Gene Expression of the D-Series Resolvin Pathway Predicts Activation of Anti-Tumor Immunity and Clinical Outcomes in Head and Neck Cancer
Abstract
:1. Introduction
2. Results
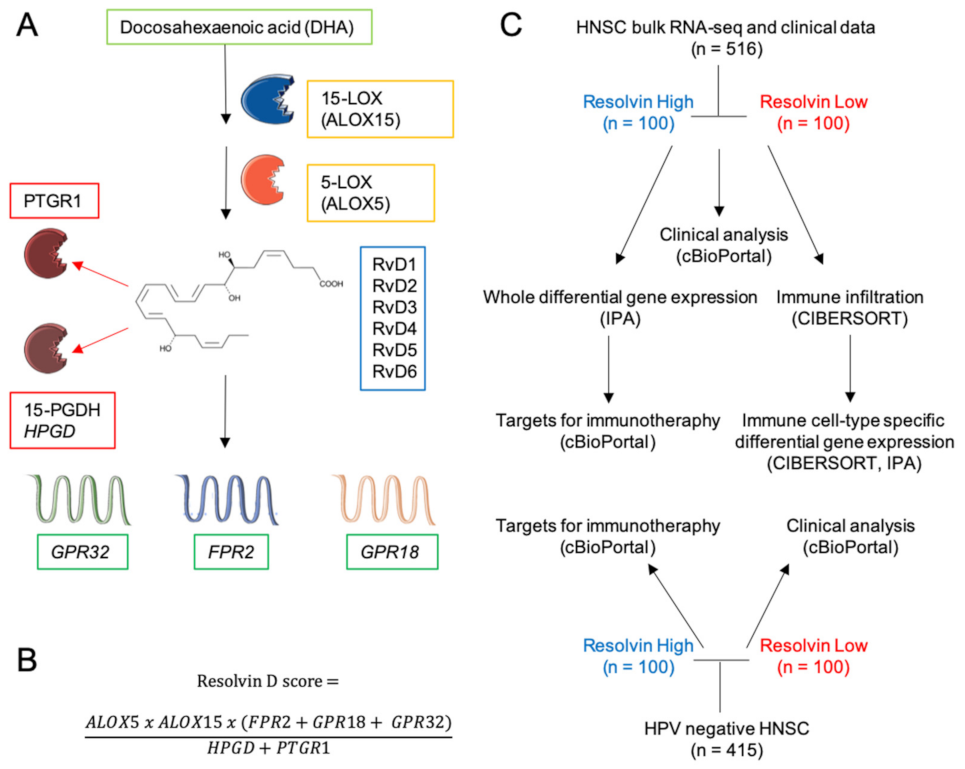
2.1. Definition of the RvD Score and Patients’ Stratification
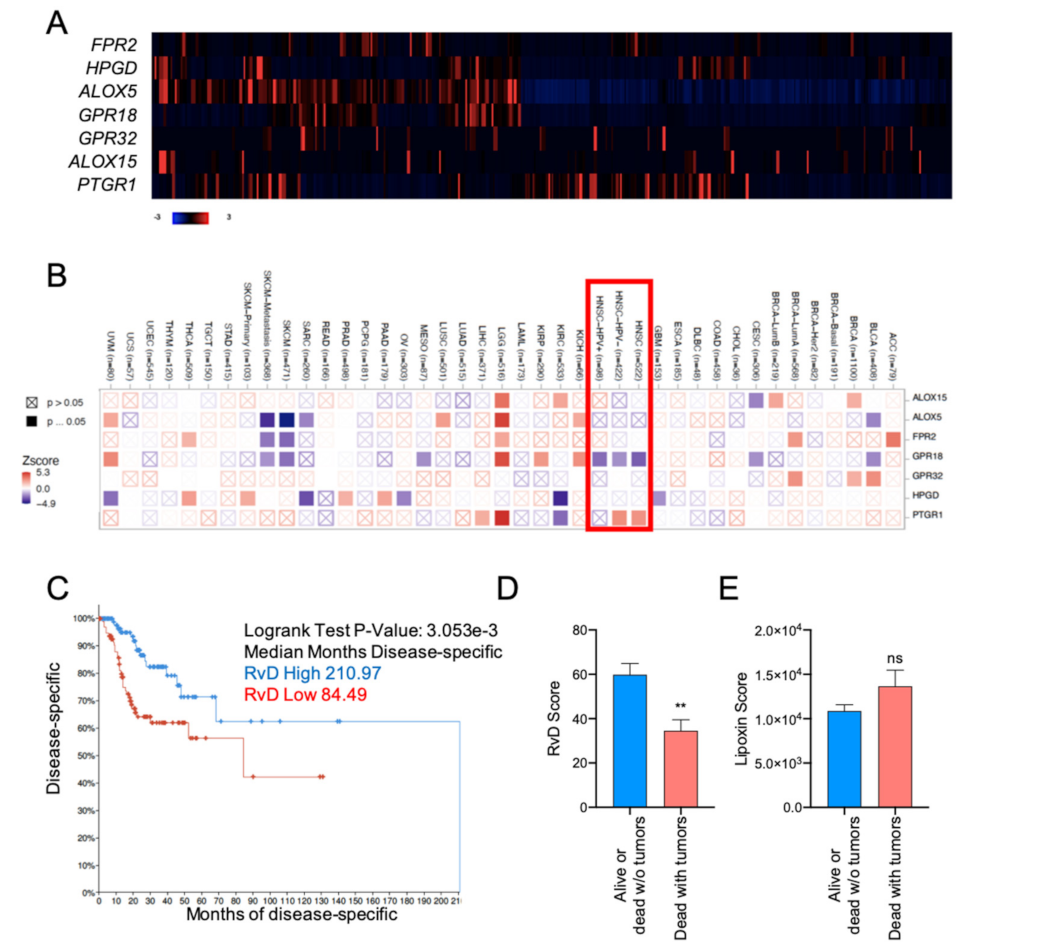
2.2. High RvD Score Predicts Better Prognosis in HNSC
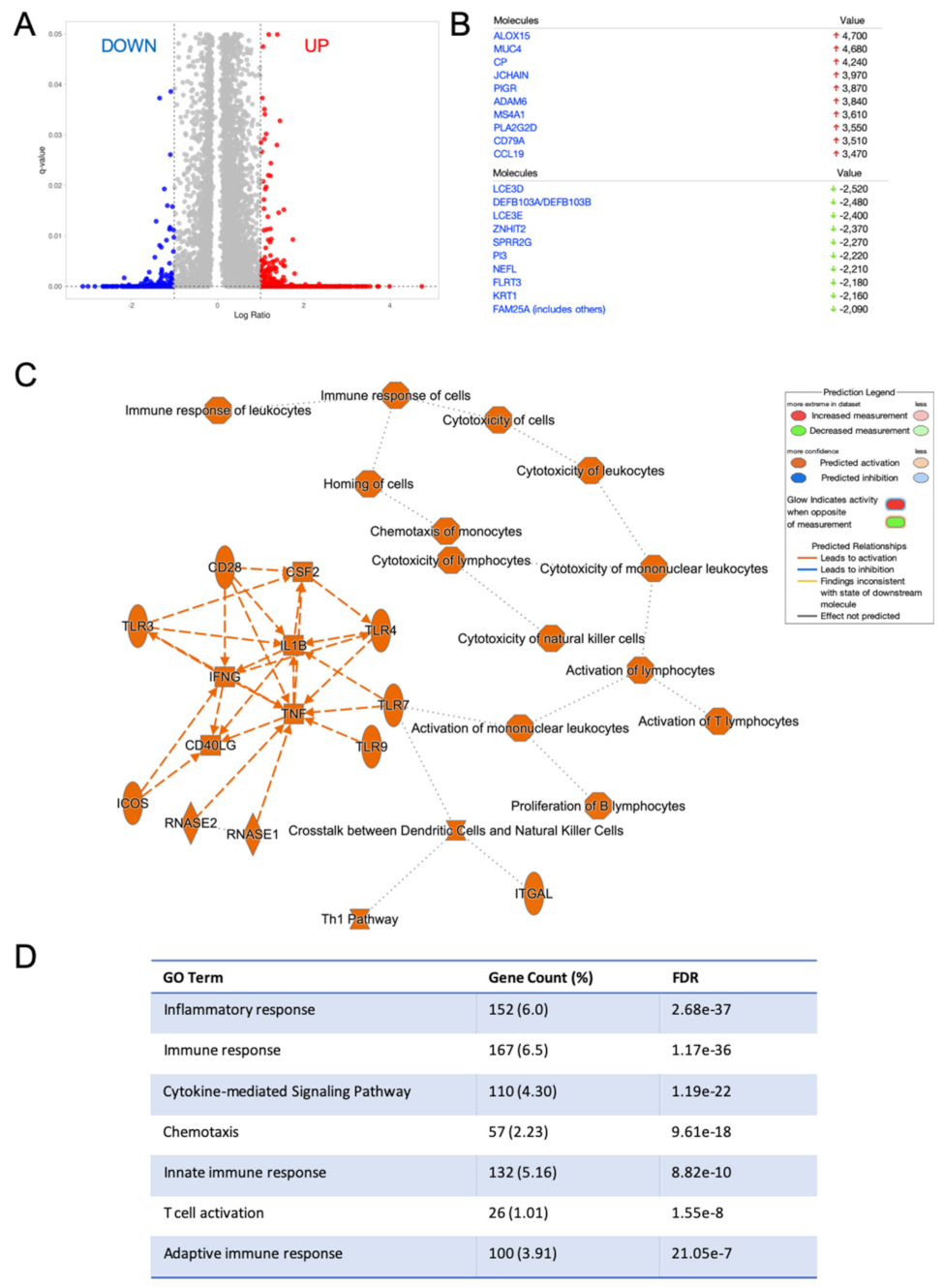
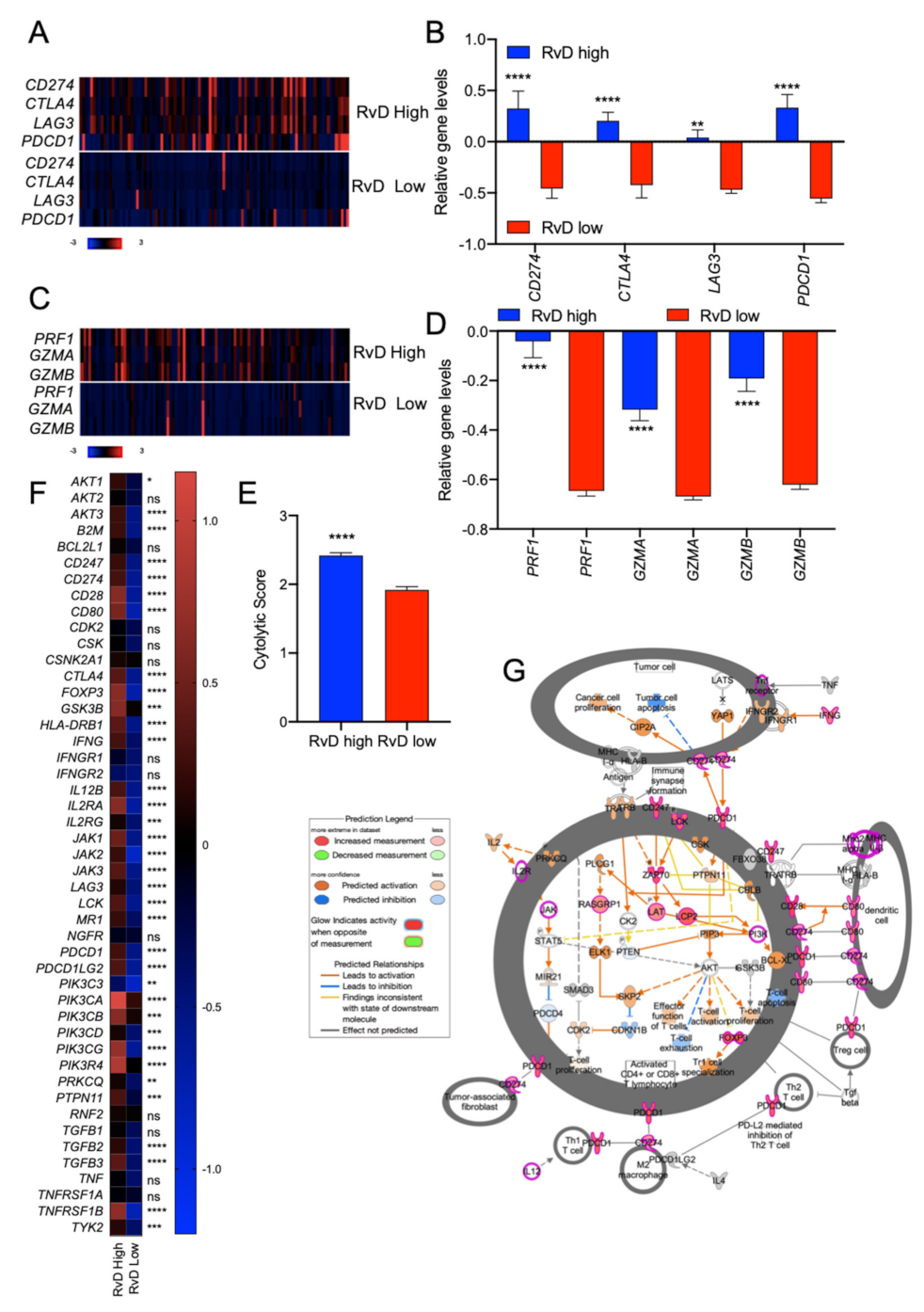
2.3. HNSC with High RvD Score Showed Transcriptional Fingerprints of Enhanced Anti-Tumor Immunity
2.4. High RvD Score Identifies Potential Responders to ICI Therapies
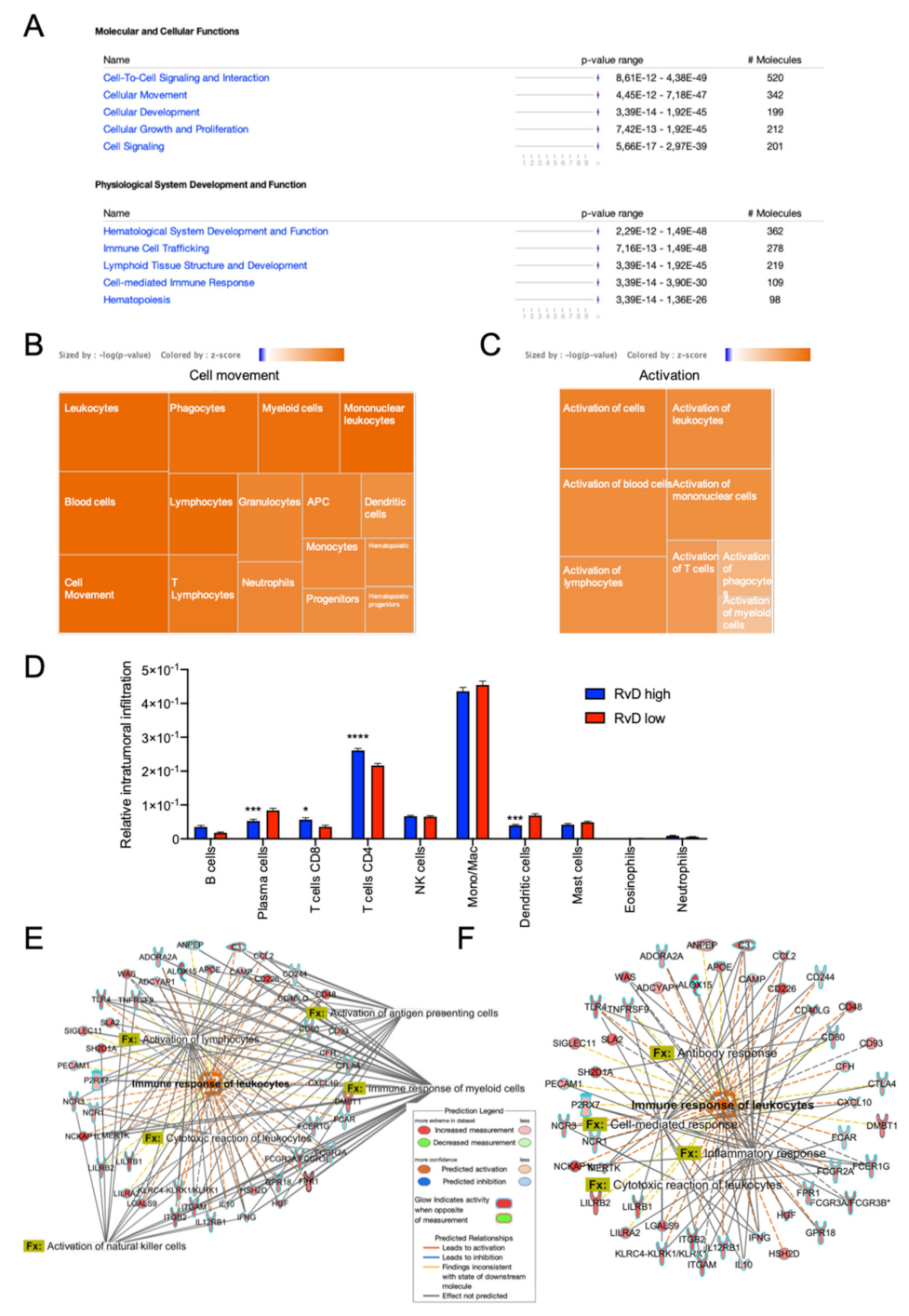
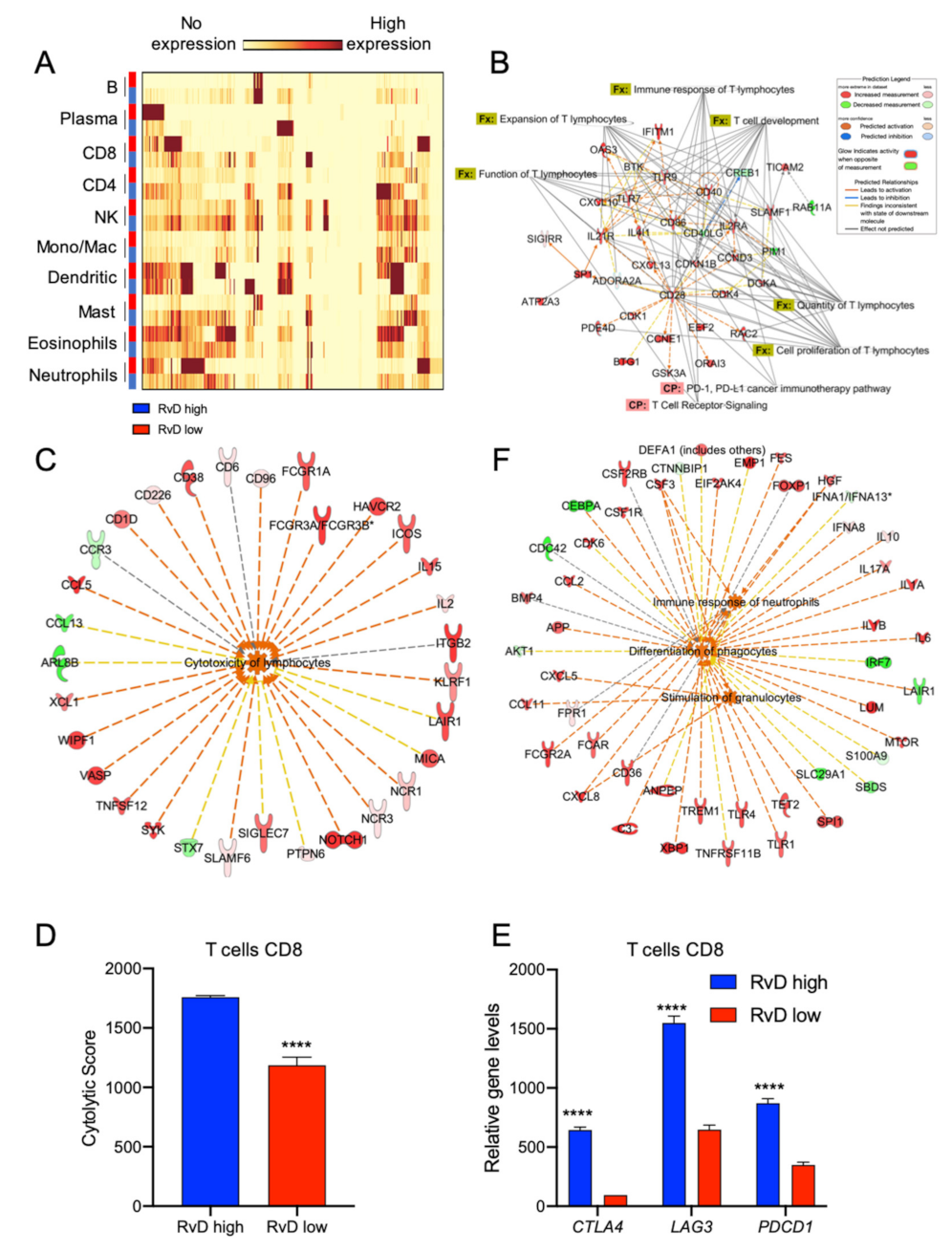
2.5. Immune Cell-Specific Gene Expression Profiles Revealed Increased Activation of Selected Leukocytes in Tumors with High RvD Score
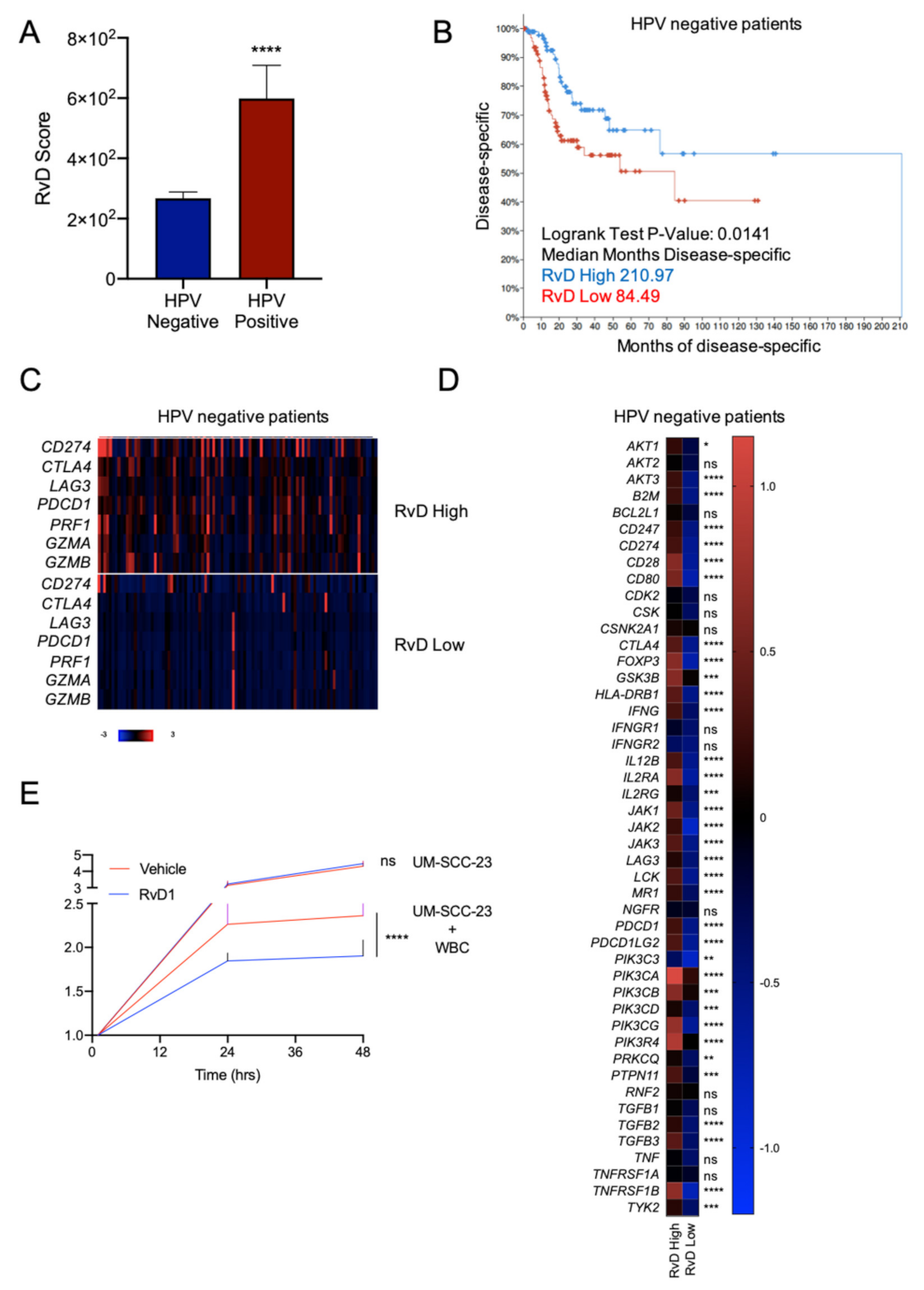
2.6. Higher RvD Scores Predicted Improved Anti-Tumor Immunity and Outcomes in HPV Negative HNSC
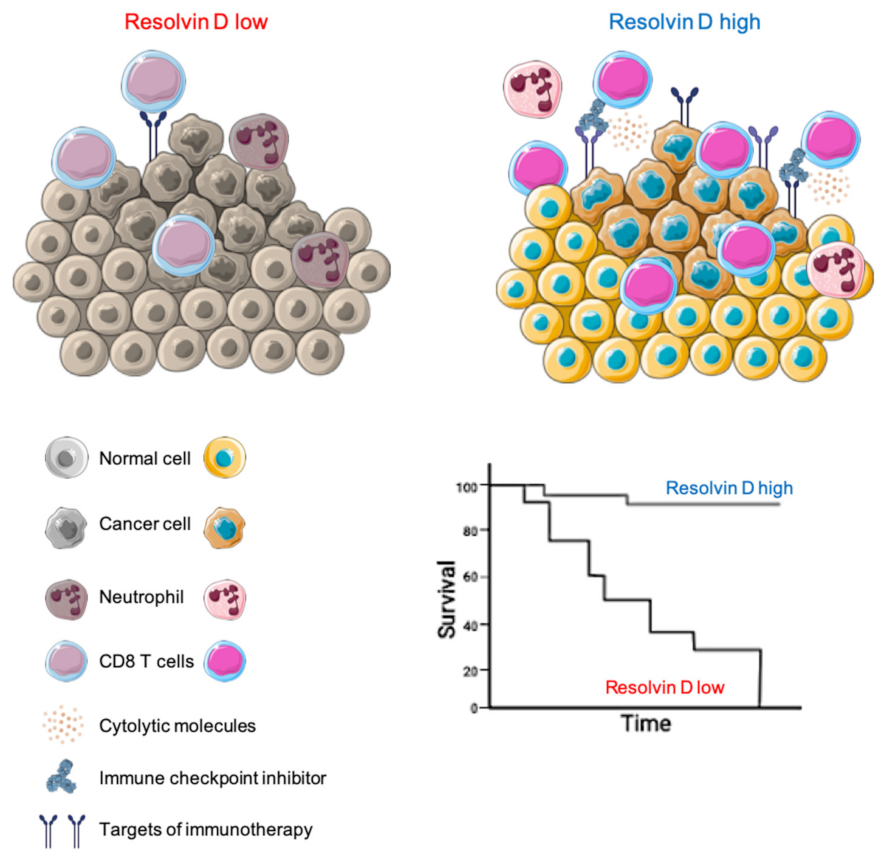
3. Discussion
4. Materials and Methods
4.1. Human Cancer Datasets and Clinical Analysis
4.2. Definition of RvD Score
4.3. Identification of Differentially Expressed Genes and Pathways in Bulk RNA-seq HNSC Dataset
4.4. Profiling of Intratumoral Immune Cell Infiltration
4.5. Leukocyte-Specific Gene Expression Profiles
4.6. Identification of Immunotherapy Responders
4.7. In Vitro Cell Growth Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Medzhitov, R. Inflammation 2010: New Adventures of an Old Flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Chiang, N.; Van Dyke, T.E. Resolving Inflammation: Dual Anti-Inflammatory and pro-Resolution Lipid Mediators. Nat. Rev. Immunol. 2008, 8, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Chiang, N.; Dalli, J. The Resolution Code of Acute Inflammation: Novel pro-Resolving Lipid Mediators in Resolution. Semin. Immunol. 2015, 27, 200–215. [Google Scholar] [CrossRef]
- Chiang, N.; Serhan, C.N. Specialized Pro-Resolving Mediator Network: An Update on Production and Actions. Essays Biochem. 2020, 64, 443–462. [Google Scholar] [CrossRef]
- Serhan, C.N.; Levy, B.D. Resolvins in Inflammation: Emergence of the pro-Resolving Superfamily of Mediators. J. Clin. Investig. 2018, 128, 2657–2669. [Google Scholar] [CrossRef]
- Chiang, N.; Serhan, C.N. Structural Elucidation and Physiologic Functions of Specialized Pro-Resolving Mediators and Their Receptors. Mol. Asp. Med. 2017, 58, 114–129. [Google Scholar] [CrossRef]
- Sun, Y.-P.; Oh, S.F.; Uddin, J.; Yang, R.; Gotlinger, K.; Campbell, E.; Colgan, S.P.; Petasis, N.A.; Serhan, C.N. Resolvin D1 and Its Aspirin-Triggered 17 R Epimer: Stereochemical assignments, anti-inflammatory properties, and enzymatic inactivation. J. Biol. Chem. 2007, 282, 9323–9334. [Google Scholar] [CrossRef]
- Mattoscio, D.; Isopi, E.; Lamolinara, A.; Patruno, S.; Medda, A.; De Cecco, F.; Chiocca, S.; Iezzi, M.; Romano, M.; Recchiuti, A. Resolvin D1 Reduces Cancer Growth Stimulating a Protective Neutrophil-Dependent Recruitment of Anti-Tumor Monocytes. J. Exp. Clin. Cancer Res. 2021, 40, 129. [Google Scholar] [CrossRef]
- Panigrahy, D.; Gartung, A.; Yang, J.; Yang, H.; Gilligan, M.M.; Sulciner, M.L.; Bhasin, S.S.; Bielenberg, D.R.; Chang, J.; Schmidt, B.A.; et al. Preoperative Stimulation of Resolution and Inflammation Blockade Eradicates Micrometastases. J. Clin. Investig. 2019, 129, 2964–2979. [Google Scholar] [CrossRef]
- Sulciner, M.L.; Serhan, C.N.; Gilligan, M.M.; Mudge, D.K.; Chang, J.; Gartung, A.; Lehner, K.A.; Bielenberg, D.R.; Schmidt, B.; Dalli, J.; et al. Resolvins Suppress Tumor Growth and Enhance Cancer Therapy. J Exp Med 2018, 215, 115–140. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, M.M.; Gartung, A.; Sulciner, M.L.; Norris, P.C.; Sukhatme, V.P.; Bielenberg, D.R.; Huang, S.; Kieran, M.W.; Serhan, C.N.; Panigrahy, D. Aspirin-Triggered Proresolving Mediators Stimulate Resolution in Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 6292–6297. [Google Scholar] [CrossRef] [PubMed]
- Vannitamby, A.; Saad, M.I.; Aloe, C.; Wang, H.; Kumar, B.; Vlahos, R.; Selemidis, S.; Irving, L.; Steinfort, D.; Jenkins, B.J.; et al. Aspirin-Triggered Resolvin D1 Reduces Proliferation and the Neutrophil to Lymphocyte Ratio in a Mutant KRAS-Driven Lung Adenocarcinoma Model. Cancers 2021, 13, 3224. [Google Scholar] [CrossRef] [PubMed]
- Orr, S.K.; Butler, K.L.; Hayden, D.; Tompkins, R.G.; Serhan, C.N.; Irimia, D. Gene Expression of Proresolving Lipid Mediator Pathways Is Associated with Clinical Outcomes in Trauma Patients. Crit. Care Med. 2015, 43, 2642–2650. [Google Scholar] [CrossRef]
- Iovoli, A.J.; Hermann, G.M.; Ma, S.J.; Platek, A.J.; Farrugia, M.K.; Yau, E.; Wooten, K.E.; Arshad, H.; Gupta, V.; Kuriakose, M.A.; et al. Association of Nonsteroidal Anti-Inflammatory Drug Use with Survival in Patients with Squamous Cell Carcinoma of the Head and Neck Treated with Chemoradiation Therapy. JAMA Netw. Open 2020, 3, e207199. [Google Scholar] [CrossRef]
- Wilson, J.C.; Murray, L.J.; Hughes, C.M.; Black, A.; Anderson, L.A. Non-Steroidal Anti-Inflammatory Drug and Aspirin Use and the Risk of Head and Neck Cancer. Br. J. Cancer 2013, 108, 1178–1181. [Google Scholar] [CrossRef]
- Becker, C.; Wilson, J.C.; Jick, S.S.; Meier, C.R. Non-Steroidal Anti-Inflammatory Drugs and the Risk of Head and Neck Cancer: A Case-Control Analysis: NSAIDs and Risk of Head and Neck Cancer. Int. J. Cancer 2015, 137, 2424–2431. [Google Scholar] [CrossRef]
- Luo, S.-D.; Wu, S.-C.; Chen, W.-C.; Wu, C.-N.; Chiu, T.-J.; Yang, Y.-H.; Li, S.-H.; Fang, F.-M.; Huang, T.-L.; Hsiao, C.-C.; et al. Low-Dose Aspirin Confers a Survival Benefit in Patients with Pathological Advanced-Stage Oral Squamous Cell Carcinoma. Sci. Rep. 2021, 11, 17161. [Google Scholar] [CrossRef]
- Shi, J.; Leng, W.; Zhao, L.; Xu, C.; Wang, J.; Chen, X.; Wang, Y.; Peng, X. Nonsteroidal Anti-Inflammatory Drugs Using and Risk of Head and Neck Cancer: A Dose-Response Meta Analysis of Prospective Cohort Studies. Oncotarget 2017, 8, 99066–99074. [Google Scholar] [CrossRef]
- Mandal, R.; Şenbabaoğlu, Y.; Desrichard, A.; Havel, J.J.; Dalin, M.G.; Riaz, N.; Lee, K.-W.; Ganly, I.; Hakimi, A.A.; Chan, T.A.; et al. The Head and Neck Cancer Immune Landscape and Its Immunotherapeutic Implications. JCI Insight 2016, 1, e89829. [Google Scholar] [CrossRef]
- Whiteside, T.L. Head and Neck Carcinoma Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2018, 24, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primer 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Lo, K.-W.; Lui, V.W.Y. Omics-Wide Quantitative B-Cell Infiltration Analyses Identify GPR18 for Human Cancer Prognosis with Superiority over CD20. Commun. Biol. 2020, 3, 234. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, gkac194. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining Cell Type Abundance and Expression from Bulk Tissues with Digital Cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Larkins, E.; Blumenthal, G.M.; Yuan, W.; He, K.; Sridhara, R.; Subramaniam, S.; Zhao, H.; Liu, C.; Yu, J.; Goldberg, K.B.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma with Disease Progression on or After Platinum-Containing Chemotherapy. Oncologist 2017, 22, 873–878. [Google Scholar] [CrossRef]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab Alone or with Chemotherapy versus Cetuximab with Chemotherapy for Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-048): A Randomised, Open-Label, Phase 3 Study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.J.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab vs Investigator’s Choice in Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck: 2-Year Long-Term Survival Update of CheckMate 141 with Analyses by Tumor PD-L1 Expression. Oral Oncol. 2018, 81, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xu, L.; Yi, M.; Yu, S.; Wu, K.; Luo, S. Novel Immune Checkpoint Targets: Moving beyond PD-1 and CTLA-4. Mol. Cancer 2019, 18, 155. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The Prognostic Landscape of Genes and Infiltrating Immune Cells across Human Cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Guo, C.; Tang, Y.; Zhang, Y.; Li, G. Mining TCGA Data for Key Biomarkers Related to Immune Microenvironment in Endometrial Cancer by Immune Score and Weighted Correlation Network Analysis. Front. Mol. Biosci. 2021, 8, 645388. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, L.; Lin, Y.; Hong, H.; Wei, Y.; Ye, L.; Wu, X. Identification of Biomarkers Related to Prognosis of Bladder Transitional Cell Carcinoma. Front. Genet. 2021, 12, 682237. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, M.; Ai, X.; Cheng, J.; Huang, Z.; Li, X.; Chen, Y. Identification of Potential Diagnostic and Prognostic Biomarkers for Colorectal Cancer Based on GEO and TCGA Databases. Front. Genet. 2021, 11, 602922. [Google Scholar] [CrossRef]
- Badoual, C.; Hans, S.; Rodriguez, J.; Peyrard, S.; Klein, C.; Agueznay, N.E.H.; Mosseri, V.; Laccourreye, O.; Bruneval, P.; Fridman, W.H.; et al. Prognostic Value of Tumor-Infiltrating CD4 + T-Cell Subpopulations in Head and Neck Cancers. Clin. Cancer Res. 2006, 12, 465–472. [Google Scholar] [CrossRef]
- Wallis, S.P.; Stafford, N.D.; Greenman, J. Clinical Relevance of Immune Parameters in the Tumor Microenvironment of Head and Neck Cancers: Clinical Relevance of Immune Parameters in the Tumor Microenvironment. Head Neck 2015, 37, 449–459. [Google Scholar] [CrossRef]
- Fialová, A.; Koucký, V.; Hajdušková, M.; Hladíková, K.; Špíšek, R. Immunological Network in Head and Neck Squamous Cell Carcinoma—A Prognostic Tool Beyond HPV Status. Front. Oncol. 2020, 10, 1701. [Google Scholar] [CrossRef]
- Codagnone, M.; Cianci, E.; Lamolinara, A.; Mari, V.C.; Nespoli, A.; Isopi, E.; Mattoscio, D.; Arita, M.; Bragonzi, A.; Iezzi, M.; et al. Resolvin D1 Enhances the Resolution of Lung Inflammation Caused by Long-Term Pseudomonas Aeruginosa Infection. Mucosal Immunol. 2018, 11, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Isopi, E.; Mattoscio, D.; Codagnone, M.; Mari, V.C.; Lamolinara, A.; Patruno, S.; D’Aurora, M.; Cianci, E.; Nespoli, A.; Franchi, S.; et al. Resolvin D1 Reduces Lung Infection and Inflammation Activating Resolution in Cystic Fibrosis. Front. Immunol. 2020, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Recchiuti, A.; Patruno, S.; Mattoscio, D.; Isopi, E.; Pomilio, A.; Lamolinara, A.; Iezzi, M.; Pecce, R.; Romano, M. Resolvin D1 and D2 Reduce SARS-CoV-2-Induced Inflammatory Responses in Cystic Fibrosis Macrophages. FASEB J. 2021, 35, e21441. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, R.; Flak, M.B.; Gonzalez-Nunez, M.; Austin-Williams, S.; Palmas, F.; Colas, R.A.; Dalli, J. Aspirin Activates Resolution Pathways to Reprogram T Cell and Macrophage Responses in Colitis-Associated Colorectal Cancer. Sci. Adv. 2022, 8, eabl5420. [Google Scholar] [CrossRef] [PubMed]
- Gooden, M.J.M.; de Bock, G.H.; Leffers, N.; Daemen, T.; Nijman, H.W. The Prognostic Influence of Tumour-Infiltrating Lymphocytes in Cancer: A Systematic Review with Meta-Analysis. Br. J. Cancer 2011, 105, 93–103. [Google Scholar] [CrossRef]
- de Ruiter, E.J.; Ooft, M.L.; Devriese, L.A.; Willems, S.M. The Prognostic Role of Tumor Infiltrating T-Lymphocytes in Squamous Cell Carcinoma of the Head and Neck: A Systematic Review and Meta-Analysis. Oncoimmunology 2017, 6, e1356148. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune Checkpoint Inhibitors: Recent Progress and Potential Biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Machiels, J.-P.; René Leemans, C.; Golusinski, W.; Grau, C.; Licitra, L.; Gregoire, V. Squamous Cell Carcinoma of the Oral Cavity, Larynx, Oropharynx and Hypopharynx: EHNS–ESMO–ESTRO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2020, 31, 1462–1475. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Bell, R.B.; Bifulco, C.B.; Burtness, B.; Gillison, M.L.; Harrington, K.J.; Le, Q.-T.; Lee, N.Y.; Leidner, R.; Lewis, R.L.; et al. The Society for Immunotherapy of Cancer Consensus Statement on Immunotherapy for the Treatment of Squamous Cell Carcinoma of the Head and Neck (HNSCC). J. Immunother. Cancer 2019, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Halder, R.C.; Almasi, A.; Sagong, B.; Leung, J.; Jewett, A.; Fiala, M. Curcuminoids and Omega-3 Fatty Acids with Anti-Oxidants Potentiate Cytotoxicity of Natural Killer Cells against Pancreatic Ductal Adenocarcinoma Cells and Inhibit Interferon Gamma Production. Front. Physiol 2015, 6, 129. [Google Scholar] [CrossRef] [PubMed]
- Chiurchiù, V.; Leuti, A.; Dalli, J.; Jacobsson, A.; Battistini, L.; Maccarrone, M.; Serhan, C.N. Proresolving Lipid Mediators Resolvin D1, Resolvin D2, and Maresin 1 Are Critical in Modulating T Cell Responses. Sci. Transl. Med. 2016, 8, 353ra111. [Google Scholar] [CrossRef]
- Mann, J.E.; Kulkarni, A.; Birkeland, A.C.; Kafelghazal, J.; Eisenberg, J.; Jewell, B.M.; Ludwig, M.L.; Spector, M.E.; Jiang, H.; Carey, T.E.; et al. The Molecular Landscape of the University of Michigan Laryngeal Squamous Cell Carcinoma Cell Line Panel. Head Neck 2019, 41, 3114–3124. [Google Scholar] [CrossRef] [PubMed]
- Citro, S.; Miccolo, C.; Medda, A.; Ghiani, L.; Tagliabue, M.; Ansarin, M.; Chiocca, S. HPV-Mediated Regulation of SMAD4 Modulates the DNA Damage Response in Head and Neck Cancer. J. Exp. Clin. Cancer Res. 2022, 41, 59. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattoscio, D.; Ferri, G.; Miccolo, C.; Chiocca, S.; Romano, M.; Recchiuti, A. Gene Expression of the D-Series Resolvin Pathway Predicts Activation of Anti-Tumor Immunity and Clinical Outcomes in Head and Neck Cancer. Int. J. Mol. Sci. 2022, 23, 6473. https://doi.org/10.3390/ijms23126473
Mattoscio D, Ferri G, Miccolo C, Chiocca S, Romano M, Recchiuti A. Gene Expression of the D-Series Resolvin Pathway Predicts Activation of Anti-Tumor Immunity and Clinical Outcomes in Head and Neck Cancer. International Journal of Molecular Sciences. 2022; 23(12):6473. https://doi.org/10.3390/ijms23126473
Chicago/Turabian StyleMattoscio, Domenico, Giulia Ferri, Claudia Miccolo, Susanna Chiocca, Mario Romano, and Antonio Recchiuti. 2022. "Gene Expression of the D-Series Resolvin Pathway Predicts Activation of Anti-Tumor Immunity and Clinical Outcomes in Head and Neck Cancer" International Journal of Molecular Sciences 23, no. 12: 6473. https://doi.org/10.3390/ijms23126473
APA StyleMattoscio, D., Ferri, G., Miccolo, C., Chiocca, S., Romano, M., & Recchiuti, A. (2022). Gene Expression of the D-Series Resolvin Pathway Predicts Activation of Anti-Tumor Immunity and Clinical Outcomes in Head and Neck Cancer. International Journal of Molecular Sciences, 23(12), 6473. https://doi.org/10.3390/ijms23126473







