May Staphylococcus lugdunensis Be an Etiological Factor of Chronic Maxillary Sinuses Infection?
Abstract
:1. Introduction
2. Results
2.1. Isolates and Patients’ Characteristics
2.2. Species Identification and Antibiotic Susceptibility
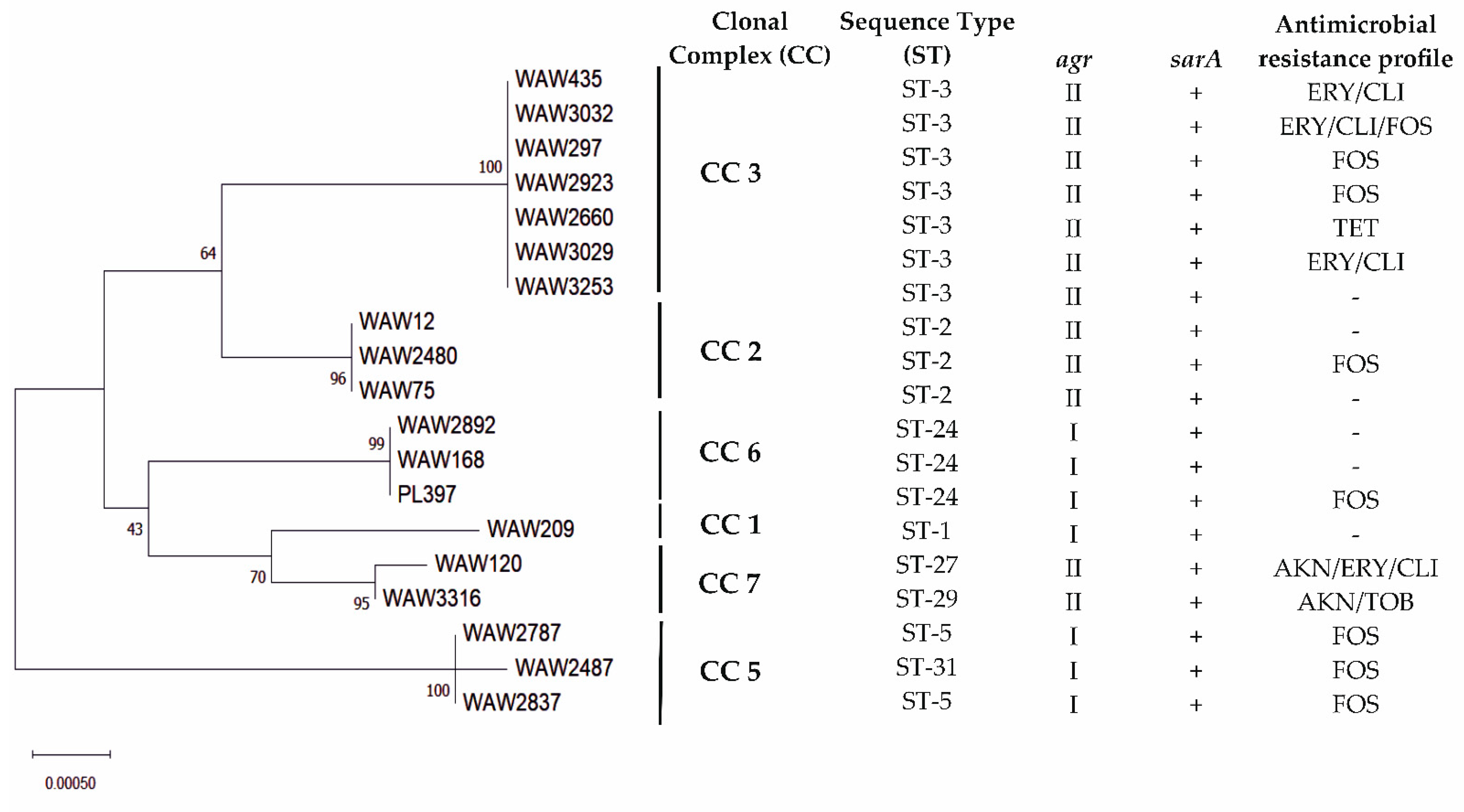
2.3. Multilocus Sequence Types
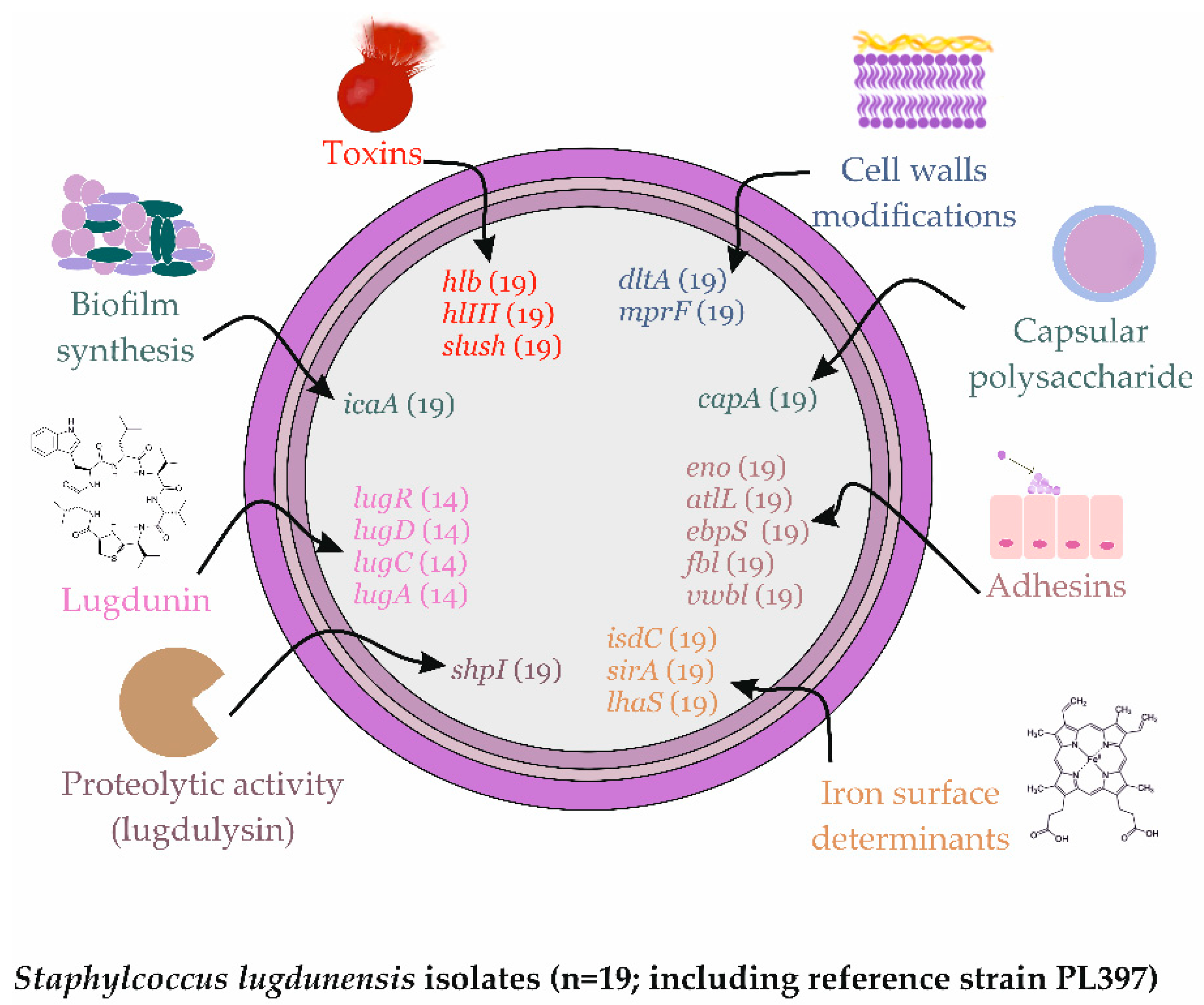
2.4. Virulence Factors Distribution
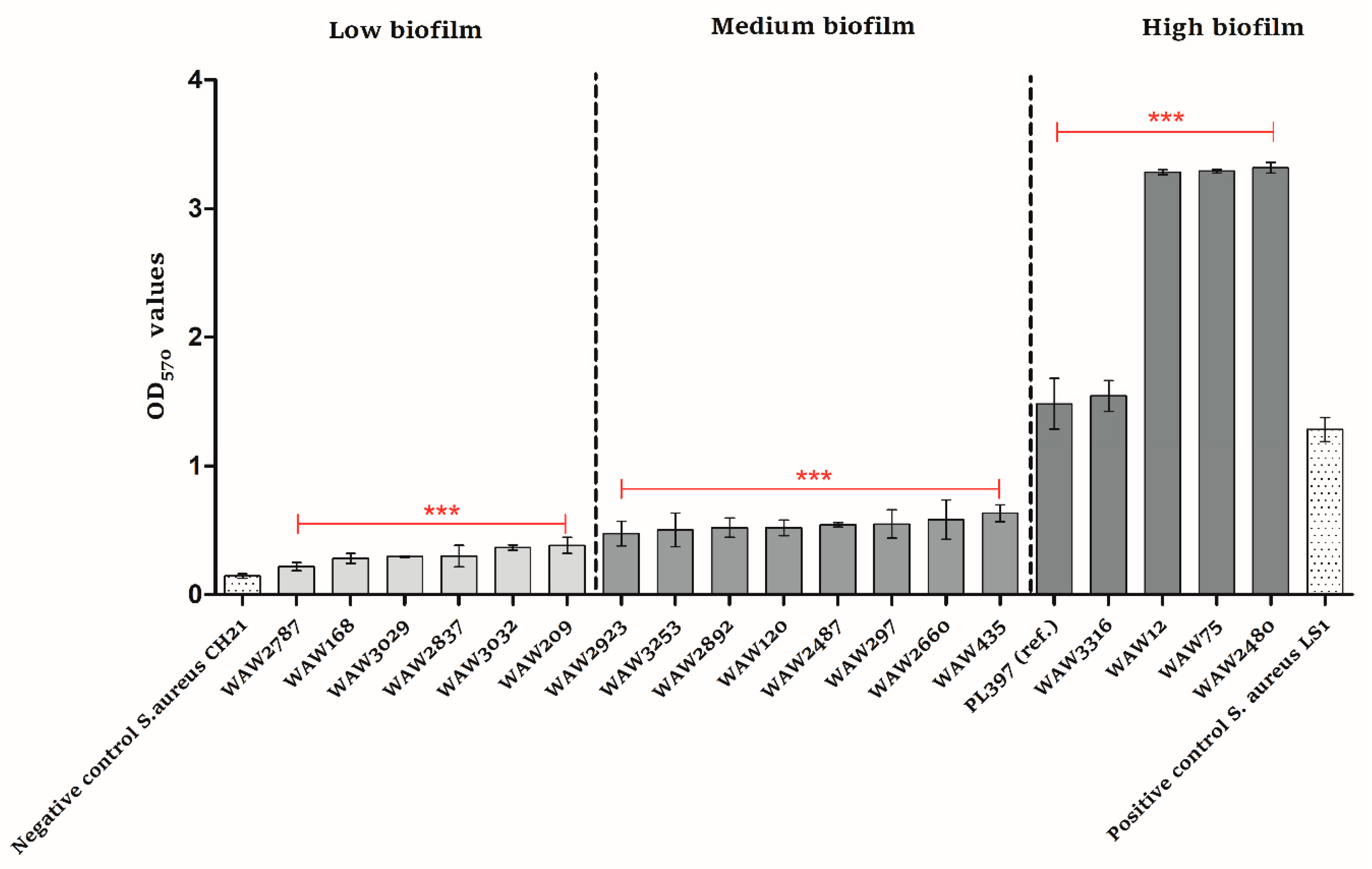
2.5. Biofilm Formation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Isolates
5.2. Genomic DNA Extraction
5.3. Species Identification
5.4. Susceptibility Testing
5.5. Phenotypic Assessment of Secreted Virulence Factors Production
5.6. Lugdunin Activity Assay
5.7. Biofilm Formation Assay
5.8. MLST Typing
5.9. Molecular Detection of Virulence Genes
5.10. Nucleotide Sequence Accession Numbers
5.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taha, L.; Stegger, M.; Söderquist, B. Staphylococcus lugdunensis: Antimicrobial susceptibility and optimal treatment options. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1449–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieber, L.; Kahlmeter, G. Staphylococcus lugdunensis in several niches of the normal skin flora. Clin. Microbiol. Infect. 2010, 16, 385–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.M.; Price, L.B.; Hungate, B.A.; Abraham, A.G.; Larsen, L.A.; Christensen, K.; Stegger, M.; Skov, R.; Andersen, P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015, 1, e1400216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilbronner, S.; Foster, T.J. Staphylococcus lugdunensis: A skin commensal with invasive pathogenic potential. Clin. Microbiol. Rev. 2021, 34, e00205-20. [Google Scholar] [CrossRef]
- Thean, Y.T.; Siew, Y.N.; He, J. Microbiological characteristics, presumptive identification, and antibiotic susceptibilities of Staphylococcus lugdunensis. J. Clin. Microbiol. 2008, 46, 2393–2395. [Google Scholar] [CrossRef] [Green Version]
- Argemi, X.; Riegel, P.; Lavigne, T.; Lefebvre, N.; Grandpré, N.; Hansmann, Y.; Jaulhac, B.; Prévost, G.; Schramm, F. Implementation of matrix-assisted laser desorption ionization-time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J. Clin. Microbiol. 2015, 53, 2030–2036. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [Green Version]
- Michalik, M.; Samet, A.; Podbielska-Kubera, A.; Savini, V.; Międzobrodzki, J.; Kosecka-Strojek, M. Coagulase-negative staphylococci (CoNS) as a significant etiological factor of laryngological infections: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 26. [Google Scholar] [CrossRef]
- Bachert, C.; Pawankar, R.; Zhang, L.; Bunnag, C.; Fokkens, W.J.; Hamilos, D.L.; Jirapongsananuruk, O.; Kern, R.; Meltzer, E.O.; Mullol, J.; et al. ICON: Chronic rhinosinusitis. World Allergy Organ. J. 2014, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Dlugaszewska, J.; Leszczynska, M.; Lenkowski, M.; Tatarska, A.; Pastusiak, T.; Szyfter, W. The pathophysiological role of bacterial biofilms in chronic sinusitis. Eur. Arch. Otorhinolaryngol. 2016, 273, 1989–1994. [Google Scholar] [CrossRef] [Green Version]
- Coffey, C.S.; Sonnenburg, R.E.; Melroy, C.T.; Dubin, M.G.; Senior, B.A. Endoscopically guided aerobic cultures in postsurgical patients with chronic rhinosinusitis. Am. J. Rhinol. 2006, 20, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Jervis-Bardy, J.; Foreman, A.; Field, J.; Wormald, P.J. Impaired mucosal healing and infection associated with Staphylococcus aureus after endoscopic sinus surgery. Am. J. Rhinol. Allergy 2009, 23, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Michalik, M.; Samet, A.; Marszałek, A.; Krawczyk, B.; Kotłowski, R.; Nowicki, A.; Anyszek, T.; Nowicki, S.; Kur, J.; Nowicki, B. Intra-operative biopsy in chronic sinusitis detects pathogenic Escherichia coli that carry fimG/H, fyuA and agn43 genes coding biofilm formation. PLoS ONE 2018, 13, e0192899. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Michalik, M.; Fordon, M.; Wysocka, M.; Samet, A.; Nowicki, B. Escherichia coli strains with virulent factors typical for uropathogens were isolated from sinuses from patients with chronic rhinosinusitis-case report. Pathogens 2020, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Both, A.; Huang, J.; Qi, M.; Lausmann, C.; Weißelberg, S.; Büttner, H.; Lezius, S.; Failla, A.V.; Christner, M.; Stegger, M.; et al. Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations. PLoS Pathog. 2021, 17, e1009304. [Google Scholar] [CrossRef] [PubMed]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [Green Version]
- Sakr, A.; Brégeon, F.; Mège, J.L.; Rolain, J.M.; Blin, O. Staphylococcus aureus nasal colonization: An update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol. 2018, 9, 2419. [Google Scholar] [CrossRef]
- Bitschar, K.; Sauer, B.; Focken, J.; Dehmer, H.; Moos, S.; Konnerth, M.; Schilling, N.A.; Grond, S.; Kalbacher, H.; Kurschus, F.C.; et al. Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 2019, 10, 2730. [Google Scholar] [CrossRef]
- Heilbronner, S. Staphylococcus lugdunensis. Trends Microbiol. 2021, 29, 1143–1145. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Huang, Y.F.; Tang, C.W.; Chen, Y.Y.; Hsieh, K.S.; Ger, L.P.; Chen, Y.S.; Liu, Y.C. Staphylococcus lugdunensis infective endocarditis: A literature review and analysis of risk factors. J. Microbiol. Immunol. Infect. 2010, 43, 478–484. [Google Scholar] [CrossRef] [Green Version]
- Non, L.R.; Santos, C.A.Q. The occurrence of infective endocarditis with Staphylococcus lugdunensis bacteremia: A retrospective cohort study and systematic review. J. Infect. 2017, 74, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Preda, M.; Mihai, M.M.; Popa, L.I.; Dițu, L.-M.; Holban, A.M.; Manolescu, L.S.C.; Popa, G.-L.; Muntean, A.-A.; Gheorghe, I.; Chifiriuc, C.M.; et al. Phenotypic and genotypic virulence features of staphylococcal strains isolated from difficult-to-treat skin and soft tissue infections. PLoS ONE 2021, 16, e0246478. [Google Scholar] [CrossRef] [PubMed]
- Chassain, B.; Lemée, L.; Didi, J.; Thiberge, J.M.; Brisse, S.; Pons, J.L.; Pestel-Caron, M. Multilocus sequence typing analysis of Staphylococcus lugdunensis implies a clonal population structure. J. Clin. Microbiol. 2012, 50, 3003–3009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argemi, X.; Hansmann, Y.; Prola, K.; Prévost, G. Coagulase-negative staphylococci pathogenomics. Int. J. Mol. Sci. 2019, 20, 1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.C.; Lee, M.H.; Yeh, C.F.; Liu, T.P.; Lin, J.F.; Ho, C.M.; Lu, J.J. Characterization of two novel variants of staphylococcal cassette chromosome mec elements in oxacillin-resistant Staphylococcus lugdunensis. J. Antimicrob. Chemother. 2017, 72, 3258–3262. [Google Scholar] [CrossRef]
- Zapotoczna, M.; Heilbronner, S.; Speziale, P.; Foster, T.J. Iron-regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J. Bacteriol. 2012, 194, 6453–6467. [Google Scholar] [CrossRef] [Green Version]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [Green Version]
- Aubourg, M.; Dhalluin, A.; Gravey, F.; Pottier, M.; Thomy, N.; Bernay, B.; Goux, D.; Martineau, M.; Giard, J.-C. Phenotypic and proteomic approaches of the response to iron-limited condition in Staphylococcus lugdunensis. BMC Microbiol. 2020, 20, 328. [Google Scholar] [CrossRef]
- Aubourg, M.; Pottier, M.; Léon, A.; Bernay, B.; Dhalluin, A.; Cacaci, M.; Torelli, R.; Ledormand, P.; Martini, C.; Sanguinetti, M.; et al. Inactivation of the response regulator AgrA has a pleiotropic effect on biofilm formation, pathogenesis and stress response in Staphylococcus lugdunensis. Microbiol. Spectr. 2022, 10, e0159821. [Google Scholar] [CrossRef] [PubMed]
- Dahyot, S.; Oxaran, V.; Niepceron, M.; Dupart, E.; Legris, S.; Destruel, L.; Didi, J.; Clamens, T.; Lesouhaitier, O.; Zerdoumi, Y.; et al. Role of the LytSR two-component regulatory system in Staphylococcus lugdunensis biofilm formation and pathogenesis. Front. Microbiol. 2020, 11, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosecka-Strojek, M.; Sabat, A.J.; Akkerboom, V.; Becker, K.; van Zanten, E.; Wisselink, G.; Miedzobrodzki, J.; Kooistra-Smid, A.; Friedrich, A.W. Development and Validation of a Reference Data Set for Assigning Staphylococcus Species Based on Next-Generation Sequencing of the 16S-23S rRNA Region. Front. Cell. Infect. Microbiol. 2019, 9, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, P.C.; Leung, A.S.; Leung, K.W.; Yuen, K.Y. Identification of slide coagulase positive, tube coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol. Pathol. MP 2001, 54, 244–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellmann, A.; Becker, K.; von Eiff, C.; Keckevoet, U.; Schumann, P.; Harmsen, D. Sequencing and staphylococci identification. Emerg. Infect. Dis. 2006, 12, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. J. Pathol. Microbiol. Immunol. 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Yeh, C.F.; Liu, T.P.; Cheng, C.W.; Chang, S.C.; Lee, M.H.; Lu, J.J. Molecular characteristics of disease-causing and commensal Staphylococcus lugdunensis isolates from 2003 to 2013 at a tertiary hospital in Taiwan. PLoS ONE 2015, 10, e0134859. [Google Scholar] [CrossRef]
- Frebourg, N.B.; Lefebvre, S.; Baert, S.; Lemeland, J.F. PCR-Based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 2000, 38, 877–880. [Google Scholar] [CrossRef] [Green Version]
- Giormezis, N.; Kolonitsiou, F.; Makri, A.; Vogiatzi, A.; Christofidou, M.; Anastassiou, E.D.; Spiliopoulou, I. Virulence factors among Staphylococcus lugdunensis are associated with infection sites and clonal spread. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 773–778. [Google Scholar] [CrossRef]
- Szabados, F.; Nowotny, Y.; Marlinghaus, L.; Korte, M.; Neumann, S.; Kaase, M.; Gatermann, S.G. Occurrence of genes of putative fibrinogen binding proteins and hemolysins, as well as of their phenotypic correlates in isolates of S. lugdunensis of different origins. BMC Res. Notes 2011, 4, 113. [Google Scholar] [CrossRef]
- Chatzigeorgiou, K.S.; Siafakas, N.; Petinaki, E.; Zerva, L. fbl gene as a species-specific target for Staphylococcus lugdunensis identification. J. Clin. Lab. Anal. 2010, 24, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Argemi, X.; Prévost, G.; Riegel, P.; Keller, D.; Meyer, N.; Baldeyrou, M.; Douiri, N.; Lefebvre, N.; Meghit, K.; Ronde Oustau, C.; et al. VISLISI trial, a prospective clinical study allowing identification of a new metalloprotease and putative virulence factor from Staphylococcus lugdunensis. Clin. Microbiol. Infect. 2017, 23, 334.e1–334.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Clinical Characteristic of Patients | Microbiological Cultures—Characteristic | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolate Name | Date of Strain Isolation | Isolation Site | Age | Gender | Underlying Diseases | Additional Patient-Associated Risk Factors | Surgery | Accompanying Bacterial Flora at Isolation Time | Isolation of S. aureus |
| WAW2480 | 6 September 2017 | left maxillary sinus | 59 | M | chronic sinusitis | none | FESS, DSN, HCNI | S. epidermidis; S. salivarius; S. mitis | no |
| WAW2487 | 8 September 2017 | right maxillary sinus | 36 | M | chronic sinusitis | sleep apnea | DSN, HCNI, UPP | E. coli | no |
| WAW2660 | 31 October 2017 | right maxillary sinus | 56 | M | chronic sinusitis | none | FESS, DSN | S. epidermidis | no |
| WAW2787 | 13 January 2018 | right maxillary sinus | 43 | M | chronic sinusitis | sleep disturbance; snoring | DSN, HCNI | no data | no data |
| WAW2837 | 8 February 2018 | left maxillary sinus | 39 | M | chronic sinusitis | nasal polyps; lower nasal concha hypertrophy | FESS | S. epidermidis; Moraxella catarrhalis; C. pseudodiphtheriticum | no |
| WAW2892 | 24 February 2018 | right maxillary sinus | 35 | M | chronic sinusitis | none | FESS | S. salivarius S. hominis | no |
| WAW2923 | 8 March 2018 | left maxillary sinus | 64 | M | chronic sinusitis | snoring | FESS, DSN, HCNI | no data | no data |
| WAW3029 | 13 April 2018 | right maxillary sinus | 46 | M | chronic sinusitis | snoring | MIST | S. epidermidis | yes B |
| WAW3032 | 13 April 2018 | right maxillary sinus | 31 | M | chronic sinusitis | recurring respiratory infections; snoring | FESS, HCNI | S. epidermidis; S. mitis; S. capitis; Pantoea agglomerans | yes A,C |
| WAW3253 | 13 June 2018 | left maxillary sinus | 44 | M | chronic sinusitis | none | FESS, DSN, HCNI (twice), CELON | S. pneumoniae; Propoonibacterium acnes; S. epidermidis | yes C |
| WAW3316 | 18 July 2018 | left maxillary sinus | 23 | M | chronic sinusitis | none | Endoscopic sinus catheterization, DSN, HCNI | S. epidermidis; C. accolens | no |
| WAW12 | 2 April 2019 | right maxillary sinus | 36 | M | chronic sinusitis | none | FESS, DSN, HCNI | C. pseudodiphtheriticum; Klebsiella oxytoca; S. epidermidis | no |
| WAW75 | 27 April 2019 | left maxillary sinus | 34 | M | chronic sinusitis | none | FESS, DSN, HCNI | S. epidermidis; E. cloacae | yes C |
| WAW120 | 20 May 2019 | left maxillary sinus | 43 | M | chronic sinusitis | nasal polyps; lower nasal concha hypertrophy | FESS, DSN, HCNI | Citrobacter freundii; S. epidermidis | yes C |
| WAW168 | 12 June 2019 | right maxillary sinus | 26 | M | chronic sinusitis | nasal polyps; lower nasal concha hypertrophy | FESS, DSN, HCNI | - | no |
| WAW209 | 31 July 2019 | right maxillary sinus | 56 | M | chronic sinusitis | nasal polyps | FESS, DSN, HCNI | S. epidermidis | no |
| WAW297 | 15 October 2019 | left maxillary sinus | 48 | M | chronic sinusitis | nasal polyps | FESS, DSN, HCNI | - | no |
| WAW435 | 25 June 2020 | right maxillary sinus | 56 | M | chronic sinusitis | nasal polyps; lower nasal concha hypertrophy; sleep apnea. | FESS | S. epidermidis | yes A |
| Virulence Factor(s) | Gene(s) | Primers | Sequence (5′–3′) | Tm (°C) | Product Size (bp) |
|---|---|---|---|---|---|
| Lugdunin | lugR | lugR_F | TGAAGTCATCATAAGTGCACACAA | 50 | 296 |
| lugR_R | ATCCTAAGGCAGAAATCCCTAAAT | ||||
| lugD | lugD_F | ACACAAGCGAAAGCGTTCAT | 48 | 717 | |
| lugD_R | GGCTACTCCCATTCCACCAA | ||||
| lugC | lugC_F | AAACGCATTCTGGACGGGAT | 50 | 994 | |
| lugC_R | TTTGGGTTGCCCGTAGTACC | ||||
| lugA | lugA_F | ACCACATAATTGCGAAGGCG | 50 | 1396 | |
| lugA_R | AGCCTCCATGTTTCCATGGTT | ||||
| Biofilm synthesis | icaA | icaA _F | ATGAAATATTTAAATTTGTTAA | 43 | 1224 |
| icaA _R | CTAATTTTTTCCTCTGTCTGG | ||||
| Adhesins/MSCRAMMs | eno | enoF | AGCTACTGCGATGTCAGCAA | 50 | 1059 |
| enoR | GCATTAGTGCCATCAGGTGC | ||||
| ebpS | ebpSF | CGTCAGCGGAACACCAAAAG | 50 | 969 | |
| ebpSR | ATTTGACTGTGACGCTCCGT | ||||
| Capsular polysaccharide biosynthesis protein | capA | capAF | ATGGAAAAAACGCTTGATT | 40 | 663 |
| capAR | CTATTTCAATTTATGGATT | ||||
| Cell walls modifications | dltA | dltAF | GACGTGCAACACCTACTGGA | 50 | 925 |
| dltAR | GATATTGAGCAAGCGCAGCC | ||||
| mprF | mprFF | TGCCACAACGACAGGTACAA | 50 | 728 | |
| mprFR | TCAATCGCTGGATGCTCGTT | ||||
| Iron surface determinants | isdC | isdCF | TCGCAGAGGGTCAGTCACTT | 50 | 429 |
| isdCR | CACTTGCTGCTGAGCCTGTA | ||||
| sirA | sirAF | ATGAATAAAGTTGTTAACATTAT | 40 | 993 | |
| sirAR | TTACTTTGATTGTTTATCA | ||||
| lhaS | lhaSF | ACCTGCCATGATTGGCTTTT | 50 | 410 | |
| lhaSR | TGTAACCTAGCCATGCACCAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosecka-Strojek, M.; Wolska-Gębarzewska, M.; Podbielska-Kubera, A.; Samet, A.; Krawczyk, B.; Międzobrodzki, J.; Michalik, M. May Staphylococcus lugdunensis Be an Etiological Factor of Chronic Maxillary Sinuses Infection? Int. J. Mol. Sci. 2022, 23, 6450. https://doi.org/10.3390/ijms23126450
Kosecka-Strojek M, Wolska-Gębarzewska M, Podbielska-Kubera A, Samet A, Krawczyk B, Międzobrodzki J, Michalik M. May Staphylococcus lugdunensis Be an Etiological Factor of Chronic Maxillary Sinuses Infection? International Journal of Molecular Sciences. 2022; 23(12):6450. https://doi.org/10.3390/ijms23126450
Chicago/Turabian StyleKosecka-Strojek, Maja, Mariola Wolska-Gębarzewska, Adrianna Podbielska-Kubera, Alfred Samet, Beata Krawczyk, Jacek Międzobrodzki, and Michał Michalik. 2022. "May Staphylococcus lugdunensis Be an Etiological Factor of Chronic Maxillary Sinuses Infection?" International Journal of Molecular Sciences 23, no. 12: 6450. https://doi.org/10.3390/ijms23126450
APA StyleKosecka-Strojek, M., Wolska-Gębarzewska, M., Podbielska-Kubera, A., Samet, A., Krawczyk, B., Międzobrodzki, J., & Michalik, M. (2022). May Staphylococcus lugdunensis Be an Etiological Factor of Chronic Maxillary Sinuses Infection? International Journal of Molecular Sciences, 23(12), 6450. https://doi.org/10.3390/ijms23126450








