Determination of Intra- and Extracellular Metabolic Adaptations of 3D Cell Cultures upon Challenges in Real-Time by NMR
Abstract
1. Introduction
- To employ diffusion-weighted NMR spectroscopy to separate between intra- and extracellular metabolic contributions in living human 3D cell cultures in a perfused bioreactor system and to investigate in vitro intracellular metabolic fingerprints;
- To collect from the bioreactor outflowing supernatant and to perform extracellular metabolic footprint characterisation providing complementary insights on the metabolic ongoing processes.
2. Results
2.1. 3D Cell Culture Characterisation
2.1.1. Evaluation of Scaffold Volume
2.1.2. Cellular Volume Evaluation with 23Na NMR
2.1.3. Cell Viability
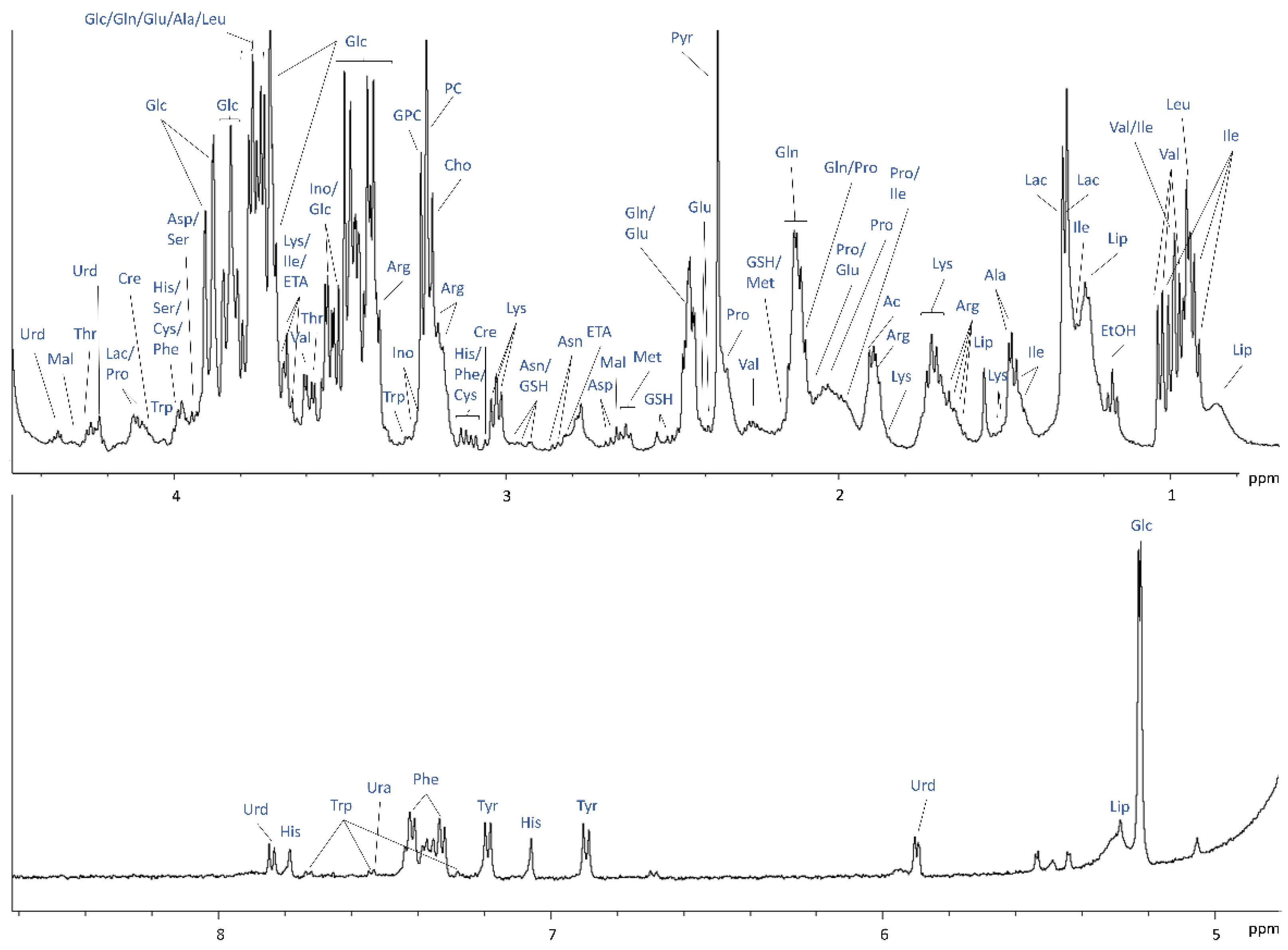
2.2. 1H NMR of Cells and Medium without Separation in the Bioreactor for Protocol Optimisation and Metabolite Assignments
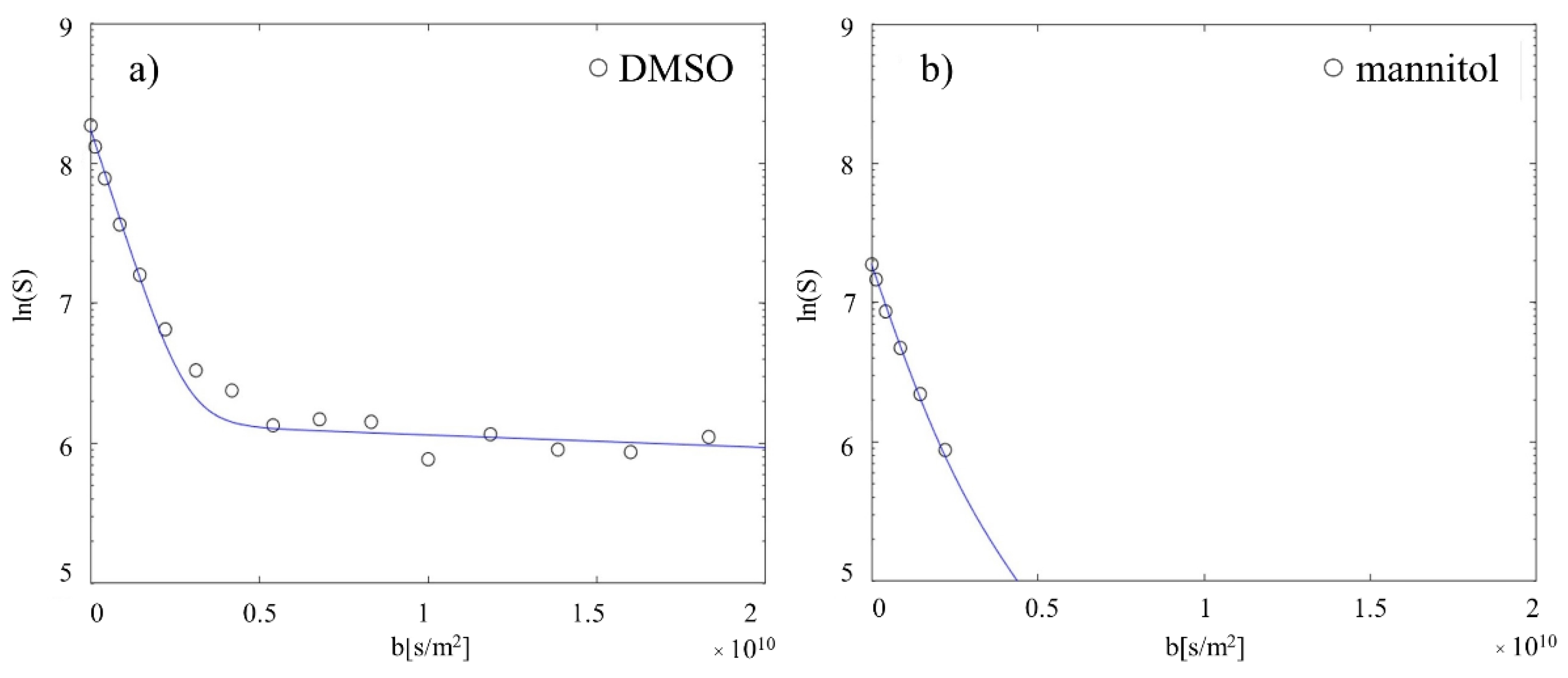
2.3. Validation of Diffusion Technique and Quantification Method
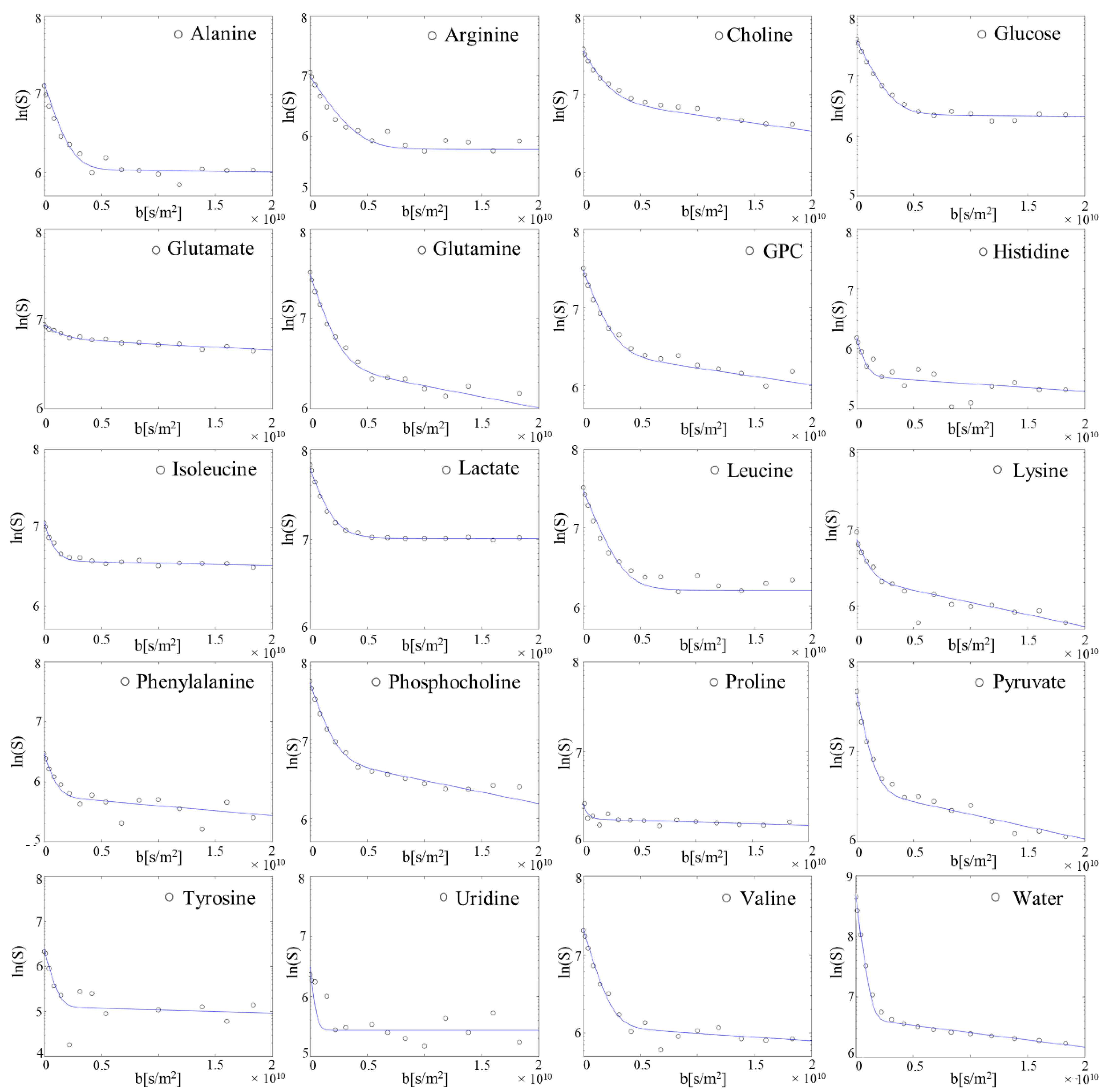
2.4. 1H NMR Diffusion Measurements
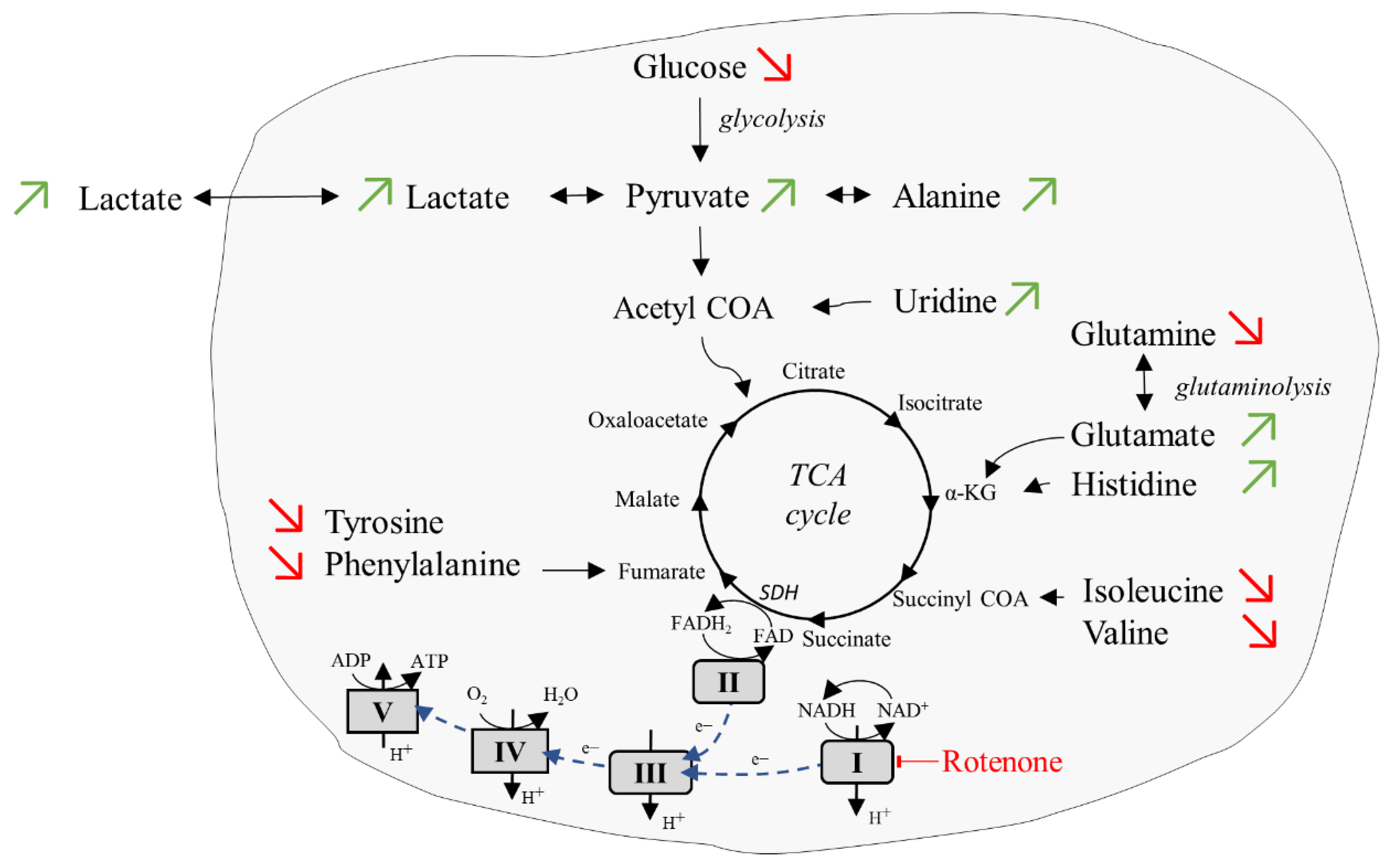
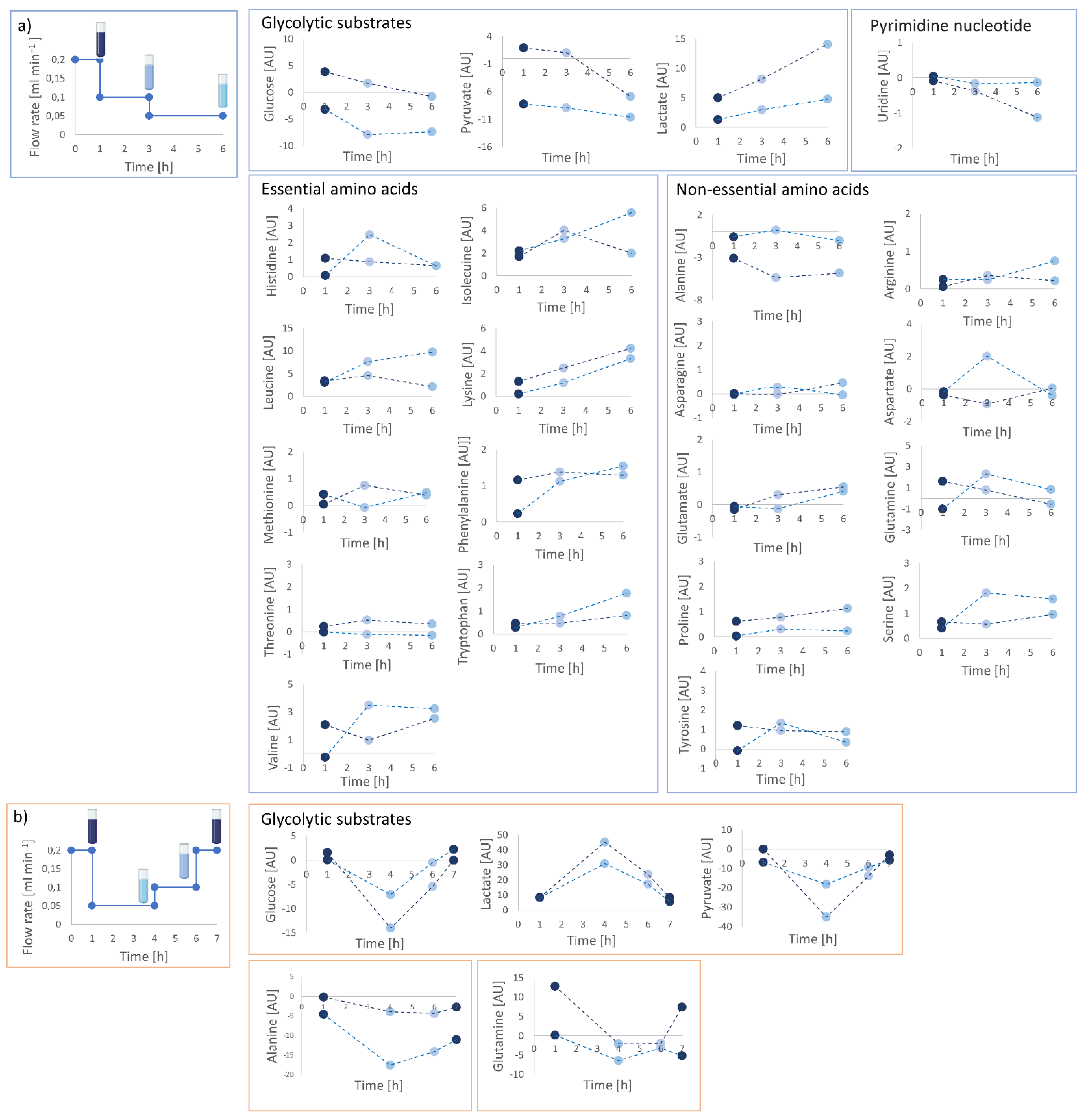
2.5. Glycolytic Stress Test
2.6. Supernatant Collection and Extracellular Footprint Determination
3. Discussion
4. Materials and Methods
4.1. From 2D to 3D Cell Culture
4.1.1. 2D Cell Culture
4.1.2. 3D Cell Culture Preparation
4.2. Preparation for the NMR Measurement
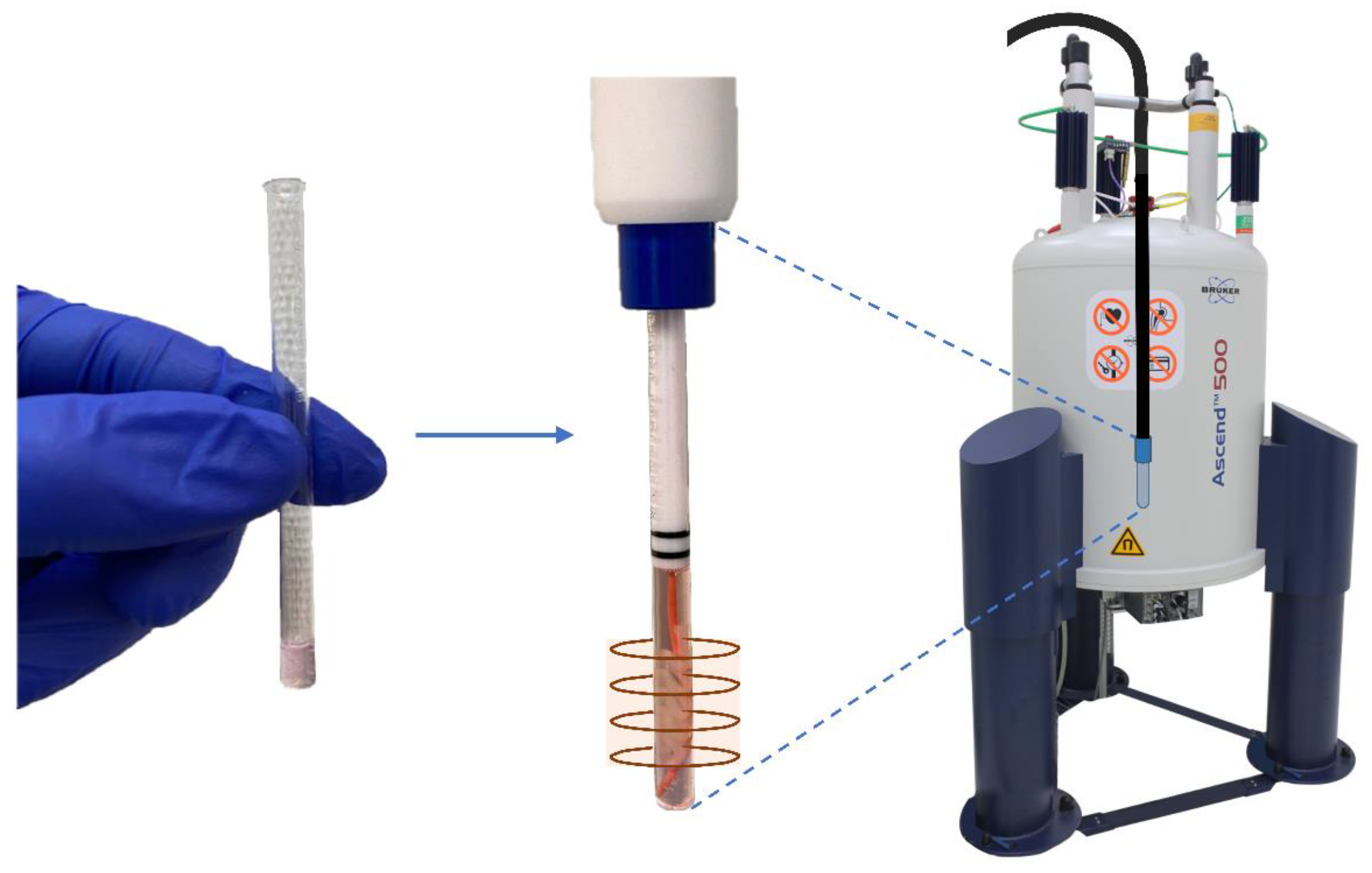
4.3. NMR Bioreactor Setup
4.4. Determination of NMR Sensitive Region
4.5. Determination of Scaffold Volume
4.6. 1H NMR for Metabolic Analysis
4.7. 1H NMR for Diffusion Measurements
4.8. Supernatant Collection and Extracellular Footprint Determination
4.9. 31P and 23Na NMR Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MEM | Minimal Essential Medium |
| LDH | Lactate dehydrogenase |
| SNR | Signal-to-Noise Ratio |
| DMSO | Dimethyl sulfoxide |
| ERETIC | Electronic Reference To access In vivo Concentrations |
| AA | Amino Acids |
| PBS | Phosphate Buffered Saline |
| CI | Complex I |
| CII | Complex II |
| SDH | Succinate dehydrogenase |
| PDH | Pyruvate dehydrogenase |
| TCA | Tricarboxylic Acid Cycle |
| FBCO04 | Control Fibroblasts N. 04 |
| FID | Free Induction Decay |
| EDTA | Ethylenediaminetetraacetic acid |
References
- Hertig, D.; Felser, A.; Diserens, G.; Kurth, S.; Vermathen, P.; Nuoffer, J.M. Selective galactose culture condition reveals distinct metabolic signatures in pyruvate dehydrogenase and complex I deficient human skin fibroblasts. Metabolomics 2019, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Yépez, V.A.; Kremer, L.S.; Iuso, A.; Gusic, M.; Kopajtich, R.; Koňaříkova, E.; Nadel, A.; Wachutka, L.; Prokisch, H.; Gagneur, J. OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. PLoS ONE 2018, 11, e0199938. [Google Scholar] [CrossRef] [PubMed]
- Barderas, M.G.; Laborde, C.M.; Posada, M.; de la Cuesta, F.; Zubiri, I.; Vivanco, F.; Alvarez-Llamas, G. Metabolomic Profiling for Identification of Novel Potential Biomarkers in Cardiovascular Diseases. J. Biomed. Biotechnol. 2011, 2011, 790132. [Google Scholar] [CrossRef] [PubMed]
- Morvan, D.; Demidem, A. NMR metabolomics of fibroblasts with inherited mitochondrial Complex I mutation reveals treatment-reversible lipid and amino acid metabolism alterations. Metabolomics 2018, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhao, L.; Tang, S.; Zhou, Q.; Lin, Q.; Li, X.; Zheng, H.; Gao, H. Metabolic effects of basic fibroblast growth factor in streptozotocin-induced diabetic rats: A 1H NMR-based metabolomics investigation. Sci. Rep. 2016, 6, 36474. [Google Scholar] [CrossRef]
- Chen, W.W.; Freinkman, E.; Wang, T.; Birsoy, K.; Sabatini, D.M. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell 2016, 166, 1324–1337. [Google Scholar] [CrossRef]
- Pan, D.; Lindau, C.; Lagies, S.; Wiedemann, N.; Kammerer, B. Metabolic profiling of isolated mitochondria and cytoplasm reveals compartment-specific metabolic responses. Metabolomics 2018, 14, 59. [Google Scholar] [CrossRef]
- Hertig, D.; Maddah, S.; Memedovski, R.; Kurth, S.; Moreno, A.; Pennestri, M.; Felser, A.; Felser, A.; Nuoffer, J.M.; Vermathen, P. Live monitoring of cellular metabolism and mitochondrial respiration in 3D cell culture system using NMR spectroscopy. Analyst 2021, 146, 4326–4339. [Google Scholar] [CrossRef]
- Van Zijl, P.C.M.; Moonen, C.T.W.; Faustino, P.; Pekar, J.; Kaplan, O.; Cohen, J.S. Complete separation of intracellular and extracellular information in NMR spectra of perfused cells by diffusion-weighted spectroscopy. Proc. Natl. Acad. Sci. USA 1991, 88, 3228–3232. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef]
- Sellick, C.A.; Hansen, R.; Maqsood, A.R.; Dunn, W.B.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Effective quenching processes for physiologically valid metabolite profiling of suspension cultured Mammalian cells. Anal. Chem. 2009, 81, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.; Davey, H.M.; Broadhurst, D.; Heald, J.K.; Rowland, J.J.; Oliver, S.G.; Kell, D.B. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 2003, 21, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, V.; Olsson, L.; Nielsen, J. Metabolic footprinting in microbiology: Methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008, 26, 490–497. [Google Scholar] [CrossRef]
- Aasebø, E.; Brenner, A.K.; Birkeland, E.; Tvedt, T.H.A.; Selheim, F.; Berven, F.S.; Bruserud, Ø. The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers 2021, 13, 1509. [Google Scholar] [CrossRef] [PubMed]
- Najac, C.; Branzoli, F.; Ronen, I.; Valette, J. Brain intracellular metabolites are freely diffusing along cell fibers in grey and white matter, as measured by diffusion-weighted MR spectroscopy in the human brain at 7 T. Brain Struct. Funct. 2016, 221, 1245–1254. [Google Scholar] [CrossRef]
- Pilatus, U.; Shim, H.; Artemov, D.; Davis, D.; van Zijl, P.C.M.; Glickson, J.D. Intracellular Volume and Apparent Diffusion Constants of Perfused Cancer Cell Cultures, as Measured by NMR. Magn. Reson. Med. 1997, 37, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H. Amino acid metabolism in mammalian cell cultures. Science 1959, 130, 432–437. [Google Scholar] [CrossRef]
- 31095-MEM|Thermo Fisher Scientific-CH. Available online: https://www.thermofisher.com/ch/en/home/technical-resources/media-formulation.196.html (accessed on 29 March 2022).
- Villereal, M.L. Sodium Fluxes in Human Fibroblasts: Kinetics of Serum-Dependent and Serum-Independent Pathways. J. Cell. Physiol. 1981, 108, 251–259. [Google Scholar] [CrossRef]
- Mancuso, A.; Fernandez, E.J.; Blanch, H.W.; Clark, D.S. A Nuclear Magnetic Resonance Technique for Determining Hybridoma Cell Concentration in Hollow Fiber Bioreactors. Bio/Technology 1990, 8, 1282–1285. [Google Scholar]
- Thoumine, O.; Cardoso, O.; Meister, J.J. Changes in the mechanical properties of fibroblasts during spreading: A micromanipulation study. Eur. Biophys. J. 1999, 28, 222–234. [Google Scholar] [CrossRef]
- Frisch, T.; Thoumine, O. Predicting the kinetics of cell spreading. J. Biomech. 2002, 35, 1137–1141. [Google Scholar] [CrossRef]
- Stefan, D.; Di Cesare, F.; Andrasescu, A.; Popa, E.; Lazariev, A.; Vescovo, E.; Strbak, O.; Williams, S.; Starcuk, Z.; Cabanas, M.; et al. Quantitation of magnetic resonance spectroscopy signals: The jMRUI software package. Meas. Sci. Technol. 2009, 20, 1–9. [Google Scholar] [CrossRef]
- Leekumjorn, S.; Sum, A.K. Molecular study of the diffusional process of DMSO in double lipid bilayers. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1751–1758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Notman, R.; Noro, M.; O’Malley, B.; Anwar, J. Molecular Basis for Dimethylsulfoxide (DMSO) Action on Lipid Membranes. J. Am. Chem. Soc. 2006, 128, 13982–13983. [Google Scholar] [CrossRef]
- Malek, A.M.; Goss, G.G.; Jiang, L.; Izumo, S.; Alper, S.L. Mannitol at Clinical Concentrations Activates Multiple Signaling Pathways and Induces Apoptosis in Endothelial Cells. Stroke 1998, 29, 2631–2640. [Google Scholar] [CrossRef]
- Pfeuffer, J.; Flögel, U.; Dreher, W.; Leibfritz, D. Restricted diffusion and exchange of intracellular water: Theoretical modelling and diffusion time dependence of 1H NMR measurements on perfused glial cells. NMR Biomed. 1998, 11, 19–31. [Google Scholar] [CrossRef]
- Mills, R. Self-Diffusion in Normal and Heavy Water in the Range 1–45°. J. Phys. Chem. 1973, 77, 685–688. [Google Scholar] [CrossRef]
- Tanaka, K. Measurements of Self-diffusion Coefficients of Water in Pure Water and in Aqueous Electrolyte Solutions. J. Chem. Soc. 1975, 71, 1127. [Google Scholar] [CrossRef]
- Cooper, G.M. The Molecular Composition of Cells. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Zhang, Y.; Guo, S.; Xie, C.; Fang, J. Uridine Metabolism and Its Role in Glucose, Lipid, and Amino Acid Homeostasis. Biomed. Res. Int. 2020, 2020, 7091718. [Google Scholar] [CrossRef]
- Rhee, S. Fibroblasts in three dimensional matrices: Cell migration and matrix remodeling. Exp. Mol. Med. 2009, 41, 858–865. [Google Scholar] [CrossRef]
- Jin, T.; Li, L.; Siow, R.C.M.; Liu, K.K. Collagen matrix stiffness influences fibroblast contraction force. Biomed. Phys. Eng. Express 2016, 2, 047002. [Google Scholar] [CrossRef]
- Duarte, I.F.; Lamego, I.; Marques, J.; Marques, M.P.P.; Blaise, B.J.; Gil, A.M. Nuclear Magnetic Resonance (NMR) Study of the Effect of Cisplatin on the Metabolic Profile of MG-63 Osteosarcoma Cells. J. Prot. Res. 2010, 9, 5877–5886. [Google Scholar] [CrossRef]
- Hoerr, V.; Duggan, G.E.; Zbytnuik, L.; Poon, K.K.H.; Große, G.; Neugebauer, U.; Methling, K.; Löffler, B.; Vogel, H.J. Characterisation and prediction of the mechanism of action of antibiotics through NMR metabolomics. BMC Microbiol. 2016, 16, 82. [Google Scholar] [CrossRef]
- Czajkowska, J.; Junka, A.; Hoppe, J.; Toporkiewicz, M.; Pawlak, A.; Migdal, P.; Oleksy-Wawrzyniak, M.; Fijałkowski, K.; Smiglak, M.; Markowska-Szczupak, A. The Co-Culture of Staphylococca Biofilm and Fibroblast Cell Line: The Correlation of Biological Phenomena with Metabolic NMR Footprint. Int. J. Mol. Sci. 2021, 22, 5826. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Stevens, A.P.; Dettmer, K.; Gottfried, E.; Hoves, S.; Kreutz, M.; Holler, E.; Canelas, A.B.; Kema, I.; Oefner, P.J. Quantitative profiling of tryptophan metabolites in serum, urine, and cell culture supernatants by liquid chromatography–tandem mass spectrometry. Anal. Bioanal. Chem. 2011, 401, 3249–3261. [Google Scholar] [CrossRef]
- Dekker, P.; Meissner, A.; Dirks, R.W.; Slagboom, E.; van Heemst, D.; Deelder, A.M.; Tanke, H.J.; Westendorp, R.G.J.; Maier, A.B. Human in vivo longevity is reflected in vitro by differential metabolism as measured by 1H-NMR profiling of cell culture supernatants. Mol. BioSyst. 2012, 8, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Aranibar, N.; Borys, M.; Mackin, N.A.; Ly, V.; Abu-Absi, N.; Abu-Absi, S.; Niemitz, M.; Schilling, B.; Li, Z.J.; Brock, B.; et al. NMR-based metabolomics of mammalian cell and tissue cultures. J. Biomol. NMR 2011, 49, 195–206. [Google Scholar] [CrossRef]
- Silva, A.C.; Teixeira, A.P.; Alves, P.M. Impact of Adenovirus infection in host cell metabolism evaluated by 1H-NMR spectroscopy. J. Biotech. 2016, 231, 16–23. [Google Scholar] [CrossRef]
- Glunde, K.; Ackerstaff, E.; Natarajan, K.; Artemov, D.; Bhujwalla, Z.M. Real-Time Changes in 1H and 31P NMR Spectra of Malignant Human Mammary Epithelial Cells During Treatment with the Anti-inflammatory Agent Indomethacin. Magn. Reson. Med. 2002, 48, 819–825. [Google Scholar] [CrossRef]
- Mardor, Y.; Kaplan, O.; Sterin, M.; Ruiz-Cabello, J.; Ash, E.; Roth, Y.; Ringel, I.; Cohen, J.S. Noninvasive Real-Time Monitoring of Intracellular Cancer Cell Metabolism and Response to Lonidamine Treatment Using Diffusion Weighted Proton Magnetic Resonance Spectroscopy. Can. Res. 2000, 60, 5179–5186. [Google Scholar]
- Flögel, U.; Niendorf, T.; Serkowa, N.; Brand, A.; Henke, J.; Leibfritz, D. Changes in organic solutes, volume, energy state, and metabolism associated with osmotic stress in a glial cell line: A multinuclear NMR study. Neurochem. Res. 1995, 20, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Dall’Asta, V.; Rossi, P.A.; Bussolati, O.; Gazzola, G.C. Regulatory volume decrease of cultured human fibroblasts involves changes in intracellular amino-acid pool. Biochim. Biophys. Acta 1994, 1220, 139–145. [Google Scholar] [CrossRef]
- Aguilar, J.A.; Nilsson, M.; Bodenhausen, G.; Morris, G.A. Spin echo NMR spectra without J modulation. Chem. Comumun. 2012, 48, 811–813. [Google Scholar] [CrossRef] [PubMed]
- Vermathen, M.; Paul, L.E.H.; Diserens, G.; Vermathen, P.; Furrer, J. 1H HR-MAS NMR Based Metabolic Profiling of Cells in Response to Treatment with a Hexacationic Ruthenium Metallaprism as Potential Anticancer Drug. PLoS ONE 2015, 10, e0128478. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Brun, A.; Rott, K.H.; Cobra, P.F.; Tonelli, M.; Eghbalnia, H.R.; Caviedes-Vidal, E.; Karasov, W.H.; Markley, J.L. NMR-Based Identification of Metabolites in Polar and Non-Polar Extracts of Avian Liver. Metabolites 2017, 7, 61. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Akoka, S.; Barantin, L.; Trierweiler, M. Concentration Measurement by Proton NMR Using the ERETIC Method. Anal. Chem. 1999, 71, 2554–2557. [Google Scholar] [CrossRef]
- Knop, R.H.; Chen, C.W.; Mitchell, J.B.; Russo, A.; McPherson, S.; Cohen, J.S. Metabolic Studies of Mammalian Cells by 31P-NMR Using a Continuous Perfusion Technique. Biochim. Biophys. Acta 1984, 804, 275–284. [Google Scholar] [CrossRef]
- Mancuso, A.; Zhu, A.; Beardsley, N.J.; Glickson, J.D.; Wehrli, S.; Pickup, S. Artificial Tumor Model Suitable for Monitoring 31P and 13C NMR Spectroscopic Changes During Chemotherapy-Induced Apoptosis in Human Glioma Cell. Magn. Reson. Med. 2005, 54, 67–78. [Google Scholar] [CrossRef]
- Ugurbil, K.; Guernsey, D.L.; Brown, T.R.; Glynn, P.; Tobkes, N.; Edelman, I.S. 31P NMR studies of intact anchorage-dependent mouse embryo fibroblasts. Proc. Natl. Acad. Sci. USA 1981, 78, 4843–4847. [Google Scholar] [CrossRef]
- Narayan, K.S.; Moress, E.A.; Chatham, C.; Barker, P.B. 31P NMR of Mammalian Cells Encapsulated in Alginate Gels Utilizing a New Phosphate-free Perfusion Medium. NMR Biomed. 1990, 3, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Milkevitch, M.; Browining, E.A.; Delikatny, E.J. Perfused Cells, Tissues and Organs by Magnetic Resonance Spectroscopy. eMagRes 2016, 5, 1333–1346. [Google Scholar]
- Simpson, N.E.; Han, Z.; Berendzen, K.M.; Sweeney, C.A.; Oca-Cossio, J.A.; Constantinidis, I.; Stacpoole, P.W. Magnetic resonance spectroscopic investigation of mitochondrial fuel metabolism and energetics in cultured human fibroblasts: Effects of pyruvate dehydrogenase complex deficiency and dichloroacetate. Mol. Genet. Metab. 2006, 89, 97–105. [Google Scholar] [CrossRef] [PubMed]

| Dfast 1 | Dslow 2 | Extra [%] | Intra [%] | Extra [mM] | Intra [mM] | Perfused MEM [mM] | |
|---|---|---|---|---|---|---|---|
| Ala | 1.78 ± 0.7 | 8.65 ± 4.4 | 93.72 ± 2.72 | 6.28 ± 2.72 | 0.24 ± 0.04 | 1.11 ± 0.39 | 0.1 |
| Arg | 1.72 ± 0.7 | 8.97 ± 7.6 | 90.13 ± 5.48 | 9.87 ± 5.48 | 0.68 ± 0.14 | 5.87 ± 4.44 | 0.6 |
| Cho | 1.10 ± 0.4 | 5.83 ± 1.6 | 78.50 ± 3.48 | 21.50 ± 3.48 | 0.11 ± 0.02 | 1.89 ± 0.14 | - |
| Glc | 1.24 ± 0.4 | 4.60 ± 4.4 | 92.57 ± 2.26 | 7.43 ± 2.26 | 5.1 ± 0.69 | 24.30 ± 7.74 | 5.55 |
| Glu | 0.80 ± 0.7 | 1.37 ± 1.0 | 46.50 ± 9.88 | 53.50 ± 9.88 | 0.16 ± 0.05 | 12.55 ± 3.34 | 0.1 |
| Gln | 1.43 ± 0.4 | 15.03 ± 8.0 | 86.97 ± 4.22 | 13.04 ± 4.22 | 1.23 ± 0.17 | 13.96 ± 6.38 | 1.95 |
| GPC | 1.39 ± 0.6 | 8.11 ± 6.5 | 90.54 ± 4.03 | 9.46 ± 4.03 | 0.11 ± 0.02 | 0.75 ± 0.38 | - |
| His | 1.85 ± 0.8 | 4.72 ± 5.7 | 86.18 ± 9.53 | 13.82 ± 9.53 | 0.11 ± 0.03 | 1.24 ± 1.01 | 0.2 |
| Ile | 1.67 ± 0.3 | 4.44 ± 3.9 | 87.38 ± 9.20 | 12.61 ± 9.20 | 0.31 ± 0.08 | 3.00 ± 2.32 | 0.4 |
| Lac | 1.24 ± 0.2 | 1.18 ± 0.6 | 80.44 ± 5.98 | 19.56 ± 5.98 | 1.17 ± 0.77 | 16.55 ± 4.43 | - |
| Leu | 1.23 ± 0.4 | 5.19 ± 5.1 | 94.17 ± 1.35 | 5.83 ± 1.35 | 0.33 ± 0.05 | 1.58 ± 0.7 | 0.4 |
| Lys | 1.47 ± 0.3 | 5.32 ± 3.3 | 86.90 ± 7.33 | 13.10 ± 7.33 | 0.32 ± 0.07 | 3.93 ± 2.89 | 0.4 |
| PC | 1.46 ± 0.5 | 8.95 ± 8.3 | 90.60 ± 2.76 | 9.40 ± 2.76 | 0.21 ± 0.04 | 1.60 ± 0.63 | - |
| Phe | 1.97 ± 0.6 | 5.90 ± 2.3 | 89.80 ± 6.45 | 10.20 ± 6.45 | 0.18 ± 0.04 | 1.33 ± 0.6 | 0.19 |
| Pro | 1.43 ± 0.9 | 2.22 ± 1.8 | 56.88 ± 10.47 | 43.12 ± 10.47 | 0.26 ± 0.08 | 13.66 ± 2.97 | 0.1 |
| Pyr | 1.67 ± 0.4 | 13.04 ± 5.1 | 92.11 ± 3.86 | 7.89 ± 3.86 | 0.51 ± 0.02 | 3.03 ± 1.22 | 0.98 |
| Tyr | 1.96 ± 1.0 | 4.07 ± 4.4 | 92.05 ± 4.20 | 7.95 ± 4.20 | 0.14 ± 0.02 | 0.90 ± 0.55 | 0.2 |
| Urd | 1.89 ± 0.6 | 3.10 ± 3.5 | 95.10 ± 3.46 | 4.90 ± 3.46 | 0.07 ± 0.02 | 0.23 ± 0.14 | 0.19 |
| Val | 1.60 ± 0.5 | 9.28 ± 9.3 | 91.25 ± 5.17 | 8.75 ± 5.17 | 0.40 ± 0.14 | 2.52 ± 1.36 | 0.39 |
| Metabolite | Before Rotenone Addition (Basal Condition) | After Rotenone Addition (Inhibition Condition) | ||||||
|---|---|---|---|---|---|---|---|---|
| Extra [%] | Intra [%] | Extra [mM] | Intra [mM] | Extra [%] | Intra [%] | Extra [mM] | Intra [mM] | |
| Ala | 97.3 | 2.7 | 0.24 | 0.67 | 89.8 | 10.2 | 0.22 | 2.5 |
| Glc | 91.3 | 8.7 | 4.7 | 36.7 | 94.8 | 5.2 | 4.4 | 20.7 |
| Glu | 55.7 | 44.3 | 0.21 | 15.6 | 42.4 | 57.6 | 0.13 | 16.6 |
| Gln | 84.3 | 15.7 | 1.1 | 18.4 | 93.5 | 6.5 | 1.4 | 8.5 |
| His | 91.4 | 8.6 | 0.11 | 0.91 | 89.6 | 10.4 | 0.09 | 1.1 |
| Ile | 82.4 | 17.6 | 0.28 | 4.89 | 84.9 | 15.1 | 0.26 | 4.1 |
| Lac | 78.5 | 21.5 | 0.77 | 19.2 | 78.1 | 21.9 | 0.97 | 24.4 |
| Leu | 92.6 | 7.4 | 0.32 | 2.4 | 89.6 | 10.4 | 0.28 | 3.1 |
| Lys | 80.6 | 19.4 | 0.32 | 6.8 | 92.5 | 7.5 | 0.23 | 1.7 |
| Phe | 89.3 | 10.7 | 0.18 | 1.8 | 93.5 | 6.5 | 0.15 | 0.88 |
| Pyr | 95.2 | 4.8 | 0.53 | 2.5 | 90.5 | 9.5 | 0.5 | 4.7 |
| Tyr | 93.4 | 6.6 | 0.13 | 0.96 | 94.3 | 5.7 | 0.13 | 0.78 |
| Urd | 97.2 | 2.8 | 0.07 | 0.21 | 91.4 | 8.6 | 0.05 | 0.62 |
| Val | 89.7 | 10.3 | 0.31 | 3.4 | 90.8 | 9.2 | 0.26 | 2.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urzì, C.; Hertig, D.; Meyer, C.; Maddah, S.; Nuoffer, J.-M.; Vermathen, P. Determination of Intra- and Extracellular Metabolic Adaptations of 3D Cell Cultures upon Challenges in Real-Time by NMR. Int. J. Mol. Sci. 2022, 23, 6555. https://doi.org/10.3390/ijms23126555
Urzì C, Hertig D, Meyer C, Maddah S, Nuoffer J-M, Vermathen P. Determination of Intra- and Extracellular Metabolic Adaptations of 3D Cell Cultures upon Challenges in Real-Time by NMR. International Journal of Molecular Sciences. 2022; 23(12):6555. https://doi.org/10.3390/ijms23126555
Chicago/Turabian StyleUrzì, Christian, Damian Hertig, Christoph Meyer, Sally Maddah, Jean-Marc Nuoffer, and Peter Vermathen. 2022. "Determination of Intra- and Extracellular Metabolic Adaptations of 3D Cell Cultures upon Challenges in Real-Time by NMR" International Journal of Molecular Sciences 23, no. 12: 6555. https://doi.org/10.3390/ijms23126555
APA StyleUrzì, C., Hertig, D., Meyer, C., Maddah, S., Nuoffer, J.-M., & Vermathen, P. (2022). Determination of Intra- and Extracellular Metabolic Adaptations of 3D Cell Cultures upon Challenges in Real-Time by NMR. International Journal of Molecular Sciences, 23(12), 6555. https://doi.org/10.3390/ijms23126555






