Genome-Wide Identification of the LHC Gene Family in Kiwifruit and Regulatory Role of AcLhcb3.1/3.2 for Chlorophyll a Content
Abstract
:1. Introduction
2. Results
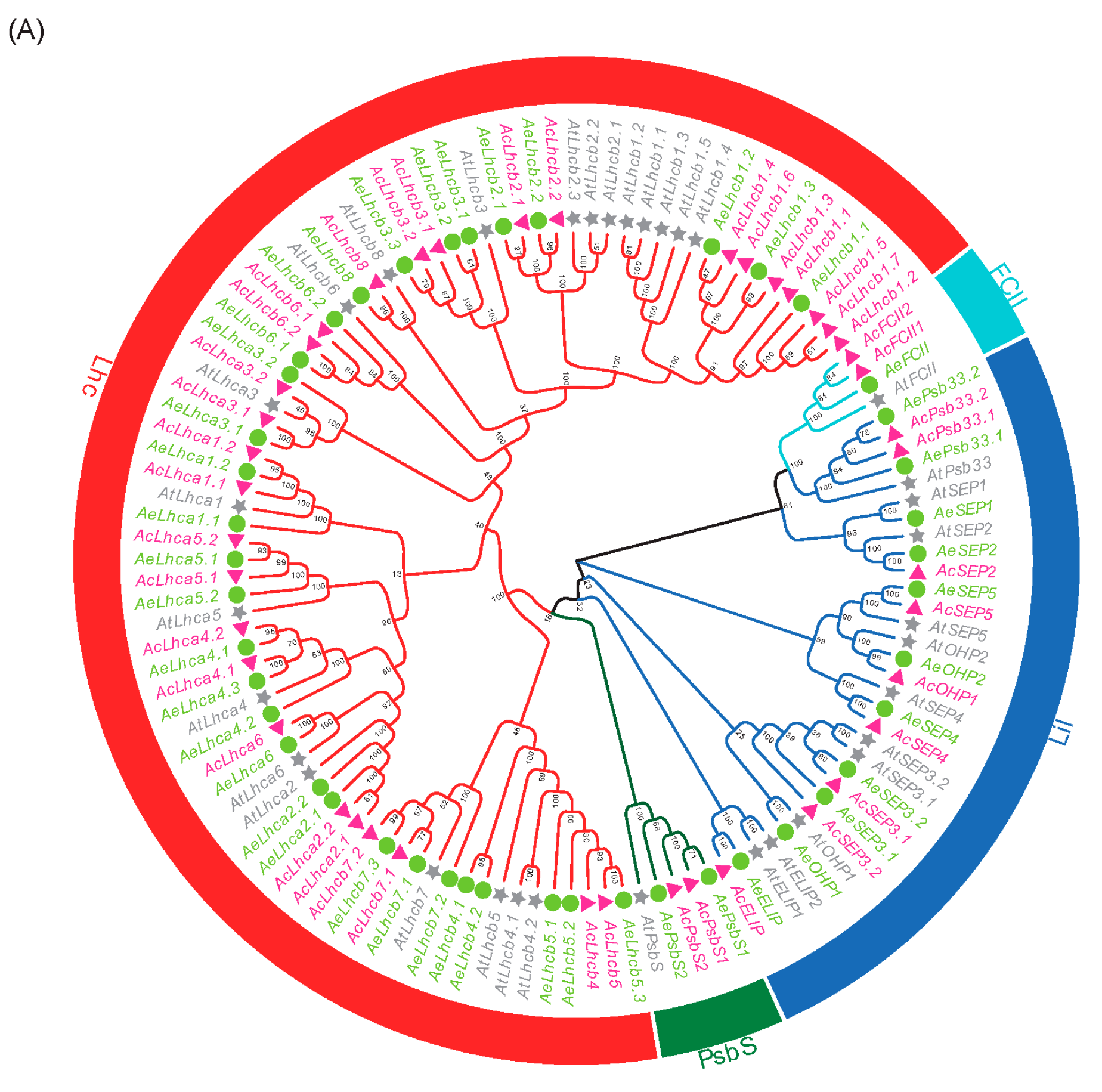
2.1. Genome-Wide Identification and Phylogenetic Analysis of Kiwifruit LHCs
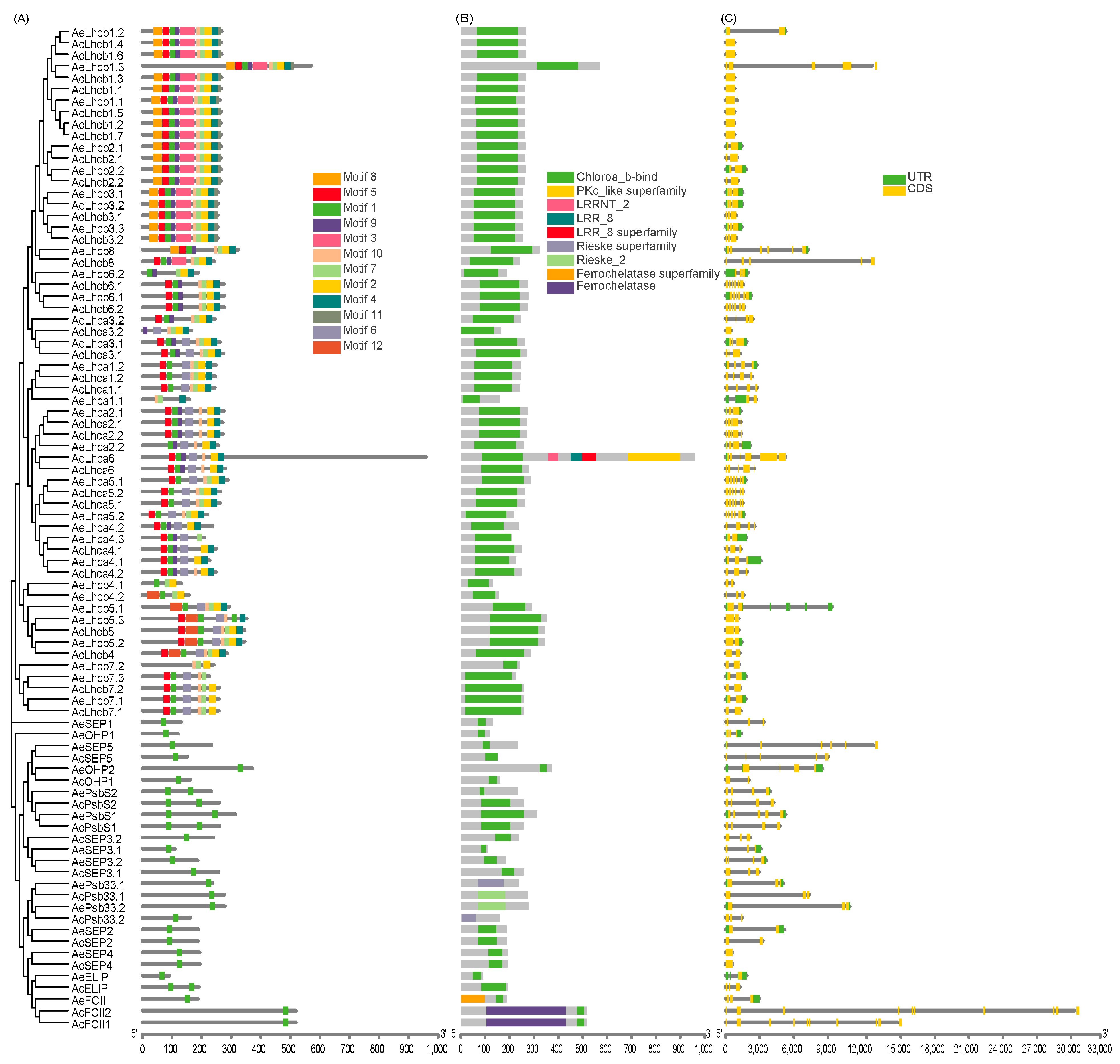
2.2. Multiple Sequence Alignment and Analysis of Kiwifruit LHCs Structure
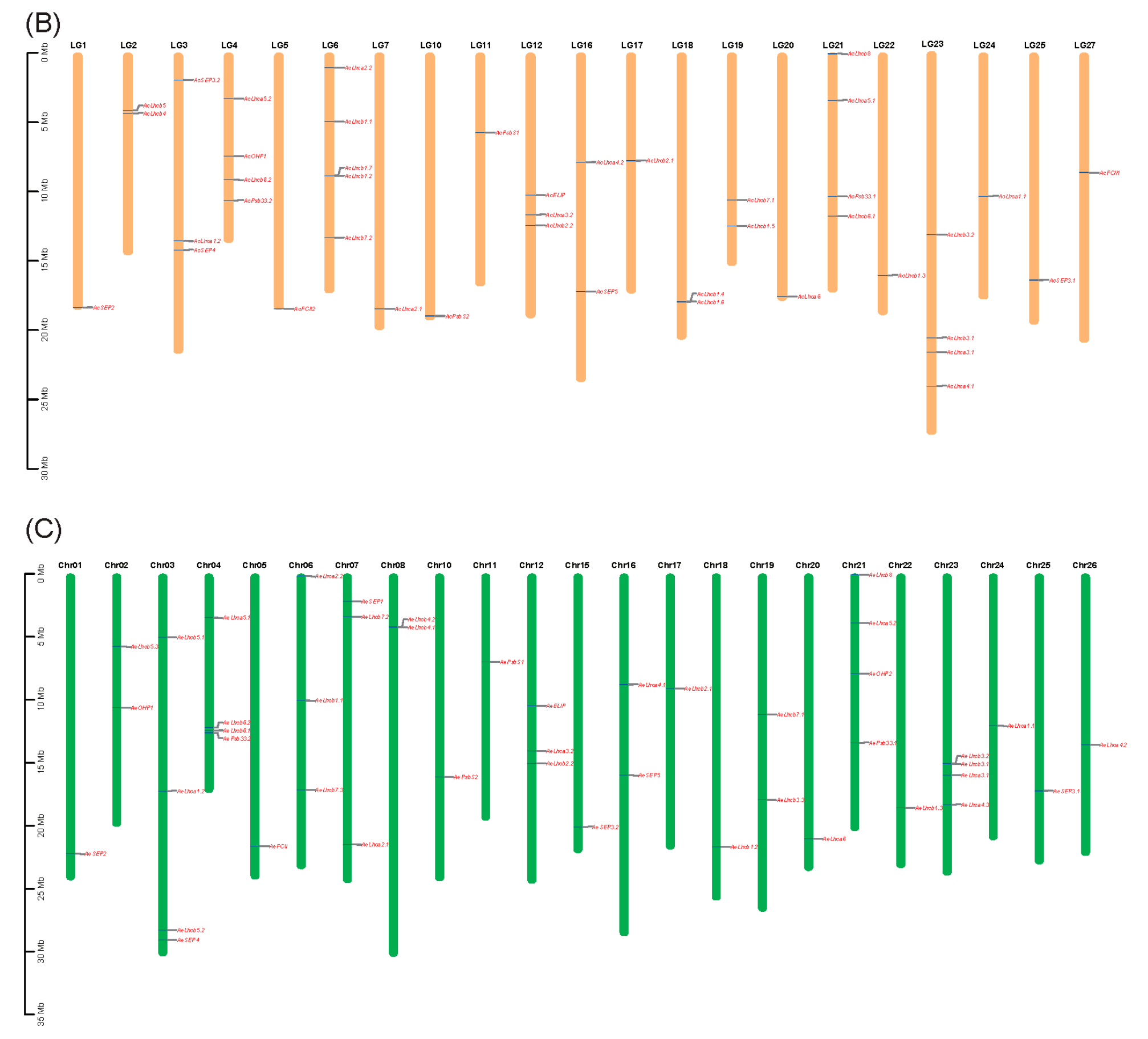
2.3. Synteny Analysis and cis-Element of Kiwifruit LHCs
2.4. AcLHC Genes Regulated Kiwifruit Responses to Biotic and Abiotic Stresses
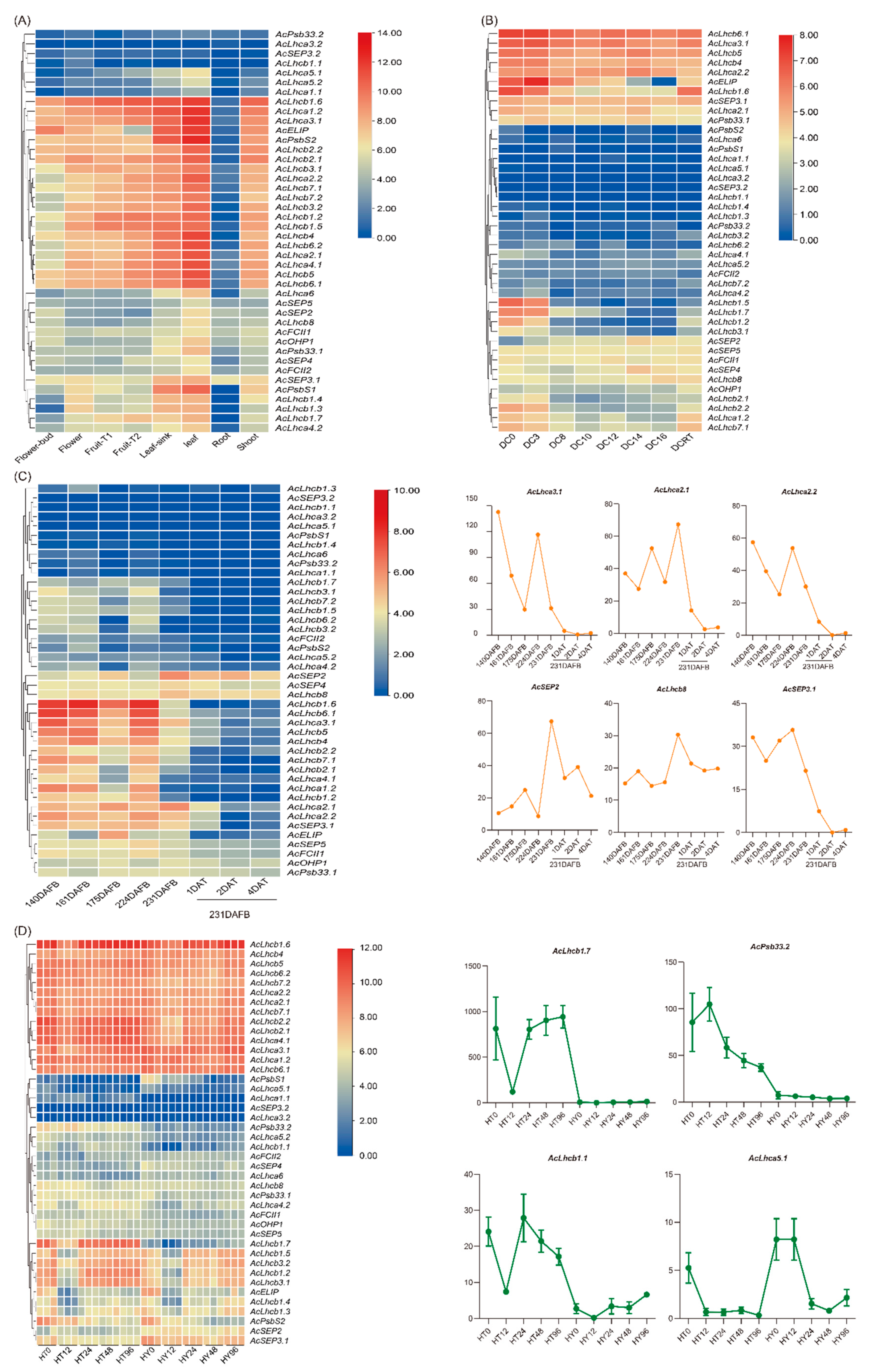
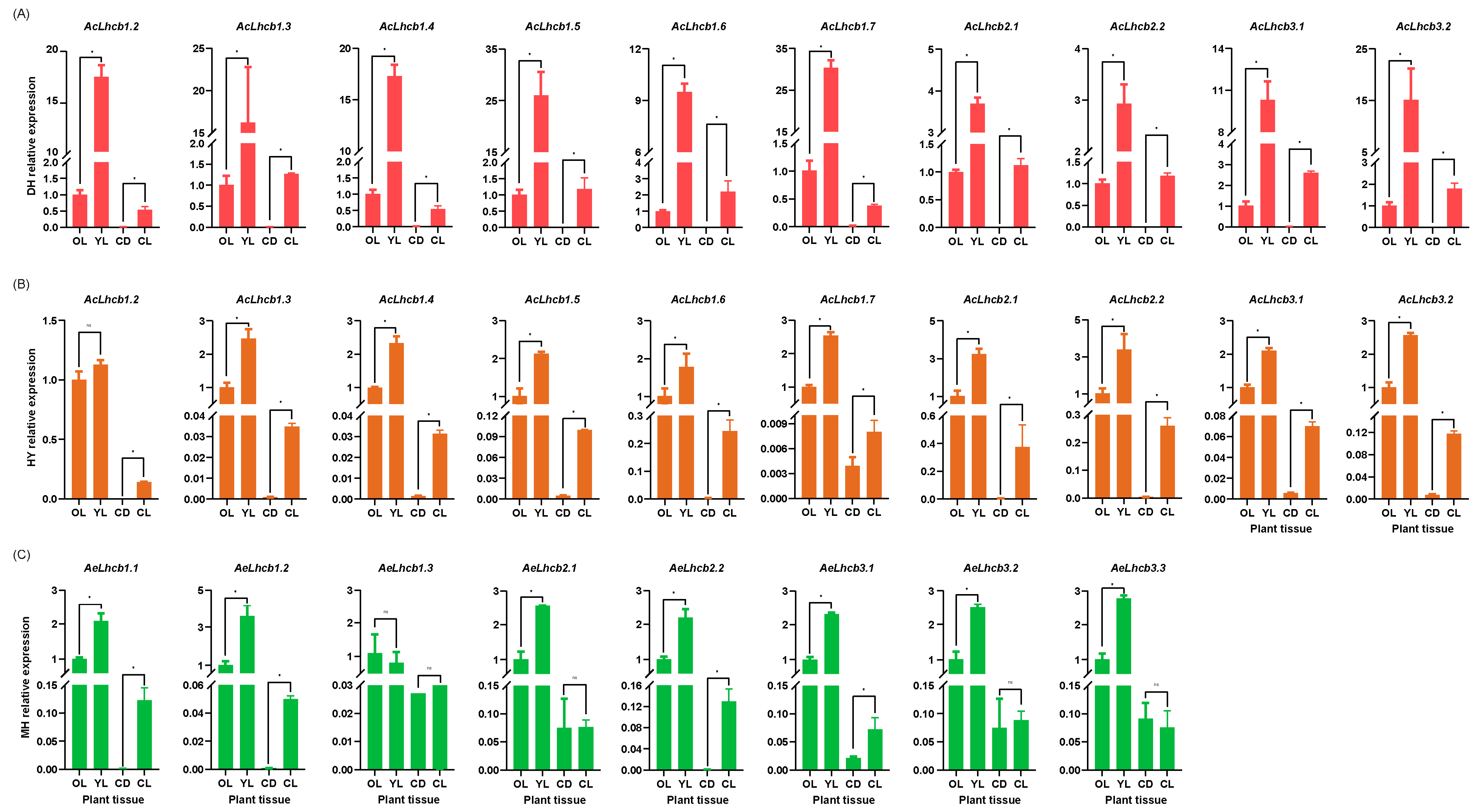
2.5. RT-qPCR Validation of Kiwifruit LHCs in Different Tissues
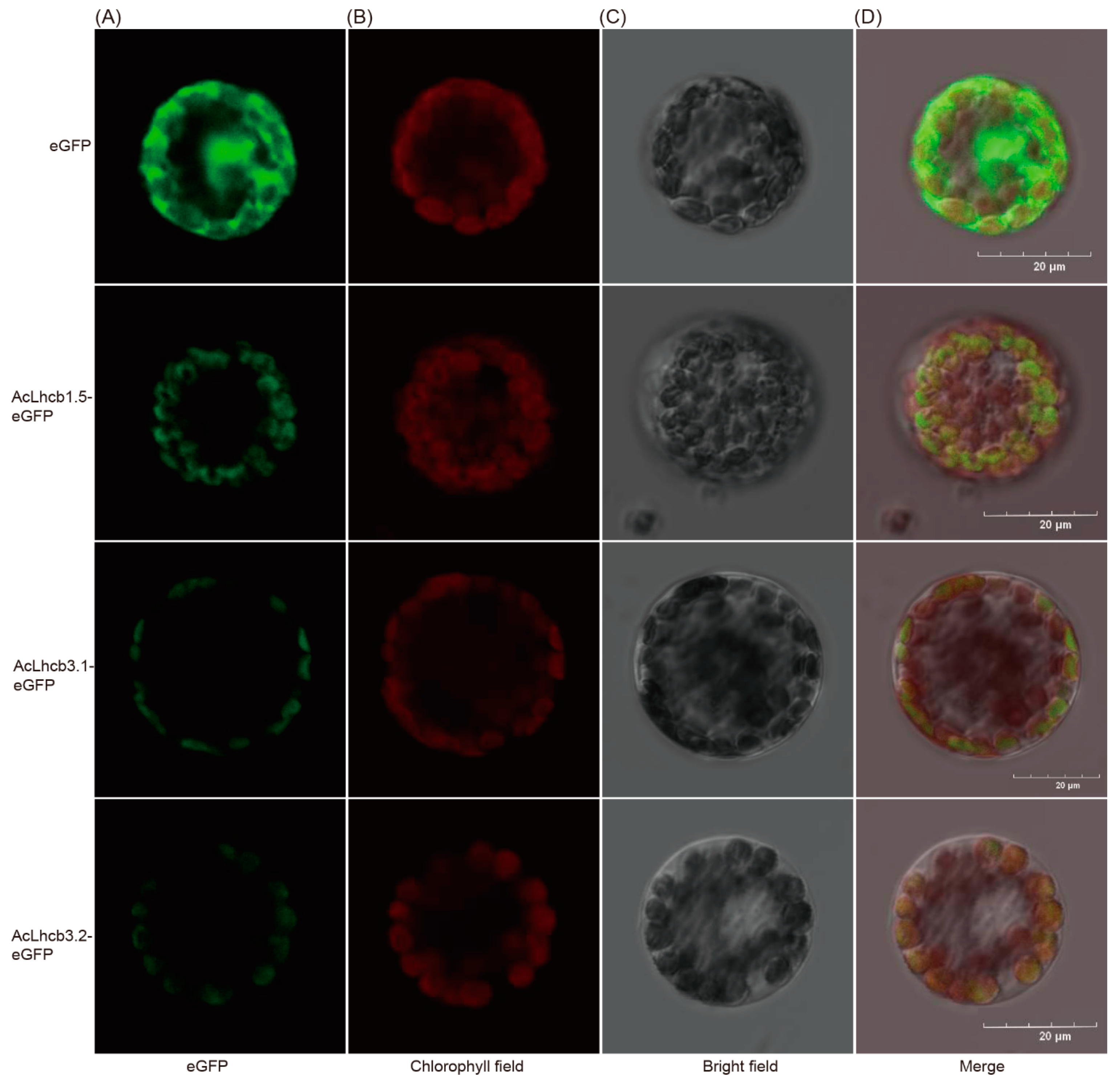
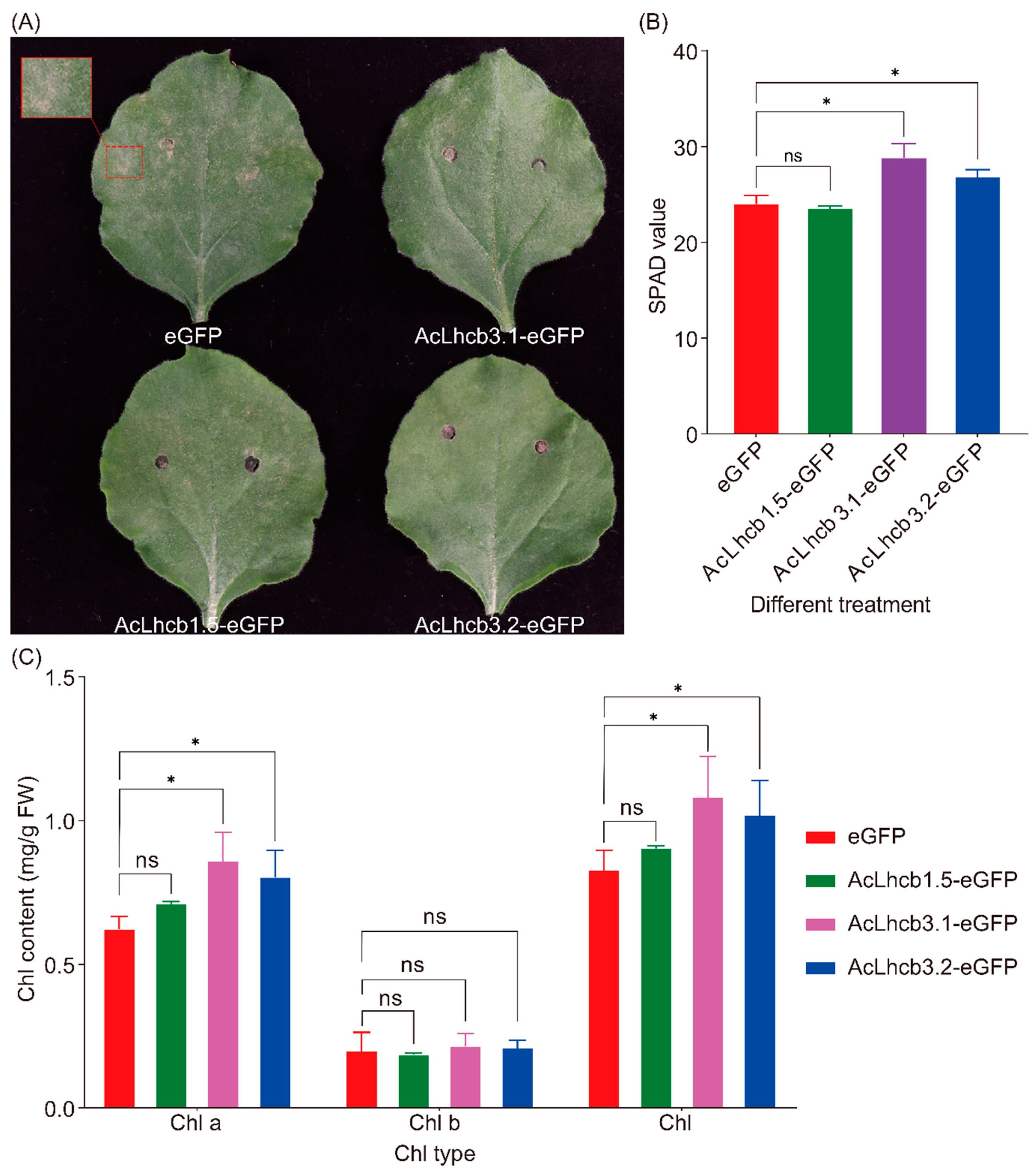
2.6. Subcellular Localization of Kiwifruit LHC and Transient Transformation of AcLhcb Genes in Tobacco Leaves
3. Discussion
4. Materials and Methods and Analysis
4.1. Genome-Wide Identification of LHC Genes in Kiwifruit
4.2. Analysis of Kiwifruit LHC Protein Structure
4.3. Gene Structure, Motif Features, and cis-Elements Analysis
4.4. Phylogenetic Analysis of LHCs
4.5. Chromosomal Location, Gene Duplication, and Synteny Analysis
4.6. Expression Analysis of Kiwifruit LHCs
4.7. Plant Materials and Treatments
4.8. RNA Extraction and cDNA Synthesis
4.9. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.10. Subcellular Localization of AcLHCs and Transformation
4.11. Transient Over-Expression Analysis in Tobacco Leaf
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kirilovsky, D.; Büchel, C. Chapter Nine—Evolution and function of light-harvesting antenna in oxygenic photosynthesis. Adv. Bot. Res. 2019, 91, 247–293. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Connecting Chlorophyll Metabolism with Accumulation of the Photosynthetic Apparatus. Trends Plant Sci. 2021, 26, 484–495. [Google Scholar] [CrossRef]
- De Bianchi, S.; Dall’Osto, L.; Tognon, G.; Morosinotto, T.; Bassi, R. Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 2008, 20, 1012–1028. [Google Scholar] [CrossRef]
- Chen, Y.E.; Ma, J.; Wu, N.; Su, Y.Q.; Zhang, Z.W.; Yuan, M.; Zhang, H.Y.; Zeng, X.Y.; Yuan, S. The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 2018, 130, 267–276. [Google Scholar] [CrossRef]
- Zou, Z.; Li, M.; Jia, R.; Zhao, H.; He, P.; Zhang, Y.; Guo, A. Genes encoding light-harvesting chlorophyll a/b-binding proteins in papaya (Carica papaya L.) and insight into lineage-specific evolution in Brassicaceae. Gene 2020, 748, 144685. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, H.; Guo, Y.; Zou, Z. Light-harvesting chlorophyll a/b-binding protein-coding genes in jatropha and the comparison with castor, cassava and arabidopsis. Peer J. 2020, 8, e8465. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J. Genomics analysis of the light-harvesting chlorophyll a/b-binding (Lhc) superfamily in cassava (Manihot esculenta Crantz). Gene 2019, 702, 171–181. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal. Behav. 2010, 5, 1537–1542. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, C.; Wang, X.; Ma, Q.; Fan, S.; Zhang, C. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2.3 on chlorophyll a synthesis. Plant Biol. (Stuttg) 2021, 23, 831–842. [Google Scholar] [CrossRef]
- Zou, Z.; Huang, Q.; An, F.J.A.B. Genome-wide Identification, Classification and Expression Analysis of Lhc Supergene Family in Castor Bean (Ricinus communis L.). Agric. Biotechnol. 2013, 2, 44–49. [Google Scholar]
- Deng, Y.S.; Kong, F.Y.; Zhou, B.; Zhang, S.; Yue, M.M.; Meng, Q.W. Heterology expression of the tomato LeLhcb2 gene confers elevated tolerance to chilling stress in transgenic tobacco. Plant Physiol. Biochem. 2014, 80, 318–327. [Google Scholar] [CrossRef]
- Meng, L.; Fan, Z.; Zhang, Q.; Wang, C.; Gao, Y.; Deng, Y.; Zhu, B.; Zhu, H.; Chen, J.; Shan, W.; et al. BEL1-LIKE HOMEODOMAIN 11 regulates chloroplast development and chlorophyll synthesis in tomato fruit. Plant J. 2018, 94, 1126–1140. [Google Scholar] [CrossRef]
- Zhang, M.; Senoura, T.; Yang, X.; Chao, Y.; Nishizawa, N.K. Lhcb2 gene expression analysis in two ecotypes of Sedumalfredii subjected to Zn/Cd treatments with functional analysis of SaLhcb2 isolated from a Zn/Cd hyperaccumulator. Biotechnol. Lett. 2011, 33, 1865–1871. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liu, R.; Yan, L.; Liu, Z.Q.; Jiang, S.C.; Shen, Y.Y.; Wang, X.F.; Zhang, D.P. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 2012, 63, 1095–1106. [Google Scholar] [CrossRef]
- Wang, S.-S.; Song, Z.-B.; Sun, Z.; Zhang, J.; Mei, Y.; Nian, H.-J.; Li, K.-Z.; Chen, L.-M. Effects of Formaldehyde Stress on Physiological Characteristics and Gene Expression Associated with Photosynthesis in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2012, 30, 1291–1302. [Google Scholar] [CrossRef]
- Peterson, R.B.; Schultes, N.P. Light-harvesting complex B7 shifts the irradiance response of photosynthetic light-harvesting regulation in leaves of Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 311–318. [Google Scholar] [CrossRef]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef]
- Jiang, Q.; Xu, Z.S.; Wang, F.; Li, M.Y.; Ma, J.; Xiong, A.S. Effects of abiotic stresses on the expression of Lhcb1 gene and photosynthesis of Oenanthe javanica and Apium graveolens. Biol. Plant 2014, 58, 256–264. [Google Scholar] [CrossRef]
- Yadavalli, V.; Neelam, S.; Rao, A.S.V.C.; Reddy, A.R.; Subramanyam, R. Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 2012, 169, 753–759. [Google Scholar] [CrossRef]
- Song, H.; Li, Y.; Xu, X.; Zhang, J.; Zheng, S.; Hou, L.; Xing, G.; Li, M. Analysis of genes related to chlorophyll metabolism under elevated CO2 in cucumber (Cucumis sativus L.). Sci. Hortic. 2020, 261, 108988. [Google Scholar] [CrossRef]
- Zhao, Y.; Qiang, C.; Wang, X.; Chen, Y.; Deng, J.; Jiang, C.; Sun, X.; Chen, H.; Li, J.; Piao, W.; et al. New alleles for chlorophyll content and stay-green traits revealed by a genome wide association study in rice (Oryza sativa). Sci. Rep. 2019, 9, 2541. [Google Scholar] [CrossRef]
- Koziol, A.G.; Borza, T.; Ishida, K.; Keeling, P.; Lee, R.W.; Durnford, D.G. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 2007, 143, 1802–1816. [Google Scholar] [CrossRef]
- Lin, W.; Guo, X.; Pan, X.; Li, Z. Chlorophyll Composition, Chlorophyll Fluorescence, and Grain Yield Change in esl Mutant Rice. Int. J. Mol. Sci. 2018, 19, 2945. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Yu, Y.; Kou, X.; Periakaruppan, R.; Chen, X.; Li, X. STAY-GREEN and light-harvesting complex II chlorophyll a/b binding protein are involved in albinism of a novel albino tea germplasm ‘Huabai 1’. Sci. Hortic. 2022, 293, 110653. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Luo, J.; Wang, H.; Dong, Q.; Wang, Y.; Yang, K.; Mao, K.; Ma, F. Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress. Plant Physiol. Biochem. 2020, 154, 517–529. [Google Scholar] [CrossRef]
- de Bianchi, S.; Betterle, N.; Kouril, R.; Cazzaniga, S.; Boekema, E.; Bassi, R.; Dall’Osto, L. Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 2011, 23, 2659–2679. [Google Scholar] [CrossRef]
- Tao, J.; Jia, H.; Wu, M.; Zhong, W.; Jia, D.; Wang, Z.; Huang, C. Genome-wide identification and characterization of the TIFY gene family in kiwifruit. BMC Genom. 2022, 23, 179. [Google Scholar] [CrossRef]
- Drummond, L. Chapter Three—The Composition and Nutritional Value of Kiwifruit. Adv. Food Nutr. Res. 2013, 68, 33–57. [Google Scholar] [CrossRef]
- Tu, M.; Wu, Y.; Li, J.; Chen, D.; Jiang, G.; Song, H.; Yin, X.; Liu, X.; Li, M.; Sun, S. Transcriptome analysis reveals the roles of chlorophyll a/b-binding proteins (CABs) and stay-green (SGR) in chlorophyll degradation during fruit development in kiwifruit. N. Z. J. Crop Hortic. Sci. 2020, 49, 106–126. [Google Scholar] [CrossRef]
- Majoros, W.H.; Holt, C.; Campbell, M.S.; Ware, D.; Yandell, M.; Reddy, T.E. Predicting gene structure changes resulting from genetic variants via exon definition features. Bioinformatics 2018, 34, 3616–3623. [Google Scholar] [CrossRef]
- Salazar, J.; Zapata, P.; Silva, C.; González, M.; Pacheco, I.; Bastías, M.; Meneses, C.; Jorquera, C.; Moreno, I.; Shinya, P.; et al. Transcriptome analysis and postharvest behavior of the kiwifruit ‘Actinidia deliciosa’ reveal the role of ethylene-related phytohormones during fruit ripening. Tree Genet. Genomes 2021, 17, 8. [Google Scholar] [CrossRef]
- Cao, W.; Zhang, H.; Zhou, Y.; Zhao, J.; Lu, S.; Wang, X.; Chen, X.; Yuan, L.; Guan, H.; Wang, G.; et al. Suppressing chlorophyll degradation by silencing OsNYC3 improves rice resistance to Rhizoctonia solani, the causal agent of sheath blight. Plant Biotechnol. J. 2022, 20, 335–349. [Google Scholar] [CrossRef]
- Cortes, A.J.; Blair, M.W. Genotyping by Sequencing and Genome-Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant Sci. 2018, 9, 128. [Google Scholar] [CrossRef]
- Wu, X.; Islam, A.; Limpot, N.; Mackasmiel, L.; Mierzwa, J.; Cortes, A.J.; Blair, M.W. Genome-Wide SNP Identification and Association Mapping for Seed Mineral Concentration in Mung Bean (Vigna radiata L.). Front. Genet. 2020, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Paz, J.; Cortes, A.J.; Hernandez-Varela, C.A.; Mejia-de-Tafur, M.S.; Rodriguez-Medina, C.; Baligar, V.C. Rootstock-Mediated Genetic Variance in Cadmium Uptake by Juvenile Cacao (Theobroma cacao L.) Genotypes, and Its Effect on Growth and Physiology. Front. Plant Sci. 2021, 12, 777842. [Google Scholar] [CrossRef]
- Tang, Y.; Wen, X.; Lu, C. Differential changes in degradation of chlorophyll–protein complexes of photosystem I and photosystem II during flag leaf senescence of rice. Plant Physiol. Biochem. 2005, 43, 193–201. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Tang, W.; Sun, X.; Yue, J.; Tang, X.; Jiao, C.; Yang, Y.; Niu, X.; Miao, M.; Zhang, D.; Huang, S.; et al. Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping. Gigascience 2019, 8, giz027. [Google Scholar] [CrossRef]
- Saito, A.; Iino, T.; Sonoike, K.; Miwa, E.; Higuchi, K. Remodeling of the major light-harvesting antenna protein of PSII protects the young leaves of barley (Hordeum vulgare L.) from photoinhibition under prolonged iron deficiency. Plant Cell Physiol. 2010, 51, 2013–2030. [Google Scholar] [CrossRef]
- Kong, F.; Zhou, Y.; Sun, P.; Cao, M.; Li, H.; Mao, Y. Identification of light-harvesting chlorophyll a/b-binding protein genes of Zostera marina L. and their expression under different environmental conditions. J. Ocean Univ. China 2016, 15, 152–162. [Google Scholar] [CrossRef]
- Liu, R.; Xu, Y.H.; Jiang, S.C.; Lu, K.; Lu, Y.F.; Feng, X.J.; Wu, Z.; Liang, S.; Yu, Y.T.; Wang, X.F.; et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 2013, 64, 5443–5456. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Rochaix, J.-D.; Bassi, R. LHC-like proteins involved in stress responses and biogenesis/repair of the photosynthetic apparatus. Biochem. J. 2019, 476, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Forth, D.; Pyke, K.A. The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J. Exp. Bot. 2006, 57, 1971–1979. [Google Scholar] [CrossRef]
- Blair, M.W.; Cortes, A.J.; Farmer, A.D.; Huang, W.; Ambachew, D.; Penmetsa, R.V.; Carrasquilla-Garcia, N.; Assefa, T.; Cannon, S.B. Uneven recombination rate and linkage disequilibrium across a reference SNP map for common bean (Phaseolus vulgaris L.). PLoS ONE 2018, 13, e0189597. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Hernandez, F.; Cortes, A.J. Last-Generation Genome-Environment Associations Reveal the Genetic Basis of Heat Tolerance in Common Bean (Phaseolus vulgaris L.). Front. Genet. 2019, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 257. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Li, D.; Zhang, Q.; Li, L.; Zhong, C.; Liu, Y.; Huang, H. Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 2018, 16, 1424–1433. [Google Scholar] [CrossRef]
- Liu, X.; Yu, F.; Yang, G.; Liu, X.; Peng, S. Identification of TIFY gene family in walnut and analysis of its expression under abiotic stresses. BMC Genom. 2022, 23, 190. [Google Scholar] [CrossRef]
- Yuan, Y.; Mei, L.; Wu, M.; Wei, W.; Shan, W.; Gong, Z.; Zhang, Q.; Yang, F.; Yan, F.; Zhang, Q.; et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Tan, X.L.; Fan, Z.Q.; Kuang, J.F.; Lu, W.J.; Reiter, R.J.; Lakshmanan, P.; Su, X.G.; Zhou, J.; Chen, J.Y.; Shan, W. Melatonin delays leaf senescence of Chinese flowering cabbage by suppressing ABFs-mediated abscisic acid biosynthesis and chlorophyll degradation. J. Pineal. Res. 2019, 67, e12570. [Google Scholar] [CrossRef]
- Berkman, S.J.; Roscoe, E.M.; Bourret, J.C. Comparing self-directed methods for training staff to create graphs using Graphpad Prism. J. Appl. Behav. Anal. 2019, 52, 188–204. [Google Scholar] [CrossRef]
- Cortes, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern Strategies to Assess and Breed Forest Tree Adaptation to Changing Climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Abid, M.; Tu, J.; Gao, P.; Wang, Z.; Huang, H. Genome-Wide Identification of the LHC Gene Family in Kiwifruit and Regulatory Role of AcLhcb3.1/3.2 for Chlorophyll a Content. Int. J. Mol. Sci. 2022, 23, 6528. https://doi.org/10.3390/ijms23126528
Luo J, Abid M, Tu J, Gao P, Wang Z, Huang H. Genome-Wide Identification of the LHC Gene Family in Kiwifruit and Regulatory Role of AcLhcb3.1/3.2 for Chlorophyll a Content. International Journal of Molecular Sciences. 2022; 23(12):6528. https://doi.org/10.3390/ijms23126528
Chicago/Turabian StyleLuo, Juan, Muhammad Abid, Jing Tu, Puxing Gao, Zupeng Wang, and Hongwen Huang. 2022. "Genome-Wide Identification of the LHC Gene Family in Kiwifruit and Regulatory Role of AcLhcb3.1/3.2 for Chlorophyll a Content" International Journal of Molecular Sciences 23, no. 12: 6528. https://doi.org/10.3390/ijms23126528
APA StyleLuo, J., Abid, M., Tu, J., Gao, P., Wang, Z., & Huang, H. (2022). Genome-Wide Identification of the LHC Gene Family in Kiwifruit and Regulatory Role of AcLhcb3.1/3.2 for Chlorophyll a Content. International Journal of Molecular Sciences, 23(12), 6528. https://doi.org/10.3390/ijms23126528





