. Strains and Inoculation Experiments
F. fujikuroi strain FF43 was isolated from infected rice spikelet in Zhejiang, China. The tested rice variety was Zhongzao 39, which was a bakanae disease-susceptible variety. The artificial inoculation of the bakanae disease pathogen referred to Zhao Yuan [71]. Rice seeds were sterilized with 3% H2O2 for 2 h, soaked for 2 days, and germinated for 1 day. The bakanae disease pathogen and sprouted seeds were inoculated into sterilized vermiculite respectively, and the vermiculite without pathogen was used to grow rice as a control. The treated rice seeds were placed in an incubator with 12 h of light and 12 h of darkness at 30 °C. They were watered each morning and evening, two milliliters at a time. There were three replicates for each treatment. DPS was used to analyze the growth differences of rice seedlings inoculated by wild-type or deletion mutant strains.
5. Conclusions
We identified 44 bZIP TFs by analyzing the whole genome data of F. fujikuroi. Through gene structure analysis, phylogenetic analysis of FfbZIP TFs, and the phenotype and pathogenic analysis of deletion mutants, it was confirmed that these TFs took part in F. fujikuroi growth, nutrient absorption and utilization, stress resistance and pathogenicity. Our research results provided help for an in-depth understanding of the regulatory mechanism of F. fujikuroi growth, stress management and pathogenicity, and provided a reference for the research of other phytopathogenic fungi.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23126658/s1.
Author Contributions
Conceptualization, K.Z., S.H. and L.L.; methodology, K.Z. and L.L.; software, K.Z. and L.L.; validation, K.Z.; formal analysis, K.Z.; investigation, K.Z.; resources, K.Z., S.H. and L.L.; data curation, K.Z.; writing—original draft preparation, K.Z.; writing—review and editing, K.Z.; visualization, K.Z; supervision, S.H. and L.L.; project administration, S.H. and L.L.; funding acquisition, S.H. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
The research in this paper was funded by Sinograin II: Technological Innovation to support environmentally friendly food production and food safety under a changing climate—Opportunities and challenges for Norway-China cooperation (CHN-2152, 14-0039 SINOGRAIN project), Key Research and Development Program of Zhejiang Province (2018C02G3010454), Technology Research Program of Zhejiang Province (2022C02014), Science and Technology Agricultural Development Project in Shanghai (2019-02-08-00-08-F01127), and the Natural Science Foundation of Zhejiang Province (LY21C140001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in insert article or supplementary material here.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gupta, A.; Solanki, I.; Bashyal, B.; Singh, Y. Bakanae of rice: An emerging disease in Asia. J. Anim. Plant Sci. 2015, 25, 1499–1514. [Google Scholar]
- Hwang, I.S.; Kang, W.R.; Hwang, D.J.; Bae, S.C.; Yun, S.H.; Ahn, I.P. Evaluation of bakanae disease progression caused by Fusarium fujikuroi in Oryza sativa L. J. Microbiol. 2013, 51, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.S.; Ahn, I.P. Multi-homologous recombination-based gene manipulation in the rice pathogen Fusarium fujikuroi. Plant Pathol. J. 2016, 32, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Sieber, C.; Bargen, K.V.; Studt, L.; Niehaus, E.M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef]
- Zhang, J.; Qu, S.; Guo, Z.; Du, X.; Du, B.; Zhang, Z. Biological functions of plant bZIP transcription factors. Northwest Bot. 2011, 44, 1066–1075. [Google Scholar]
- Yang, Z.; Wang, X.; Li, X.; Yang, C. Research progress on transcription factors in higher plants. Hereditary 2004, 26, 403–408. [Google Scholar]
- Weirauch, M.T.; Hughes, T.R. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell Biochem. 2011, 10, 25–73. [Google Scholar]
- Traven, A.; Jelicic, B.; Sopta, M. Yeast Gal4: A transcriptional paradigm revisited. EMBO J. 2006, 7, 496–499. [Google Scholar] [CrossRef]
- Yin, W.; Cui, P.; Wei, W.; Lin, Y.; Luo, C. Genome-wide identification and analysis of the basic leucine zipper (bZIP) transcription factor gene family in Ustilaginoidea virens. Genome 2017, 60, 35–49. [Google Scholar] [CrossRef]
- Park, J.; Park, J.; Jang, S.; Kim, S.; Kong, S.; Choi, J.; Ahn, K.; Kim, J.; Lee, S.; Kim, S.; et al. FTFD: An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 2008, 24, 1024–1025. [Google Scholar] [CrossRef]
- Zg, E.; Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 2014, 13, 3025. [Google Scholar]
- Ali, Z.; Sarwat, S.S.; Karim, I.; Faridi, R.; Jaskani, M.J.; Khan, A.A. Functions of plant’s bZIP transcription factors. Pak. J. Agric. Sci. 2016, 53, 303–314. [Google Scholar]
- Lin, C.H.; Yang, S.L.; Chung, K.R. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant-Microbe Interact. 2009, 22, 942. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Yamashino, T.; Mizuno, T. Characterization of bZip-Type Transcription Factor AtfA with Reference to Stress Responses of Conidia of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2008, 72, 2756–2760. [Google Scholar] [CrossRef]
- Hagiwara, D.; Kondo, A.; Fujioka, T.; Abe, K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 2008, 54, 325–338. [Google Scholar] [CrossRef]
- Balázs, A.; Pócsi, I.; Hamari, Z.; Leiter, E.; Emri, T.; Miskei, M.; Oláh, J.; Tóth, V.; Hegedus, N.; Prade, R.A.; et al. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genom. 2010, 283, 289–303. [Google Scholar] [CrossRef]
- Mulder, H.J.; Saloheimo, M.; Penttil, M.; Madrid, S.M. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genom. 2004, 271, 130–140. [Google Scholar] [CrossRef]
- Richie, D.L.; Hartl, L.; Aimanianda, V.; Winters, M.S.; Fuller, K.K.; Miley, M.D.; White, S.; McCarthy, J.W.; Latgé, J.P.; Feldmesser, M.; et al. A Role for the Unfolded Protein Response (UPR) in Virulence and Antifungal Susceptibility in Aspergillus fumigatus. PLos Pathog. 2009, 5, e1000258. [Google Scholar] [CrossRef]
- Cui, R.; Zhang, Y.; Chen, X.; Gu, C.; Zhang, Q. The latest research progress of plant bZIP involved in the regulation of stress response. Biotechnol. Bull. 2019, 35, 143–155. [Google Scholar]
- Ma, X.; Hu, X.; Zhang, Y.; Yan, H. bZIP transcription factor in plant abiotic stress. Mol. Plant Breed. 2020, 18, 126–133. [Google Scholar]
- Pérez-Pérez, W.D.; Carrasco-Navarro, U.; García-Estrada, C.; Kosalková, K.; Gutiérrez-Ruíz, M.C.; Barrios-González, J.; Fierro, F. bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum. Microb. Cell Factories 2022, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Di, R.; Falkowski, P.G. Light-harvesting complex gene regulation by a MYB-family transcription factor in the marine diatom, Phaeodactylum tricornutum. Photosynth. Res. 2022, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Gaslonde, T.; Roullier, C.; Wang, H.; Bodin, J.; Porée, F.H.; Ruprich-Robert, G.; Chapeland-Leclerc, F. Functional characterization of the GATA-type transcription factor PaNsdD in the filamentous fungus Podospora anserina and its interplay with the sterigmatocystin pathway. Appl. Environ. Microbiol. 2022, 88, e0237821. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, Y.; Tian, S. Function of pH-dependent transcription factor PacC in regulating development, pathogenicity, and mycotoxin biosynthesis of phytopathogenic fungi. FEBS J. 2022, 289, 1723–1730. [Google Scholar] [CrossRef]
- Gai, Y. Comparative Genome and Function of bZIP Transcription Factor of Alternaria sp.; Zhejiang University: Hangzhou, China, 2019. [Google Scholar]
- Lu, J. Functional Study of the bZIP Transcription Factor PsBZPl of Phytophthora sojae; Nanjing Agricultural University: Nanjing, China, 2012. [Google Scholar]
- Chen, J. The Functional Study of the bZIP Transcription Factor Ss-Adal in Sclerotinia Sclerotiorum; Jilin University: Changchun, China, 2014. [Google Scholar]
- Xu, R. Identification of P450 and bZIP Genes in the Biosynthesis of Ochratoxin A in Aspergillus niger; Tianjin University of Science and Technology: Tianjin, China , 2019. [Google Scholar]
- Zhao, J.; Peng, M.; Chen, W.; Xing, X.; Shan, Y.; Fan, Z.; Shi, Y.; Li, H.; Yang, X.; Li, H.; et al. Transcriptome analysis and functional validation identify a putative bZIP transcription factor, Fpkapc, that regulates development, stress responses, and virulence in Fusarium pseudograminearum. Phytopathology 2022, 29, PHYTO12210520R. [Google Scholar]
- Shin, J.; Bui, D.C.; Kim, S.; Jung, S.Y.; Nam, H.J.; Lim, J.Y.; Choi, G.J.; Lee, Y.W.; Kim, J.E.; Son, H. The novel bZIP transcription factor Fpo1 negatively regulates perithecial development by modulating carbon metabolism in the ascomycete fungus Fusarium graminearum. Environ. Microbiol. 2020, 22, 2596–2612. [Google Scholar] [CrossRef]
- Chen, L.; Ma, Y.; Zhao, J.; Geng, X.; Chen, W.; Ding, S.; Li, H.; Li, H. The bZIP transcription factor FpAda1 is essential for fungal growth and conidiation in Fusarium pseudograminearum. Curr. Genet. 2020, 66, 507–515. [Google Scholar] [CrossRef]
- Hussain, S.; Tai, B.; Hussain, A.; Jahan, I.; Yang, B.; Xing, F. Genome-wide identification and expression analysis of the basic leucine zipper (bZIP) transcription factor gene family in Fusarium graminearum. Genes 2022, 13, 607. [Google Scholar] [CrossRef]
- Wagner, D.; Schmeinck, A.; Mos, M.; Morozov, I.Y.; Caddick, M.X.; Tudzynski, B. The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specific loci. Eukaryot. Cell 2010, 9, 1588–1601. [Google Scholar] [CrossRef]
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. The importance of being flexible: The case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 2009, 10, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Pousada, C.; Menezes, R.A.; Pimentel, C. The Yap family and its role in stress response. Yeast 2010, 27, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Deppmann, C.D.; Alvania, R.S.; Taparowsky, E.J. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol. Biol. Evol. 2006, 23, 1480–1492. [Google Scholar] [CrossRef]
- Rosonina, E.; Duncan, S.M.; Manley, J.L. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes 2012, 26, 350–355. [Google Scholar] [CrossRef]
- Guo, L.; Ghassemian, M.; Komives, E.A.; Russell, P. Cadmium-induced proteome remodeling regulated by Spc1/Sty1 and Zip1 in fission yeast. Toxicol. Sci. 2012, 129, 200–212. [Google Scholar] [CrossRef][Green Version]
- Seong, K.H.; Maekawa, T.; Ishii, S. Inheritance and memory of stress-induced epigenome change: Roles played by the ATF-2 family of transcription factors. Genes Cells 2012, 17, 249–263. [Google Scholar] [CrossRef]
- Cakir, B. Bax induces activation of the unfolded protein response by inducing HAC1 mRNA splicing in Saccharomyces cerevisiae. Yeast 2012, 29, 395–406. [Google Scholar] [CrossRef]
- Jakubowski, S.J.; Kerr, J.E.; Garza, I.; Krishnamoorthy, V.; Bayliss, R.; Waksman, G.; Christie, P.J. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 2010, 71, 779–794. [Google Scholar] [CrossRef]
- Flory, M.R.; Carson, A.R.; Muller, E.G.; Aebersold, R. An Smc-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol. Cell 2004, 16, 619–630. [Google Scholar] [CrossRef]
- Xing, X. Functional characterization of the predicted Smc domain in Nesprin-2. Biochim. Biophys. Acta-Lipids Lipid Metab. 2013, 79, 42–44. [Google Scholar]
- Shi, Z.; Bai, X.; Yu, H. Research progress on molecular mechanism of cohesin maintaining genome structure. Nat. J. 2021, 43, 1–8. [Google Scholar]
- Iams, K.P.; Young, J.R.; Nene, V.; Desai, J.; Webster, P.; Musoke, A.J. Characterisation of the gene encoding a 104-kilodalton microneme-rhoptry protein of Theileria parva. Mol. Biochem. Parasitol. 1990, 39, 47–60. [Google Scholar] [CrossRef]
- Riou, J.F.; Gabillot, M.; Philippe, M.; Schrevel, J.; Riou, G. Purification and characterization of Plasmodium berghei DNA topoisomerases I and II: Drug action, inhibition of decatenation and relaxation, and stimulation of DNA cleavage. Biochemistry 1986, 25, 1471–1479. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, H.Y.; Wei, L.; Lu, D.; Du, M.; Yuan, M.; Shi, D.; Chen, X.; Wang, P.; Chen, X.L.; et al. Discovery of exolytic heparinases and their catalytic mechanism and potential application. Nat. Commun. 2021, 12, 1263. [Google Scholar] [CrossRef]
- Carlson, C.M.; Endrizzi, B.T.; Wu, J.; Ding, X.; Weinreich, M.A.; Walsh, E.R.; Wani, M.A.; Lingrel, J.B.; Hogquist, K.A.; Jameson, S.C. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 2006, 442, 299–302. [Google Scholar] [CrossRef]
- Castillo, E.A.; Ayté, J.; Chiva, C.; Moldón, A.; Carrascal, M.; Abián, J.; Jones, N.; Hidalgo, E. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 2002, 45, 243–254. [Google Scholar] [CrossRef]
- Gao, J.; Davidson, M.K.; Wahls, W.P. Distinct regions of ATF/CREB proteins Atf1 and Pcr1 control recombination hotspot ade6-M26 and the osmotic stress response. Nucleic Acids Res. 2008, 36, 2838–2851. [Google Scholar] [CrossRef]
- Peters, L.L.; Turtzo, L.C.; Birkenmeier, C.S.; Barker, J.E. Distinct fetal Ank-1 and Ank-2 related proteins and mRNAs in normal and nb/nb mice. Blood 1993, 81, 2144–2149. [Google Scholar] [CrossRef]
- Schlieker, C.; Korbel, G.A.; Kattenhorn, L.M.; Ploegh, H.L. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 2005, 79, 15582–15585. [Google Scholar] [CrossRef]
- Atambayeva, S.A.; Khailenko, V.A.; Ivashchenko, A.T. Intron and Exon Length Variation in Arabidopsis, Rice, Nematode, and Human. Mol. Biol. 2008, 42, 648. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Litou, Z.I.; Bagos, P.G.; Tsirigos, K.D.; Liakopoulos, T.D.; Hamodrakas, S.J. Prediction of cell wall sorting signals in gram-positive bacteria with a hidden markov model: Application to complete genomes. J. Bioinform. Comput. Biol. 2008, 6, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W (improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice). Springer Neth. 2008, 5, 376–377. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Anyuan, G.; Qihui, Z.; Xin, C.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar]
- Lister, W.C. Tools of biological research. Phys. Bull. 1962, 13, 23. [Google Scholar] [CrossRef]
- Yu, M.; Yu, J.; Cao, H.; Yong, M.; Liu, Y. Genome-wide identification and analysis of the GATA transcription factor gene family in Ustilaginoidea virens. Genome 2019, 12, 807–816. [Google Scholar] [CrossRef]
- Jana, T.; Lam-Tung, N.; Arndt, V.H.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 44–46. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Wang, Y.; Wei, W.; Li, C.; Liu, Y.; Qu, J.; Meng, Q.; Lin, Y.; Yin, W.; Yang, Y.; et al. The Bax inhibitor UvBI-1, a negative regulator of mycelial growth and conidiation, mediates stress response and is critical for pathogenicity of the rice false smut fungus Ustilaginoidea virens. Curr. Genet. 2019, 65, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Hou, Z. Knockout and function of FgPDE1 gene in Fusarium graminearum. China Biotechnol. 2015, 35, 59–67. [Google Scholar]
- Zhang, Y.; Qu, H. Construction and functional analysis of FvST12 gene knockout vector from Fusarium verticillium. China Agric. Sci. 2011, 44, 9. [Google Scholar]
- Coughlan, M.P. Mechanism of cellulose degradation by bacteria and fungi (Part 1). Anim. Feed. Sci. Technol. 1991, 32, 77–100. [Google Scholar] [CrossRef]
- Wang, X. Knockout and phenotypic analysis of cat1 gene of Fusarium oxysporum 4 physiological race. J. Trop. Crops 2017, 38, 8. [Google Scholar]
- Zhao, Y. Research on Biocontrol Bacteria Screening and Prevention and Control of Bakanae Disease in Rice; Guangxi University: Nanning, China, 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).