A Bursaphelenchus xylophilus Effector, BxSCD3, Suppresses Plant Defense and Contributes to Virulence
Abstract
:1. Introduction
2. Results
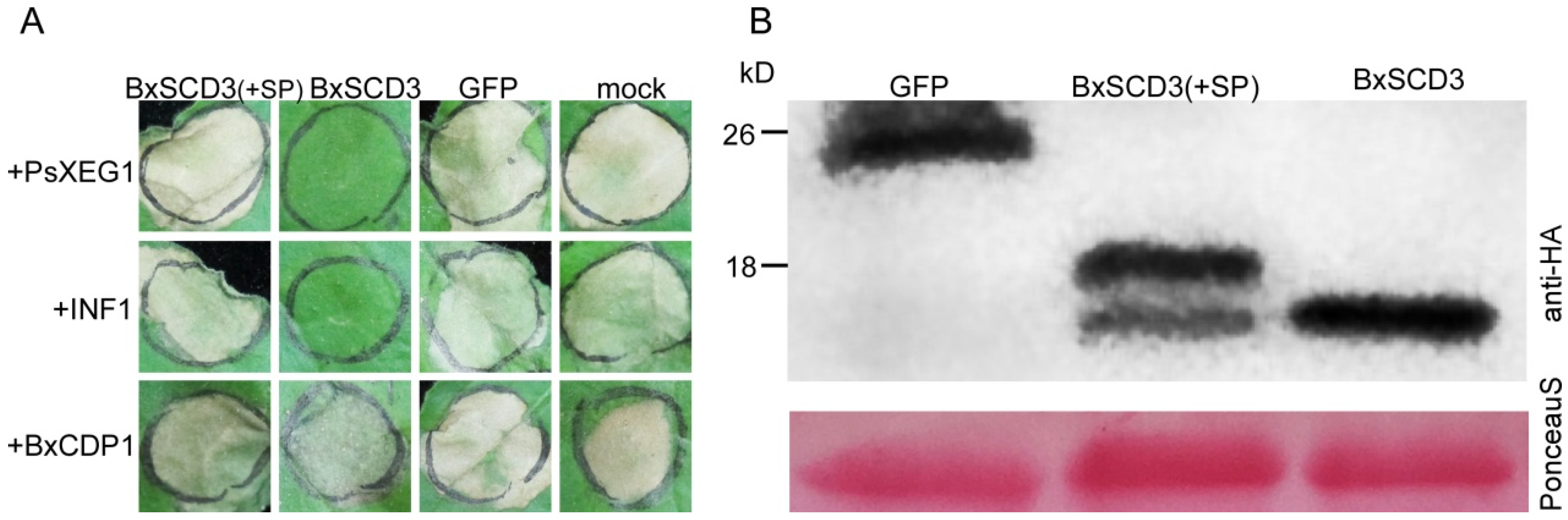
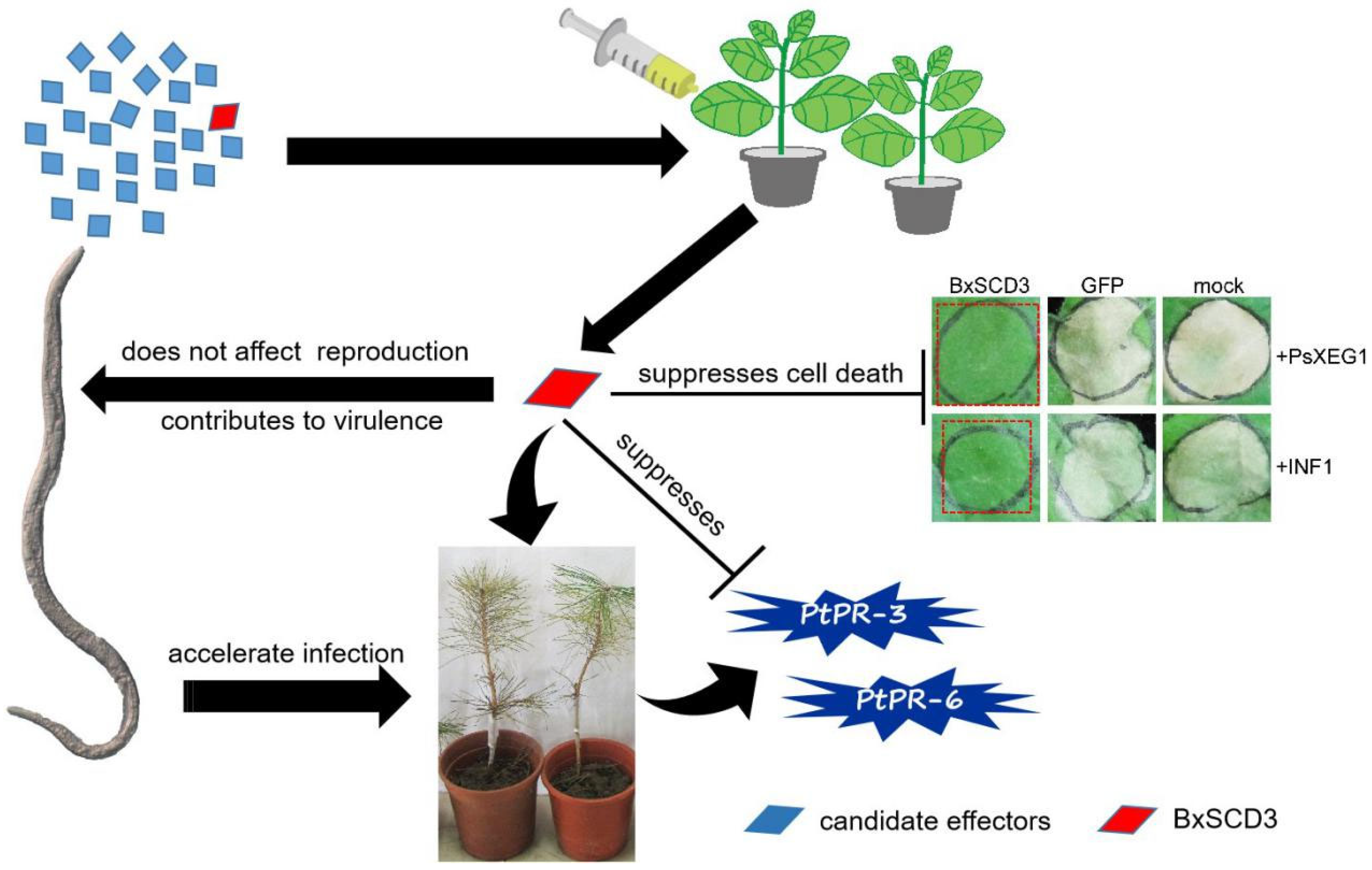
2.1. BxSCD3 Suppresses PsXEG1- and INF1-Induced Cell Death in N. benthamiana When Secreted into the Intracellular Space
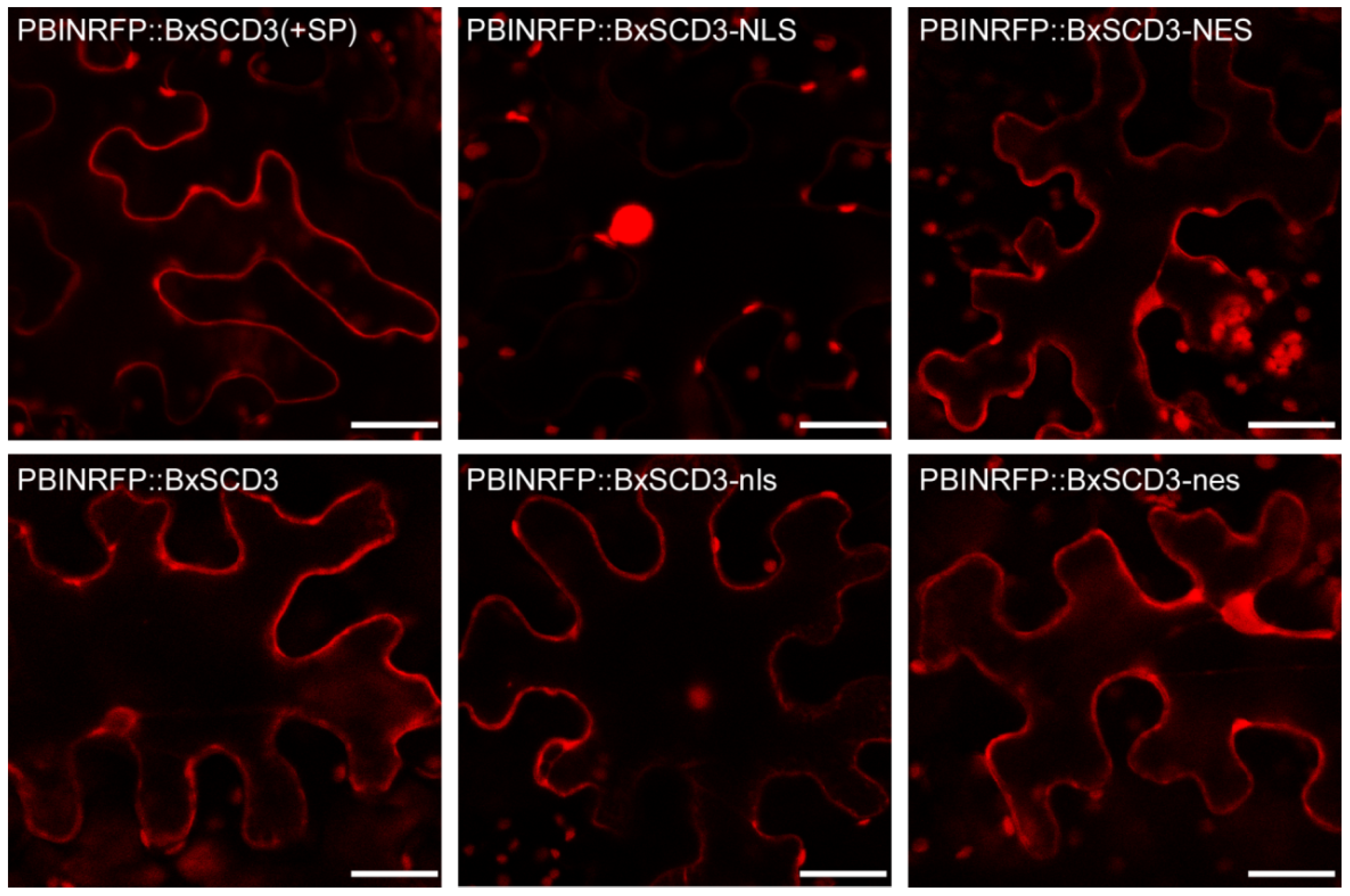
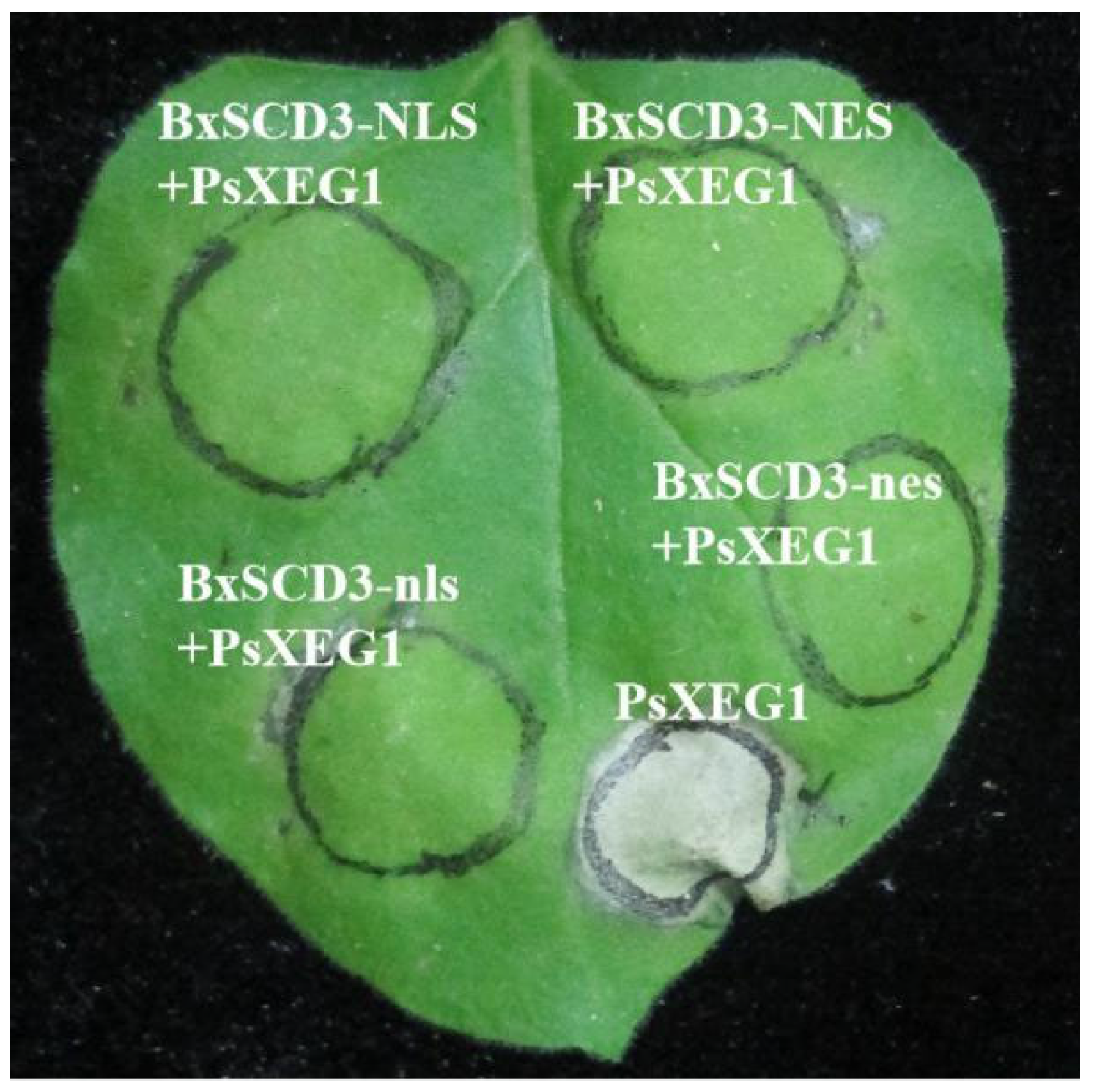
2.2. BxSCD3 Plays an Inhibitory Role in Plant Immunity Whether It Is Located in the Cytoplasm or the Nucleus
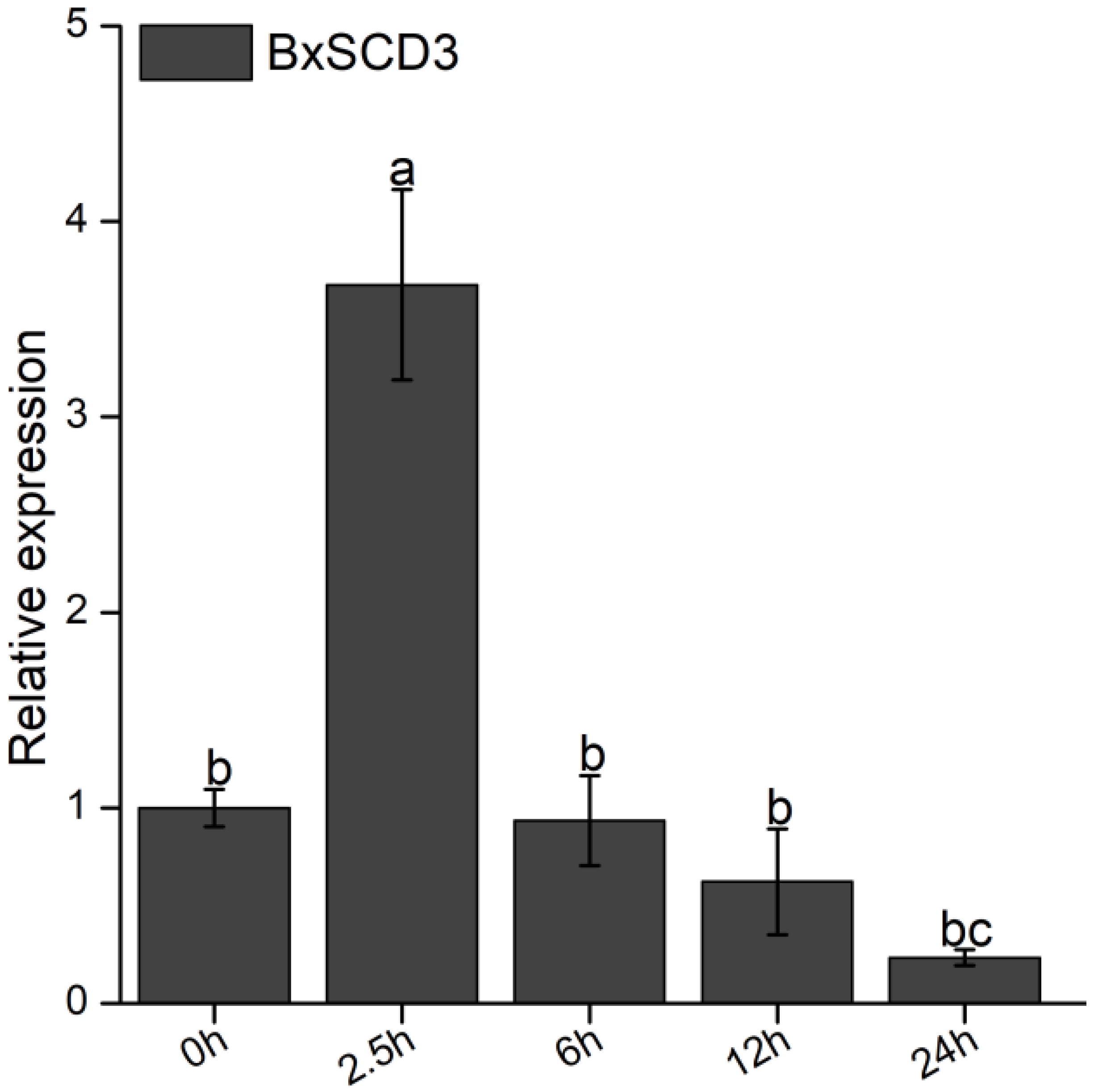
2.3. The Expression of BxSCD3 Increased Significantly at the Early Infection Stage
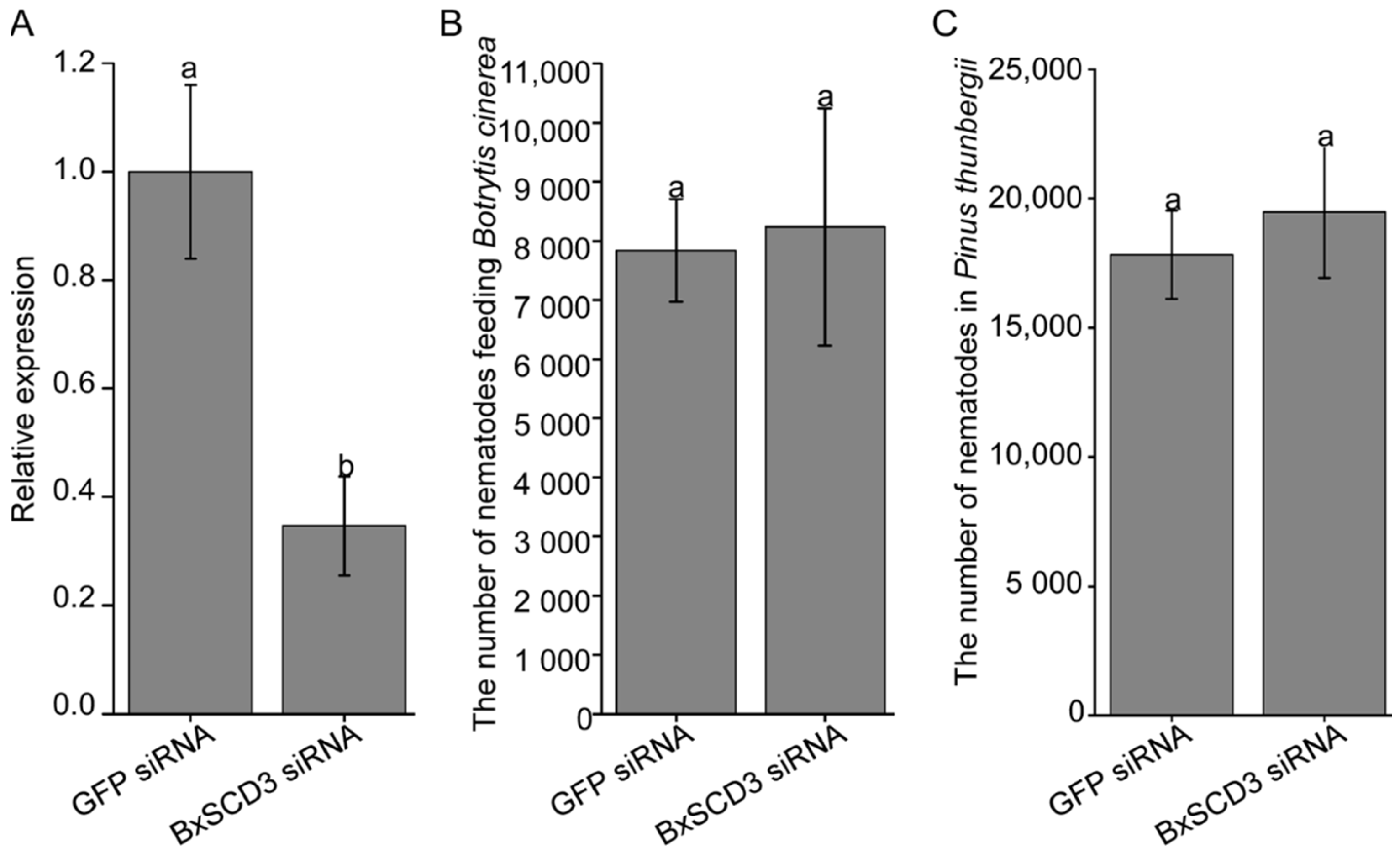
2.4. BxSCD3 Does Not Affect B. xylophilus Reproduction, Either at the Mycophagous Stage or the Phytophagous Stage
2.5. BxSCD3 Contributes to Virulence during Infection
2.6. BxSCD3 Significantly Influenced the Relative Expression Levels of Defense-Related Genes in P. thunbergii at the Early Infection Stage
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. RNA Isolation and cDNA Synthesis
4.3. Plasmid Constructs
4.4. Sequence Analysis
4.5. Agrobacterium Tumefaciens Infiltration Assays
4.6. Subcellular Localization in N. benthamiana
4.7. Western Blotting
4.8. Real-Time Quantitative PCR
4.9. In Vitro RNAi of the BxSCD3 and Inoculation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-Lopez, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.J.; Wu, X.Q.; Li, H.Y.; Wang, Y.C.; Huang, X.; Wang, Y.; Li, Y. BxCDP1 from the pine wood nematode Bursaphelenchus xylophilus is recognized as a novel molecular pattern. Mol. Plant Pathol. 2020, 21, 923–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.F.; Chen, F.M.; Xie, L.Y.; Pan, H.Y.; Ye, J.R.; Hantula, J. Genetic diversity of pine-parasitic nematodes Bursaphelenchus xylophilus and Bursaphelenchus mucronatus in China. Forest Pathol. 2017, 47, e12334. [Google Scholar] [CrossRef]
- Jones, J.T.; Li, H.; Moens, M.; Mota, M.; Kikuchi, T. Bursaphelenchus xylophilus: Opportunities in comparative genomics and molecular host-parasite interactions. Mol. Plant Pathol. 2008, 9, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.; Espada, M.; Vieira, P.; Mota, M. Pine Wilt Disease: A threat to European forestry. Eur. J. Plant Pathol. 2012, 133, 89–99. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Wang, Q.; Jing, M.; Guo, B.; Wu, J.; Wang, H.; Wang, Y.; Lin, L.; Wang, Y.; Ye, W.; et al. Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 2017, 214, 361–375. [Google Scholar] [CrossRef] [Green Version]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Jing, M.; Guo, B.; Li, H.; Yang, B.; Wang, H.; Kong, G.; Zhao, Y.; Xu, H.; Wang, Y.; Ye, W.; et al. A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant Binding immunoglobulin Proteins. Nat. Commun. 2016, 7, 11685. [Google Scholar] [CrossRef]
- Kettles, G.J.; Bayon, C.; Canning, G.; Rudd, J.J.; Kanyuka, K. Apoplastic recognition of multiple candidate effectors from the wheat pathogen Zymoseptoria tritici in the nonhost plant Nicotiana benthamiana. New Phytol. 2017, 213, 338–350. [Google Scholar] [CrossRef]
- Chen, J.; Lin, B.; Huang, Q.; Hu, L.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 2017, 13, e1006301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Chen, X.; Guo, Y.; Zhang, X.; Zhang, H.; Li, F.; Huang, G.; Meng, Y.; Shan, W. Phytophthora infestans RXLR effector PITG20303 targets a potato MKK1 protein to suppress plant immunity. New Phytol. 2021, 229, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, C.; Liu, Q.; Jian, H. Comparative analysis of pre- and post-parasitic transcriptomes and mining pioneer effectors of Heterodera avenae. Cell Biosci. 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goverse, A.; Smant, G. The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 2014, 52, 243–265. [Google Scholar] [CrossRef]
- Naalden, D.; Haegeman, A.; de Almeida-Engler, J.; Birhane, E.F.; Bauters, L.; Gheysen, G. The Meloidogyne graminicola effector Mg16820 is secreted in the apoplast and cytoplasm to suppress plant host defense responses. Mol. Plant Pathol. 2018, 19, 2416–2430. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C.N.; Perfus-Barbeoch, L.; Quentin, M.; Zhao, J.; Magliano, M.; Marteu, N.; Da Rocha, M.; Nottet, N.; Abad, P.; Favery, B. A root-knot nematode small glycine and cysteine-rich secreted effector, MiSGCR1, is involved in plant parasitism. New Phytol. 2018, 217, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Liu, S.; Liu, Q.; Niu, J.; Liu, P.; Zhao, J.; Jian, H. An ANNEXIN-like protein from the cereal cyst nematode Heterodera avenae suppresses plant defense. PLoS ONE 2015, 10, e0122256. [Google Scholar]
- Li, Z.; Yin, Z.; Fan, Y.; Xu, M.; Kang, Z.; Huang, L. Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAX-induced PCD. Front. Plant Sci. 2015, 6, 579. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.J.; Wu, X.Q.; Ding, X.L.; Ye, J.R. Comparative transcriptomic analysis of candidate effectors to explore the infection and survival strategy of Bursaphelenchus xylophilus during different interaction stages with pine trees. BMC Plant Biol. 2021, 1, 224–237. [Google Scholar] [CrossRef]
- Espada, M.; Silva, A.C.; Eves van den Akker, S.; Cock, P.J.; Mota, M.; Jones, J.T. Identification and characterization of parasitism genes from the pinewood nematode Bursaphelenchus xylophilus reveals a multilayered detoxification strategy. Mol. Plant Pathol. 2016, 17, 286–295. [Google Scholar] [CrossRef] [Green Version]
- Tsai, I.J.; Tanaka, R.; Kanzaki, N.; Akiba, M.; Yokoi, T.; Espada, M.; Jones, J.T.; Kikuchi, T. Transcriptional and morphological changes in the transition from mycetophagous to phytophagous phase in the plant-parasitic nematode Bursaphelenchus xylophilus. Mol. Plant Pathol. 2016, 17, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, L.J.; Wu, X.Q.; Ye, J.R. A Bursaphelenchus xylophilus effector, Bx-FAR-1, suppresses plant defense and affects nematode infection of pine trees. Eur. J. Plant Pathol. 2020, 153, 637–650. [Google Scholar] [CrossRef]
- Wen, T.Y.; Wu, X.Q.; Hu, L.J.; Qiu, Y.J.; Rui, L.; Zhang, Y.; Ding, X.L.; Ye, J.R. A novel pine wood nematode effector, BxSCD1, suppresses plant immunity and interacts with an ethylene-forming enzyme in pine. Mol. Plant Pathol. 2021, 22, 1399–1412. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.L.; Kale, S.D.; Wang, X.; Chen, Y.; Wang, Q.; Wang, X.; Jiang, R.H.; Arredondo, F.D.; Anderson, R.G.; Thakur, P.B.; et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell Rep. 2008, 20, 1118–1133. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Song, T.; Zhu, L.; Ye, W.; Wang, Y.; Shao, Y.; Dong, S.; Zhang, Z.; Dou, D.; Zheng, X.; et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 2015, 27, 2057–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, P.; Maier, T.R.; Eves-van den Akker, S.; Howe, D.K.; Zasada, I.; Baum, T.J.; Eisenback, J.D.; Kamo, K. Identification of candidate effector genes of Pratylenchus penetrans. Mol. Plant Pathol. 2018, 19, 1887–1907. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.Q.; Cao, B.; Tian, C.M.; Liang, Y.M. Comparative transcriptome analysis and identification of candidate effectors in two related rust species (Gymnosporangium yamadae and Gymnosporangium asiaticum). BMC Genom. 2017, 18, 651. [Google Scholar] [CrossRef] [Green Version]
- Tyler, B.M.; Tripathy, S.; Zhang, X.; Dehal, P.; Jiang, R.H.; Aerts, A.; Arredondo, F.D.; Baxter, L.; Bensasson, D.; Beynon, J.L. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 2006, 313, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Han, C.; Ferreira, A.O.; Yu, X.; Ye, W.; Tripathy, S. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 2011, 23, 2064–2086. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ni, H.; Du, X.; Wang, S.; Ma, X.W.; Nurnberger, T.; Guo, H.S.; Hua, C. The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 2017, 215, 368–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Li, H.; Wang, L.; Xie, S.; Zhang, Y.; Kang, R.; Zhang, M.; Zhang, P.; Li, Y.; Hu, Y.; et al. A novel effector, CsSp1, from Bipolaris sorokiniana, is essential for colonization in wheat and is also involved in triggering host immunity. Mol. Plant Pathol. 2022, 23, 218–236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, L.; Liu, Q.; Liu, P.; Li, S.; Yang, D.; Chen, Y.; Pagnotta, S.; Favery, B.; Abad, P.; et al. A MIF-like effector suppresses plant immunity and facilitates nematode parasitism by interacting with plant annexins. J. Exp. Bot. 2019, 70, 5943–5958. [Google Scholar] [CrossRef]
- Ohtsubo, N.; Mitsuhara, I.; Koga, M.; Seo, S.; Ohashi, Y. Ethylene promotes the necrotic lesion formation and basic PR gene expression in TMV-infected tobacco. Plant Cell Physiol. 1999, 40, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.N.; Wu, X.Q.; Ye, J.R.; Xue, Q. Identification of autophagy in the pine wood nematode Bursaphelenchus xylophilus and the molecular characterization and functional analysis of two novel autophagy-related genes, BxATG1 and BxATG8. Int. J. Mol. Sci. 2016, 17, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Q.; Wu, X.Q.; Zhang, W.J.; Deng, L.N.; Wu, M.M. Cathepsin L-like cysteine proteinase genes are associated with the development and pathogenicity of pine wood nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2019, 20, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.L.; Ye, J.R.; Lin, S.X.; Wu, X.Q.; Li, D.; Nian, B. Deciphering the molecular variations of pine wood nematode Bursaphelenchus xylophilus with different virulence. PLoS ONE 2016, 11, e0156040. [Google Scholar] [CrossRef] [Green Version]
- Thomas, N.P.; Soren, B.; von Gunnar, H.; Henrik, N. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; Volume 6, pp. 175–182. [Google Scholar]
- Yin, W.; Dong, S.; Zhai, L.; Lin, Y.; Zheng, X.; Wang, Y. The Phytophthora sojae Avr1d gene encodes an RxLR-dEER effector with presence and absence polymorphisms among pathogen strains. Mol. Plant-Microbe Interact. 2013, 26, 958–968. [Google Scholar] [CrossRef] [Green Version]
- Hirao, T.; Fukatsu, E.; Watanabe, A. Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization. BMC Plant Biol. 2012, 12, 13. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.J.; Wu, X.Q.; Li, H.Y.; Zhao, Q.; Wang, Y.C.; Ye, J.R. An effector, BxSapB1, induces cell death and contributes to virulence in the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant-Microbe Interact. 2019, 32, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.Z.; Wu, X.Q.; Ye, J.R.; Zhang, S.N.; Wang, C. NOS-likemediated nitric oxide is involved in Pinus thunbergii response to the invasion of Bursaphelenchus xylophilus. Plant Cell Rep. 2012, 31, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Rempel, C.B.; Hall, R. Comparison of disease measures for assessing resistance in canola (Brassica napus) to blackleg (Leptosphaeria maculans). Can. J. Bot. 1996, 74, 1930–1936. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.-J.; Wu, X.-Q.; Wen, T.-Y.; Qiu, Y.-J.; Rui, L.; Zhang, Y.; Ye, J.-R. A Bursaphelenchus xylophilus Effector, BxSCD3, Suppresses Plant Defense and Contributes to Virulence. Int. J. Mol. Sci. 2022, 23, 6417. https://doi.org/10.3390/ijms23126417
Hu L-J, Wu X-Q, Wen T-Y, Qiu Y-J, Rui L, Zhang Y, Ye J-R. A Bursaphelenchus xylophilus Effector, BxSCD3, Suppresses Plant Defense and Contributes to Virulence. International Journal of Molecular Sciences. 2022; 23(12):6417. https://doi.org/10.3390/ijms23126417
Chicago/Turabian StyleHu, Long-Jiao, Xiao-Qin Wu, Tong-Yue Wen, Yi-Jun Qiu, Lin Rui, Yan Zhang, and Jian-Ren Ye. 2022. "A Bursaphelenchus xylophilus Effector, BxSCD3, Suppresses Plant Defense and Contributes to Virulence" International Journal of Molecular Sciences 23, no. 12: 6417. https://doi.org/10.3390/ijms23126417
APA StyleHu, L.-J., Wu, X.-Q., Wen, T.-Y., Qiu, Y.-J., Rui, L., Zhang, Y., & Ye, J.-R. (2022). A Bursaphelenchus xylophilus Effector, BxSCD3, Suppresses Plant Defense and Contributes to Virulence. International Journal of Molecular Sciences, 23(12), 6417. https://doi.org/10.3390/ijms23126417





