Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism
Abstract
1. Introduction
2. Current Status of Antimicrobial Resistance in A. baumannii
Intrinsic Resistance in A. baumannii
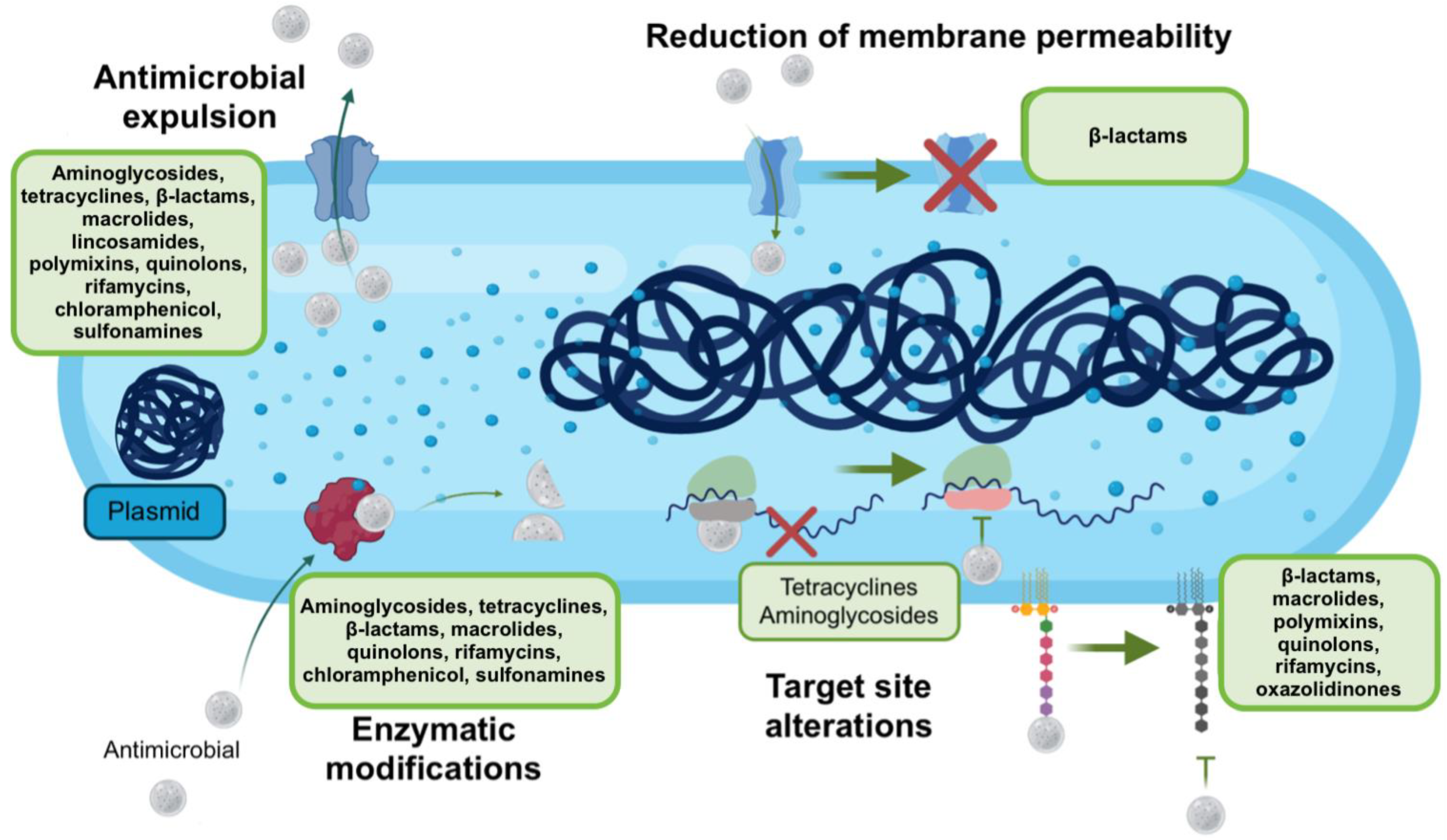
3. Mechanisms of Antimicrobial Resistance
3.1. Antimicrobial Modification
3.2. Antimicrobial Efflux and Decreased Permeability
3.3. Antimicrobial Sequestration
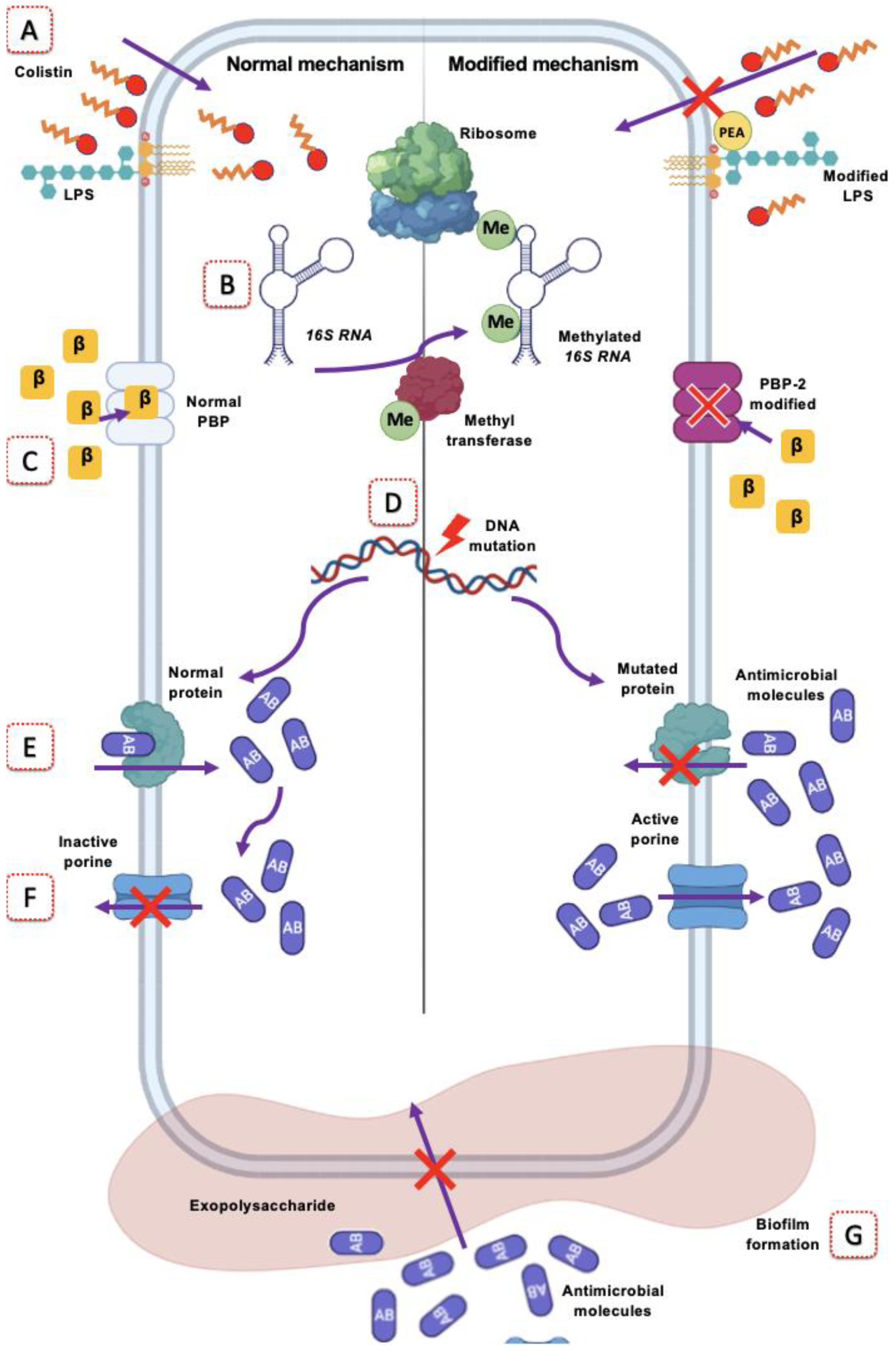
3.4. Modification of the Target Site
4. Antimicrobials Whose Effect Is Evaded by Target Site Modification in A. baumannii
4.1. Polymyxin Resistance
Colistin Resistance in A. baumannii
4.2. Resistance to β-Lactams
4.3. Rifampicin Resistance
4.4. Resistance to Fluoroquinolones
4.5. Macrolides Resistance
4.6. Tetracycline Resistance
4.7. Oxazolidinone Resistance
4.8. Aminoglycoside Resistance
5. Treatment Options for A. baumannii Infections
5.1. Current Treatment Options
- Use antimicrobial associations.
- Administer them by optimizing the pharmacokinetic/pharmacodynamic (PK/PD) ratio.
- Give preference to antimicrobials that retain some degree of in vitro activity.
- Optimize other therapeutic measures, e.g., surgical debridement or removal of infected tissues or devices.
- Be sure that it is an infection instead of a colonization before starting the treatment.
- ■
- Bacteremia: In cases associated with catheters or intravascular devices, give priority to their removal. Start the scheme with carbapenems, aminoglycosides, and rifampicin.
- ■
- Pneumonia: Give preference to combined therapy for community-acquired pneumonia; for ventilator-associated pneumonia, when the patient has endotracheal intubation and mechanical ventilatory support, the patient should be placed in a semi-sitting position between 30° and 45°, preferably in a kinetic bed, which provides position changes with head elevation, in order to reduce the production of secretions. There is not enough evidence to support the generalized use of endotracheal cannulas impregnated with antiseptics for the reduction of VAP. (GPC IMSS624-13).
- ■
- Urinary Tract Infection: Give preference in the plan to aminoglycosides and carbapenems. Use the Foley catheters for as long as necessary and remove them as soon as possible.
- ■
- Central Nervous System Infection: Give preference in the plan to carbapenems and rifampicin for systemic use. Evaluate intraventricular or intrathecal use with aminoglycosides; however, their use is controversial since there are not much data in this regard.
- ■
- Abdominal infection. In this site, it is essential to give priority to surgical treatment; start the plan with tigecycline, sulbactam, carbapenems, or aminoglycosides.
- ■
- Skin and soft tissue infection: Prioritize surgical treatment by removing dead and contaminated tissue; initiate scheme with tigecycline, carbapenems, and aminoglycosides.
- ■
- Infection or Osteoarticular: Give priority to surgical treatment by scraping bone and dead tissue; perform surgical lavage. Give preference in the plan to aminoglycosides and carbapenems.
- ○
- Sulbactam, if MIC is less than or equal to 32 mg/L;
- ○
- Meropenem or imipenem, if MIC is less than or equal to 16 mg/L;
- ○
- Colistin: Less than or equal to 1 mg/L;
- ○
- Tigecycline, if MIC is less than or equal to 4 mg/L.
5.2. Alternative Therapies without Antibiotics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Manyahi, J.; Kibwana, U.; Mgimba, E.; Majigo, M. Multi-drug resistant bacteria predict mortality in bloodstream infection in a tertiary setting in Tanzania. PLoS ONE 2020, 15, e0220424. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.D.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv. Sci. (Weinh) 2019, 7, 1901872. [Google Scholar] [CrossRef] [PubMed]
- Organización Mundial de la Salud. OMS|La Resistencia a Los Antimicrobianos. Available online: https://www.who.int/antimicrobial-resistance/es/ (accessed on 31 December 2021).
- Touchon, M.; Cury, J.; Yoon, E.J.; Krizova, L.; Cerqueira, G.C.; Murphy, C.; Feldgarden, M.; Wortman, J.; Clermont, D.; Lambert, T.; et al. The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol. Evol. 2014, 6, 2866–2882. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Giammanco, A.; Calà, C.; Fasciana, T.; Dowzicky, M.J. Global Assessment of the Activity of Tigecycline against Multidrug-Resistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. MSphere 2017, 2, e00310–e00316. [Google Scholar] [CrossRef]
- Nguyen, M.; Joshi, S.G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital acquired infections: A scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Meawed, T.E.; Ahmed, S.M.; Mowafy, S.; Samir, G.M.; Anis, R.H. Bacterial and fungal ventilator associated pneumonia in critically ill COVID-19 patients during the second wave. J. Infect. Public Health 2021, 14, 1375–1380. [Google Scholar] [CrossRef]
- Rouzé, A.; Martin-Loeches, I.; Povoa, P.; Makris, D.; Artigas, A.; Bouchereau, M.; Lambiotte, F.; Metzelard, M.; Cuchet, P.; Boulle Geronimi, C.; et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: A European multicenter cohort study. Intensive Care Med. 2021, 47, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Castanheira, M.; Arends, S.; Goossens, H.; Flamm, R.K. Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: Results from 20 years of the SENTRY Antimicrobial Surveillance Program (1997–2016). J. Antimicrob. Chemother. 2019, 74, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.G.; Brito, J.C.M.; da Cruz Nizer, W.S. Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: Two problems, one solution? Med. Hypotheses. 2020, 144, 110139. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.A.A.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life 2020, 72, 2097–2111. [Google Scholar] [CrossRef]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Clark, N.M.; Zhanel, G.G.; Lynch, J.P. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Curr. Opin. Crit. Care 2016, 22, 491–499. [Google Scholar] [CrossRef]
- Robledo, I.E.; Aquino, E.E.; Santé, M.I.; Santana, J.L.; Otero, D.M.; León, C.F.; Vázquez, G.J. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 2010, 54, 1354. [Google Scholar] [CrossRef]
- Caneiras, C.; Calisto, F.; da Silva, G.J.; Lito, L.; Melo-Cristino, J.; Duarte, A. First Description of Colistin and Tigecycline-Resistant Acinetobacter baumannii Producing KPC-3 Carbapenemase in Portugal. Antibiotics 2018, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A.; Halat, D.H. Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.A.; Amyes, S.G.B. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Benmahmod, A.B.; Said, H.S.; Ibrahim, R.H. Prevalence and Mechanisms of Carbapenem Resistance Among Acinetobacter baumannii Clinical Isolates in Egypt. Microb. Drug Resist. 2019, 25, 480–488. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, J.H.; Lee, J.J.; Park, K.S.; Karim, A.M.; Lee, C.R.; Jeong, B.C.; Lee, S.H. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci. 2015, 16, 9654–9692. [Google Scholar] [CrossRef]
- Damier-Piolle, L.; Magnet, S.; Brémont, S.; Lambert, T.; Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008, 52, 557–562. [Google Scholar] [CrossRef]
- Savari, M.; Ekrami, A.; Shoja, S.; Bahador, A. Plasmid borne Carbapenem-Hydrolyzing Class D β-Lactamases (CHDLs) and AdeABC efflux pump conferring carbapenem-tigecycline resistance among Acinetobacter baumannii isolates harboring TnAbaRs. Microb. Pathog. 2017, 104, 310–317. [Google Scholar] [CrossRef]
- Cho, Y.J.; Moon, D.C.; Jin, J.S.; Choi, C.H.; Lee, Y.C.; Lee, J.C. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. Dis. 2009, 64, 185–190. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, C.; Wu, J.; Jiang, R.; Mi, Z.; Huang, Z. A novel aminoglycoside-modifying enzyme gene aac (6’)-Ib in a pandrug-resistant Acinetobacter baumannii strain. J. Hosp. Infect. 2009, 73, 184–185. [Google Scholar] [CrossRef]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef] [PubMed]
- Appleman, M.D.; Belzberg, H.; Citron, D.M.; Heseltine, P.N.R.; Yellin, A.E.; Murray, J.; Berne, T.V. In vitro activities of nontraditional antimicrobials against multiresistant Acinetobacter baumannii strains isolated in an intensive care unit outbreak. Antimicrob. Agents Chemother. 2000, 44, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.G.; Alves, M.C.; Cruz, W.S.; Paiva, M.C. Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: A huge public health threat. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1009–1019. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standars Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI supplement M100; Clinical and Laboratory Standars Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Gordon, N.C.; Wareham, D.W. Multidrug-resistant Acinetobacter baumannii: Mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 2010, 35, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Mussi, M.A.; Limansky, A.S.; Viale, A.M. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 2005, 49, 1432–1440. [Google Scholar] [CrossRef]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
- Knight, D.; Dimitrova, D.D.; Rudin, S.D.; Bonomo, R.A.; Rathera, P.N. Mutations Decreasing Intrinsic β-Lactam Resistance Are Linked to Cell Division in the Nosocomial Pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016, 60, 3751. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef]
- Moradi, J.; Hashemi, F.B.; Bahador, A. Antibiotic Resistance of Acinetobacter baumannii in Iran: A Systemic Review of the Published Literature. Osong Public Health Res. Perspect. 2015, 6, 79–86. [Google Scholar] [CrossRef]
- Chávez, M.; Gómez, R.F.; Cabrera, C.E.; Esparza, M. Patrones de resistencia a antibióticos de Acinetobacter baumannii en un hospital de Colombia. An. Fac. Med. 2015, 76, 21–26. [Google Scholar] [CrossRef][Green Version]
- Bonnin, R.A.; Nordmann, P.; Poirel, L. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: A state of the art. Expert Rev. Anti-Infect. Ther. 2013, 11, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Mahon, C.R.; Lehman, D.C. Textbook of Diagnostic Microbiology, 6th ed.; Elsevier Saunders: St. Louis, MI, USA, 2019; pp. 247–268. [Google Scholar]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Bacterial resistance to antibiotics: Enzymatic degradation and modification. Adv. Drug Deliv. Rev. 2005, 57, 1451–1470. [Google Scholar] [CrossRef] [PubMed]
- Antunes, N.T.; Fisher, J.F. Acquired Class D β-Lactamases. Antibiotics 2014, 3, 398–434. [Google Scholar] [CrossRef]
- Li, H.; Luo, Y.F.; Williams, B.J.; Blackwell, T.S.; Xie, C.M. Structure and function of OprD protein in Pseudomonas aeruginosa: From antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 2012, 302, 63–68. [Google Scholar] [CrossRef]
- Martínez, J.L.; Muniesa, M.; Dionisio, F.; Kaur, P.; Peterson, E. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 2928. [Google Scholar] [CrossRef]
- Pagès, J.M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef]
- Kobylka, J.; Kuth, M.S.; Müller, R.T.; Geertsma, E.R.; Pos, K.M. AcrB: A mean, keen, drug efflux machine. Ann. N. Y. Acad. Sci. 2020, 1459, 38–68. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Lambert, P.A. Bacterial resistance to antibiotics: Modified target sites. Adv. Drug Deliv. Rev. 2005, 57, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 2003, 51, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Gehrlein, M.; Leying, H.; Cullmann, W.; Wendt, S.; Opferkuch, W. Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy 1991, 37, 405–412. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Domingues, S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Tipton, K.A.; Rather, P.N. Extraction and Visualization of Capsular Polysaccharide from Acinetobacter baumannii. Methods Mol. Biol. 2019, 1946, 227–231. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Basatian-Tashkan, B.; Niakan, M.; Khaledi, M.; Afkhami, H.; Sameni, F.; Bakhti, S.; Mirnejad, R. Antibiotic resistance assessment of Acinetobacter baumannii isolates from Tehran hospitals due to the presence of efflux pumps encoding genes (adeA and adeS genes) by molecular method. BMC Res. Notes 2020, 13, 1–6. [Google Scholar] [CrossRef]
- Martínez-Trejo, A. Detección del Sistema CRISPR-Cas en Cepas Clínicas de Pseudomonas aeruginosa y Acinetobacter baumannii. Master’s Thesis, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Ciudad de Mexico, Mexico, 3 August 2021. [Google Scholar]
- Singh, H.; Thangaraj, P.; Chakrabarti, A. Acinetobacter baumannii: A Brief Account of Mechanisms of Multidrug Resistance and Current and Future Therapeutic Management. J. Clin. Diagn. Res. 2013, 7, 2602. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Multidrug-Resistant Gram-Negative Pathogens: The Urgent Need for “Old” Polymyxins. Adv. Exp. Med. Biol. 2019, 1145, 9–13. [Google Scholar] [CrossRef]
- Powers, M.J.; Herrera, C.M.; Tucker, A.T.; Davies, B.W.; Trent, M.S. Isolation of Lipid Cell Envelope Components from Acinetobacter baumannii. Methods Mol. Biol. 2019, 1946, 233–252. [Google Scholar] [CrossRef]
- Boinett, C.J.; Cain, A.K.; Hawkey, J.; Hoang, N.T.D.; Khanh, N.N.T.; Thanh, D.P.; Dordel, J.; Campbell, J.I.; Lan, N.P.H.; Mayho, M.; et al. Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb. Genom. 2019, 5, e000246. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.X.; Wozniak, J.E.; Weiss, D.S. Methods to Evaluate Colistin Heteroresistance in Acinetobacter baumannii. Methods Mol. Biol. 2019, 1946, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.S.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Transcriptome Remodeling of Acinetobacter baumannii during Infection and Treatment. MBio 2017, 8, e02193-16. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Moussa, S.H.; Durand-Réville, T.F.; Tommasi, R.; Miller, A. Acinetobacter baumannii OmpA Is a Selective Antibiotic Permeant Porin. ACS Infect. Dis. 2018, 4, 373–381. [Google Scholar] [CrossRef]
- Hamilton, T.E.; Lawrence, P.J. The formation of functional penicillin-binding proteins. J. Biol. Chem. 1975, 250, 6578–6585. [Google Scholar] [CrossRef]
- Cayô, R.; Rodríguez, M.C.; Espinal, P.; Fernández-Cuenca, F.; Ocampo-Sosa, A.A.; Pascual, Á.; Ayala, J.A.; Vila, J.; Martínez-Martínez, L. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 5907–5913. [Google Scholar] [CrossRef]
- Vashist, J.; Tiwari, V.; Das, R.; Kapil, A.; Rajeswari, M.R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 2011, 133, 332. [Google Scholar]
- Vázquez-López, R.; Solano-Gálvez, S.G.; Vignon-Whaley, J.J.J.; Vaamonde, J.A.A.; Alonzo, L.A.P.; Reséndiz, A.R.; Álvarez, M.M.; López, E.N.V.; Franyuti-Kelly , G.; Álvarez-Hernández, D.A.; et al. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 2011, 63, 1061–1067. [Google Scholar] [CrossRef]
- Tupin, A.; Gualtieri, M.; Roquet-Banères, F.; Morichaud, Z.; Brodolin, K.; Leonetti, J.P. Resistance to rifampicin: At the crossroads between ecological, genomic and medical concerns. Int. J. Antimicrob. Agents 2010, 35, 519–523. [Google Scholar] [CrossRef]
- Esterly, J.S.; Richardson, C.L.; Eltoukhy, N.S.; Qi, C.; Scheetz, M.H. Genetic Mechanisms of Antimicrobial Resistance of Acinetobacter baumannii. Ann. Pharmacother. 2011, 45, 218–228. [Google Scholar] [CrossRef]
- Liu, Y.H.; Kuo, S.C.; Lee, Y.T.; Chang, I.C.Y.; Yang, S.P.; Chen, T.L.; Fung, C.P. Amino acid substitutions of quinolone resistance determining regions in GyrA and ParC associated with quinolone resistance in Acinetobacter baumannii and Acinetobacter genomic species 13TU. J. Microbiol. Immunol. Infect. 2012, 45, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ardebili, A.; Lari, A.R.; Beheshti, M.; Lari, E.R. Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran. J. Basic Med. Sci. 2015, 18, 623. [Google Scholar] [PubMed]
- Lin, Y.C.; Hsia, K.C.; Chen, Y.C.; Sheng, W.H.; Chang, S.C.; Liao, M.H.; Li, S.Y. Genetic Basis of Multidrug Resistance in Acinetobacter Clinical Isolates in Taiwan. Antimicrob. Agents Chemother. 2010, 54, 2078. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to Macrolide Antibiotics in Public Health Pathogens. Cold Spring Harb. Perspect. Med. 2016, 6, a025395. [Google Scholar] [CrossRef]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef]
- Beheshti, M.; Ardebili, A.; Beheshti, F.; Lari, A.R.; Siyadatpanah, A.; Pournajaf, A.; Gautam, D.; Dolma, K.G.; Nissapatorn, V. Tetracycline resistance mediated by tet efflux pumps in clinical isolates of Acinetobacter baumannii. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, 88. [Google Scholar] [CrossRef]
- Falagas, M.E.; Vardakas, K.Z.; Kapaskelis, A.; Triarides, N.A.; Roussos, N.S. Tetracyclines for multidrug-resistant Acinetobacter baumannii infections. Int. J. Antimicrob. Agents 2015, 45, 455–460. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Pages, J.M.; Ferrand, A. Clinical Status of Efflux Resistance Mechanisms in Gram-Negative Bacteria. Antibiotics 2021, 10, 1117. [Google Scholar] [CrossRef]
- Bozdogan, B.; Appelbaum, P.C. Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 2004, 23, 113–119. [Google Scholar] [CrossRef]
- Nie, L.; Lv, Y.; Yuan, M.; Hu, X.; Nie, T.; Yang, X.; Li, G.; Pang, J.; Zhang, J.; Li, C.; et al. Genetic basis of high level aminoglycoside resistance in Acinetobacter baumannii from Beijing, China. Acta Pharm. Sin. B 2014, 4, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, S.M.; Farshadzadeh, Z.; Janabadi, S.; Musavi, M.; Shahi, F.; Moradi, M.; Khoshnood, S. Evaluating the frequency of carbapenem and aminoglycoside resistance genes among clinical isolates of Acinetobacter baumannii from Ahvaz, south-west Iran. New Microbes New Infect. 2020, 38, 100779. [Google Scholar] [CrossRef] [PubMed]
- Tahbaz, S.V.; Azimi, L.; Lari, A.R. Characterization of aminoglycoside resistance mechanisms in Acinetobacter Baumannii isolates from burn wound colonization. Ann. Burn. Fire Disasters 2019, 32, 115. [Google Scholar]
- Rizk, M.A.; Abou El-Khier, N.T. Aminoglycoside Resistance Genes in Acinetobacter baumannii Clinical Isolates. Clin. Lab. 2019, 65, 1285–1291. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Papathanakos, G.; Andrianopoulos, I.; Papathanasiou, A.; Koulenti, D.; Gartzonika, K.; Koulouras, V. Pandrug-resistant Acinetobacter baumannii treatment: Still a debatable topic with no definite solutions. J. Antimicrob. Chemother. 2020, 75, 3081. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Golden, A.R.; Zelenitsky, S.; Wiebe, K.; Lawrence, C.K.; Adam, H.J.; Idowu, T.; Domalaon, R.; Schweizer, F.; Zhanel, M.A.; et al. Cefiderocol: A Siderophore Cephalosporin with Activity Against Carbapenem-Resistant and Multidrug-Resistant Gram-Negative Bacilli. Drugs 2019, 79, 271–289. [Google Scholar] [CrossRef]
- Farás Barletta, C.R.; Pérez Ponce, L.J.; Castro Vega, G.; Pujol Pérez, M.; Barletta Del Castillo, J.E.; Dueñas Pérez, Y. Multidrug-resistant Acinetobacter baumannii: A challange for current therapeutic. Medisur 2018, 16, 322–334. [Google Scholar]
- Bagińska, N.; Pichlak, A.; Górski, A.; Jończyk-Matysiak, E. Specific and Selective Bacteriophages in the Fight against Multidrug-resistant Acinetobacter baumannii. Virol. Sin. 2019, 34, 347. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Neshani, A.; Sedighian, H.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Jahangiri, A. Antimicrobial peptides as a promising treatment option against Acinetobacter baumannii infections. Microb. Pathog. 2020, 146, 104238. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in vivo Models. Front. Microbiol. 2021, 12, 677. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; LeBlanc, A.M.; de Souza Oliviera, R.P.; Todorov, S.D. Use of Synbiotics (Probiotics and Prebiotics) to Improve the Safety of Foods. In Practical Food Safety–Contemporary Issues and Future Directions; Bhat, R., Gomez-Lopez, V.M., Eds.; Wiley: Chichester, UK, 2014; pp. 497–531. [Google Scholar]
- Gradisteanu Pircalabioru, G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Czobor Barbu, I.; Cristescu, R.; Chifiriuc, M.-C. Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 2021, 13, 196. [Google Scholar] [CrossRef]
- Aguilar, L.; Giménez, M.J.; García-Rey, C.; Barberán, J.; Aguilar, L. Experiencia clínica con tigeciclina en el tratamiento de infecciones nosocomiales producidas por aislados con mecanismos de resistencia prevalentes. Rev. Esp. Quimioter. 2009, 22, 48–56. [Google Scholar]
| Antimicrobial Family | Antimicrobials | |
|---|---|---|
| β-lactams | Penicillins | Ampicillin a |
| Amoxicillin-clavulanate a | ||
| Ticarcillin Mezlocillin Piperacillin | ||
| Piperacillin-tazobactam | ||
| Cephalosporins | Cefoxitin Cefotetan Cefepime Ceftazidime Cephalothin Ceftriaxone Cefotaxime | |
| Monobactams | Aztreonam a | |
| Carbapenems | Ertapenem a Imipenem Meropenem | |
| Amphenicols | Chloramphenicol a | |
| Phosphonates | Fosfomycin a | |
| Sulfonamides and diaminopyrimidines | Trimetroprim a Trimethoprim/sulfamethoxasol | |
| Aminoglycosides | Amikacin Gentamicin Trobamycin | |
| Macrolides | Erythromycin Azithromycin | |
| Tetracyclines | Glycylcyclines Tigecycline Doxycycline Minocycline | |
| Fluoroquinolones | Ciprofloxacin Norfloxacin Levofloxacin Moxifloxacin Gatifloxacin | |
| Nitrofurans | Nitrofurantoin | |
| Polymyxins | Polymyxin B Colistin | |
| Criteria | Condition | Options | |||
|---|---|---|---|---|---|
| Using Tigecycline (4) as a backbone | If MIC is less than or equal to 2 mg/L (sensitive) | Associated with aminoglycosides (gentamicin or amikacin) | Associated with sulbactam | Associated with rifampicin | Associated with carbapenem |
| If MIC is equal to 4 mg/L (intermediate) | Associated with sulbactam + carbapenem | Sulbactam + fosfomycin | Sulbactam + rifampicin | Sulbactam + aminoglycoside (amikacin/gentamicin) | |
| If MIC is greater than 8 mg/L (resistant), do not use tigecycline. Substitute MINOCYCLINE | Carbapenem (imipenem or meropenem) + sulbactam + rifampin | Carbapenem + sulbactam + aminoglycosides | Carbapenem + aminoglycoside rifampicin | ||
| Using β-lactam-β-lactamase inhibitors | Do not use if the strain is carbapenem-resistant. | Meropenem/vaborbactam | Imipenem/relebactam | Ceftazidime/avibactam | Ceftolozane/tazobactam or aztreonam/avibactam |
| For carbapenem-resistant strains | A. baumannii that does not produce MBL | Ampicillin/sulbactam and trimethoprim/sulfamethoxazole | Ampicillin-sulbactam + polymyxins (polymyxin b/colistin) | Sulbactam/avibactam | Ampicillin/sulbactam with ceftazidime/avibactam |
| New alternatives | |||||
| New antimicrobials | Minocycline alone or in association | Eravacycline | Cefiderocol (resistant strains have been found and availability is limited | ||
| Based on phages and probiotics | vB_Ab-M-G7 | Bϕ-C62 | Βϕ-R2096 | Endolysins of phage ØABP-01 | Bifidobacterium brief on digestive tract infections |
| Molecule-based | DS-8587 is a new fluoroquinolone that acts by inhibiting DNA topoisomerase | BAL 30072 monosulfactam; active against many Gram-negative bacteria, including those producing metallo-β-lactamases and KPC, and has a synergistic effect with carbapenems | GC-072 in preclinical phase. It is an oxoquinolizine compound | Gallium nitrate or gallium protoporphyrin IX whose activity is to sequester Fe ions | Rose Bengal (SecA inhibitor) in combination with imipenem or meropenem |
| Bacteriocins | ST4A produced by E. mundtii | Nisin in clinical trials on pathogens associated with VAP | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Trejo, A.; Ruiz-Ruiz, J.M.; Gonzalez-Avila, L.U.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Loyola-Cruz, M.A.; Bello-López, J.M.; Castro-Escarpulli, G. Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. Int. J. Mol. Sci. 2022, 23, 6582. https://doi.org/10.3390/ijms23126582
Martínez-Trejo A, Ruiz-Ruiz JM, Gonzalez-Avila LU, Saldaña-Padilla A, Hernández-Cortez C, Loyola-Cruz MA, Bello-López JM, Castro-Escarpulli G. Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. International Journal of Molecular Sciences. 2022; 23(12):6582. https://doi.org/10.3390/ijms23126582
Chicago/Turabian StyleMartínez-Trejo, Arturo, Juan Manuel Ruiz-Ruiz, Luis Uriel Gonzalez-Avila, Andrés Saldaña-Padilla, Cecilia Hernández-Cortez, Miguel Angel Loyola-Cruz, Juan Manuel Bello-López, and Graciela Castro-Escarpulli. 2022. "Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism" International Journal of Molecular Sciences 23, no. 12: 6582. https://doi.org/10.3390/ijms23126582
APA StyleMartínez-Trejo, A., Ruiz-Ruiz, J. M., Gonzalez-Avila, L. U., Saldaña-Padilla, A., Hernández-Cortez, C., Loyola-Cruz, M. A., Bello-López, J. M., & Castro-Escarpulli, G. (2022). Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. International Journal of Molecular Sciences, 23(12), 6582. https://doi.org/10.3390/ijms23126582












