Physiological, Agronomical, and Proteomic Studies Reveal Crucial Players in Rice Nitrogen Use Efficiency under Low Nitrogen Supply
Abstract
1. Introduction
2. Results
2.1. Selection of Two Contrasting Rice Cultivars
2.2. Analysis of Variance of the Stage-Specific Experiment at Low-N and Optimum-N
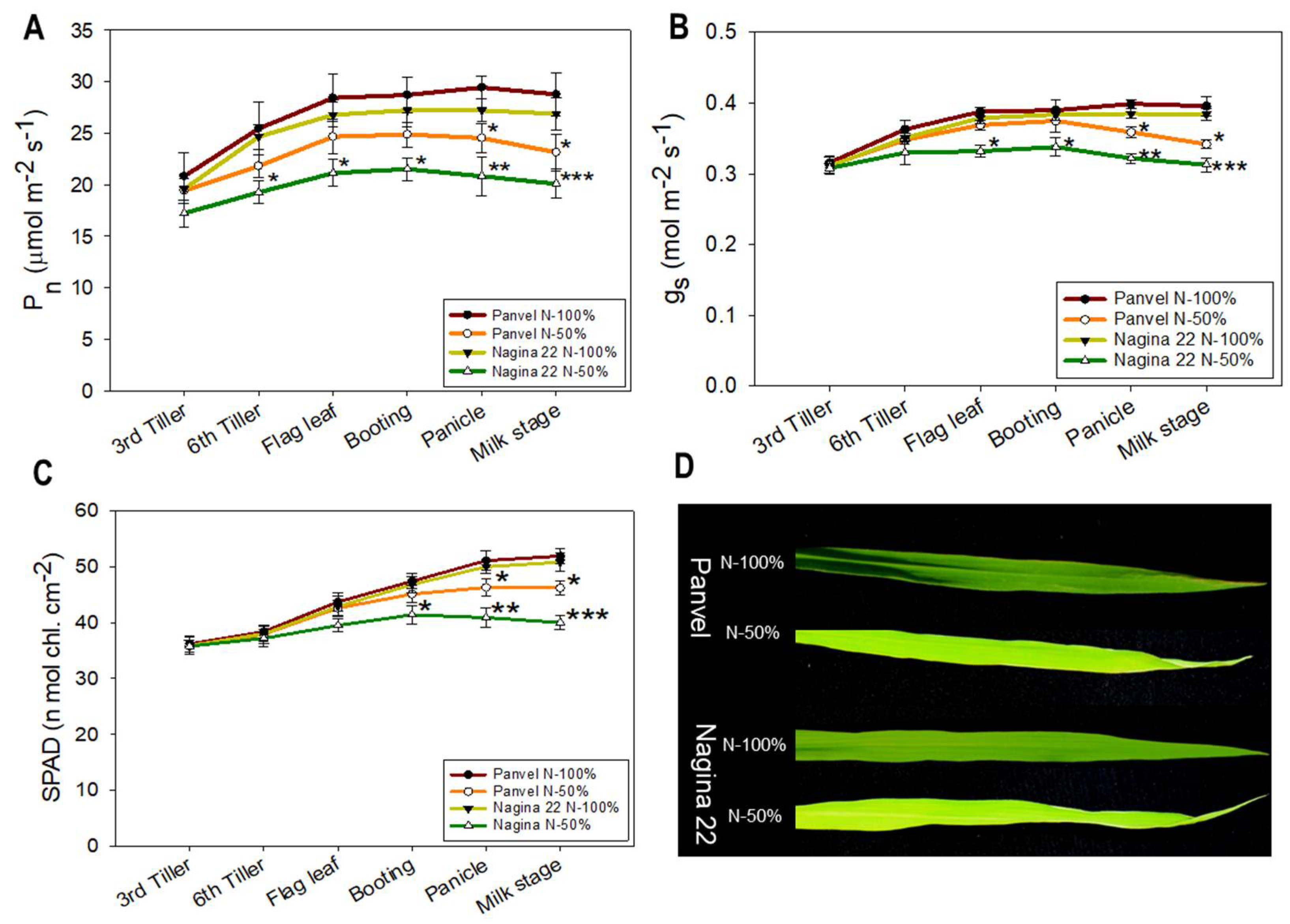
2.3. Panvel and Nagina 22 Show Physiological Differences at Low N and Optimum N Supply
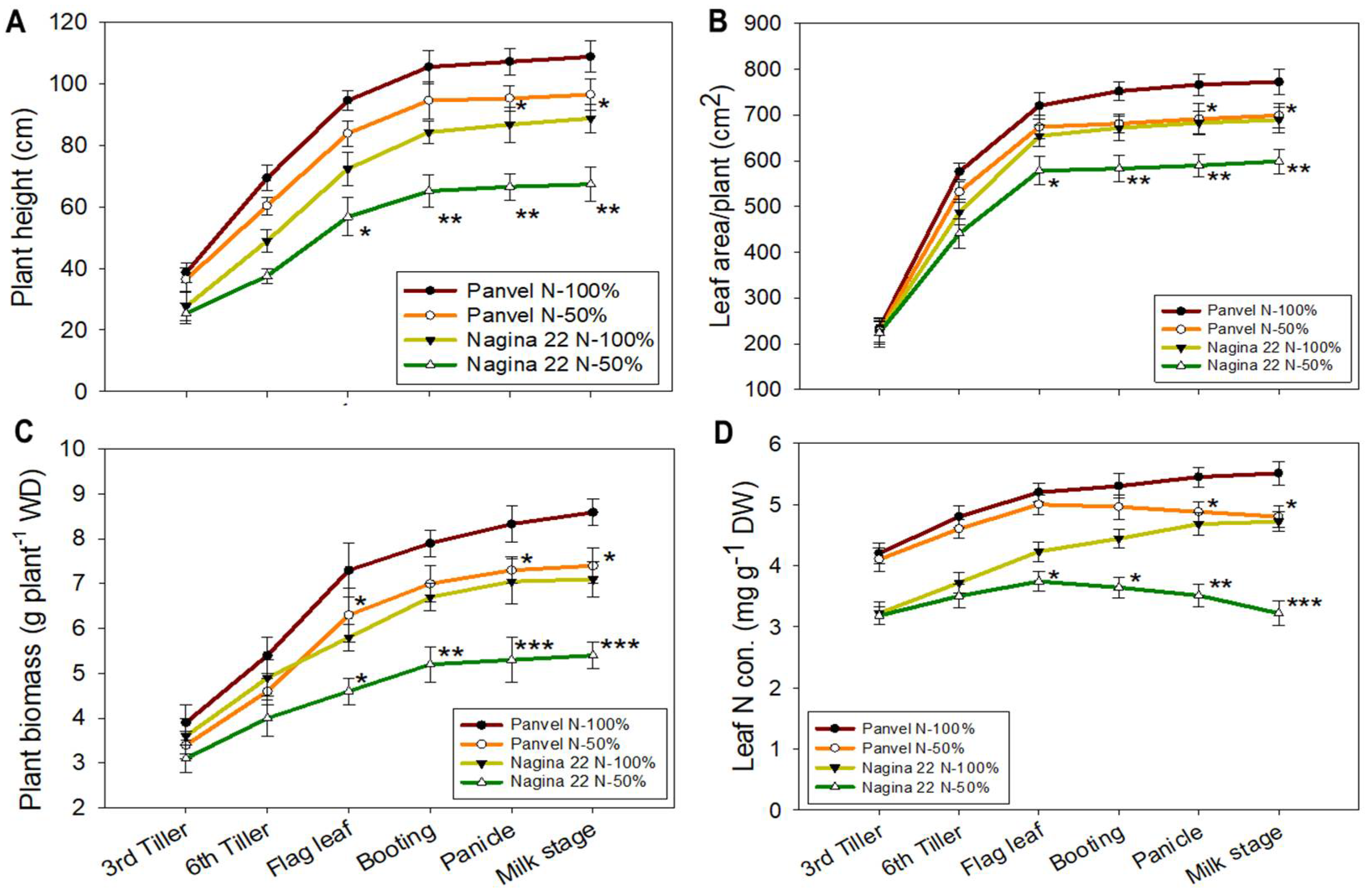
2.4. Plant Growth and LNC Vary between Rice Cultivars at a Low-N Supply
2.5. Rubisco Activity and Soluble Protein Differ between Panvel and Nagina 22
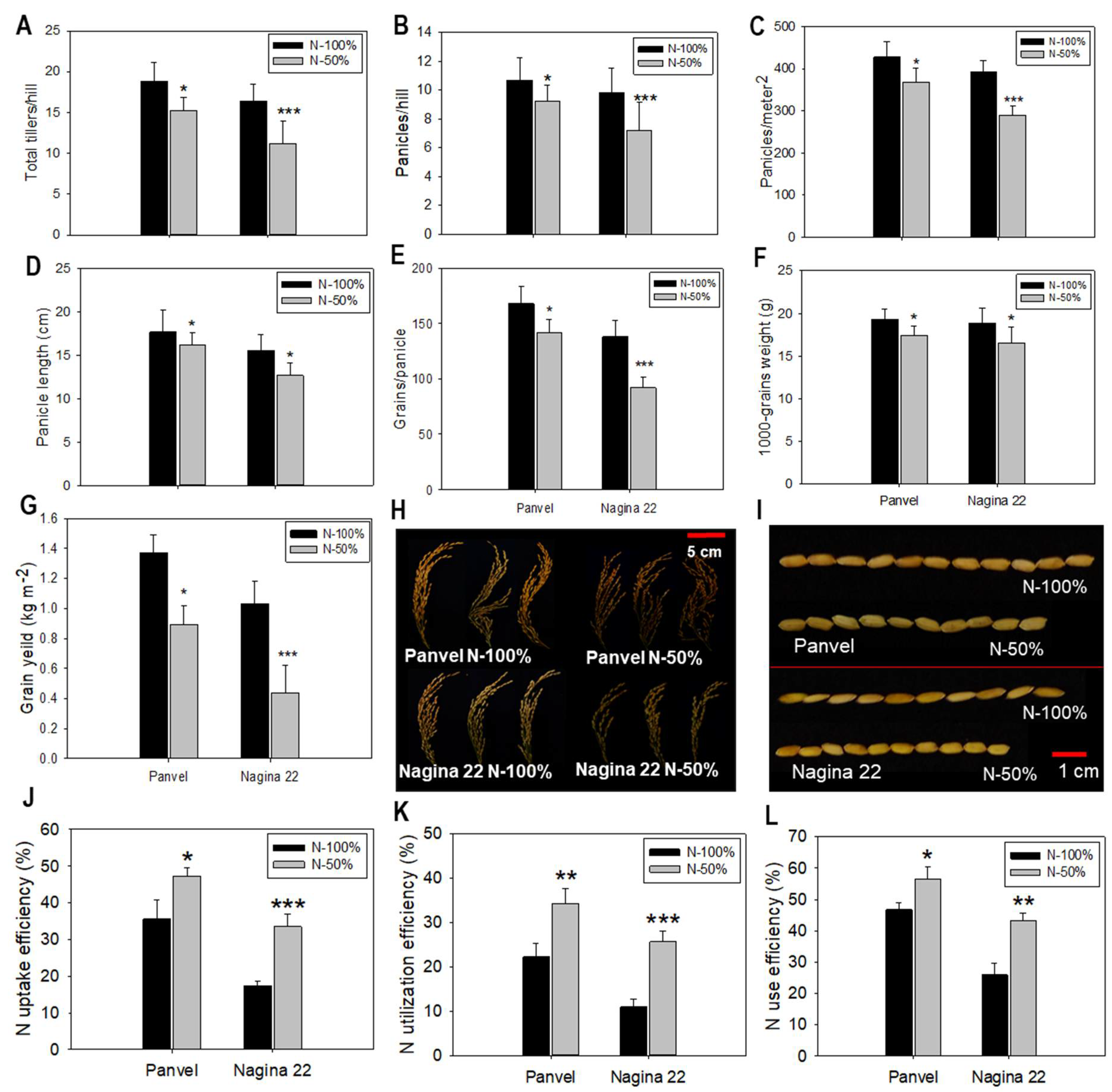
2.6. Agronomic Parameters and N Use Efficiency Are Enhanced in Panvel
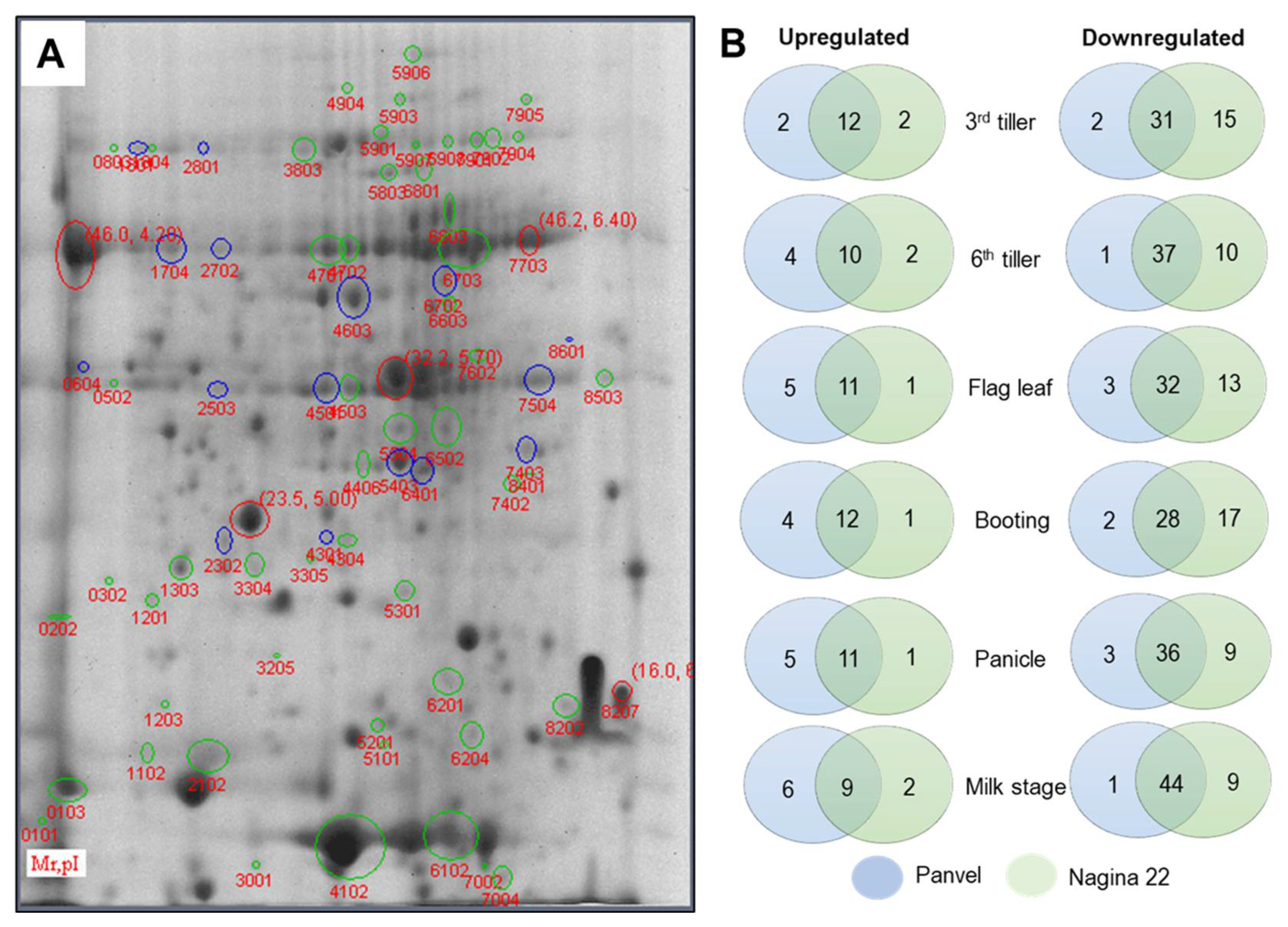
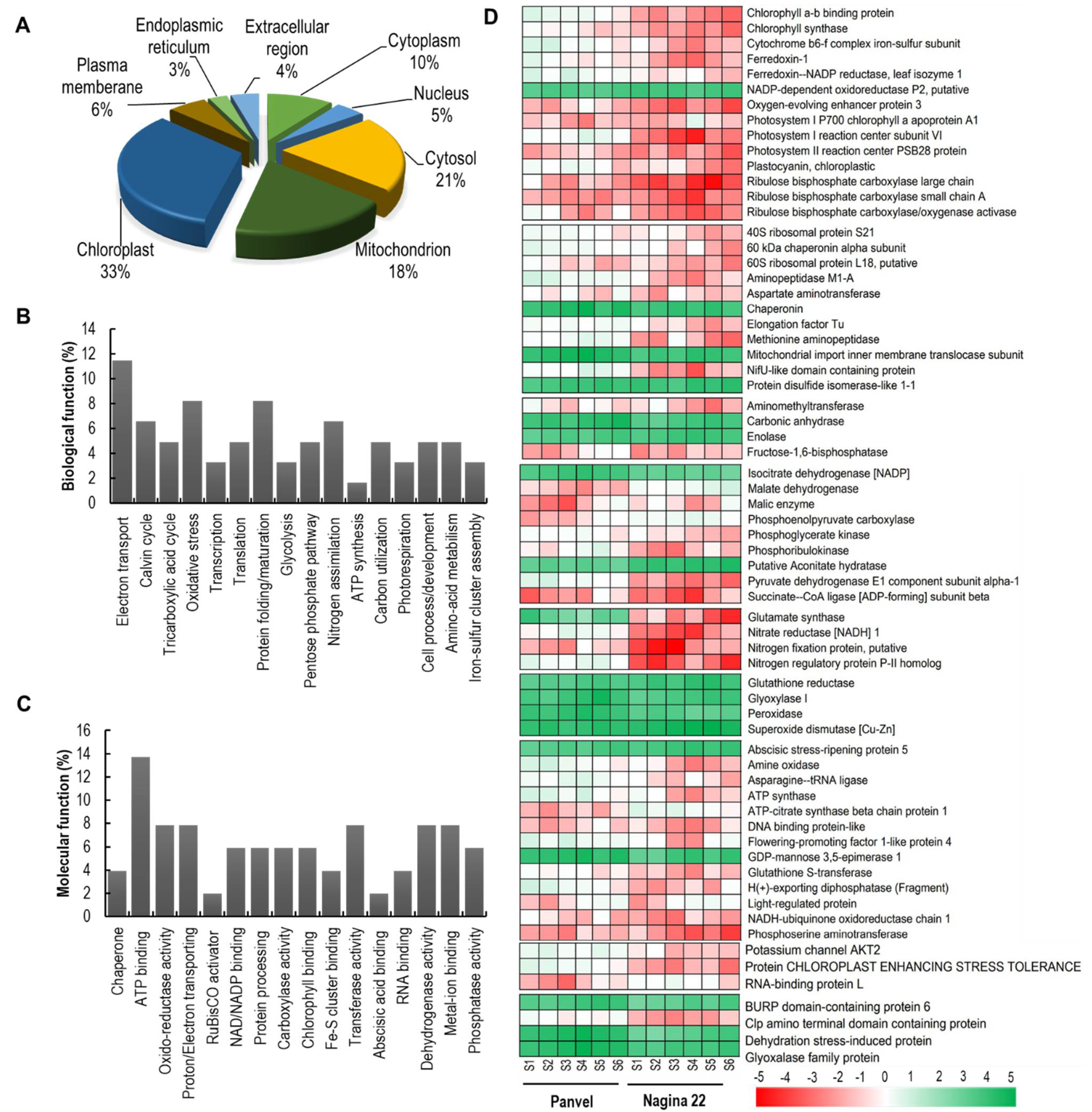
2.7. Differentially Expressed Proteins in Panvel and Nagina 22 Rice Cultivars
2.8. Spatial Distribution and Cellular and Molecular Functions of DEPs
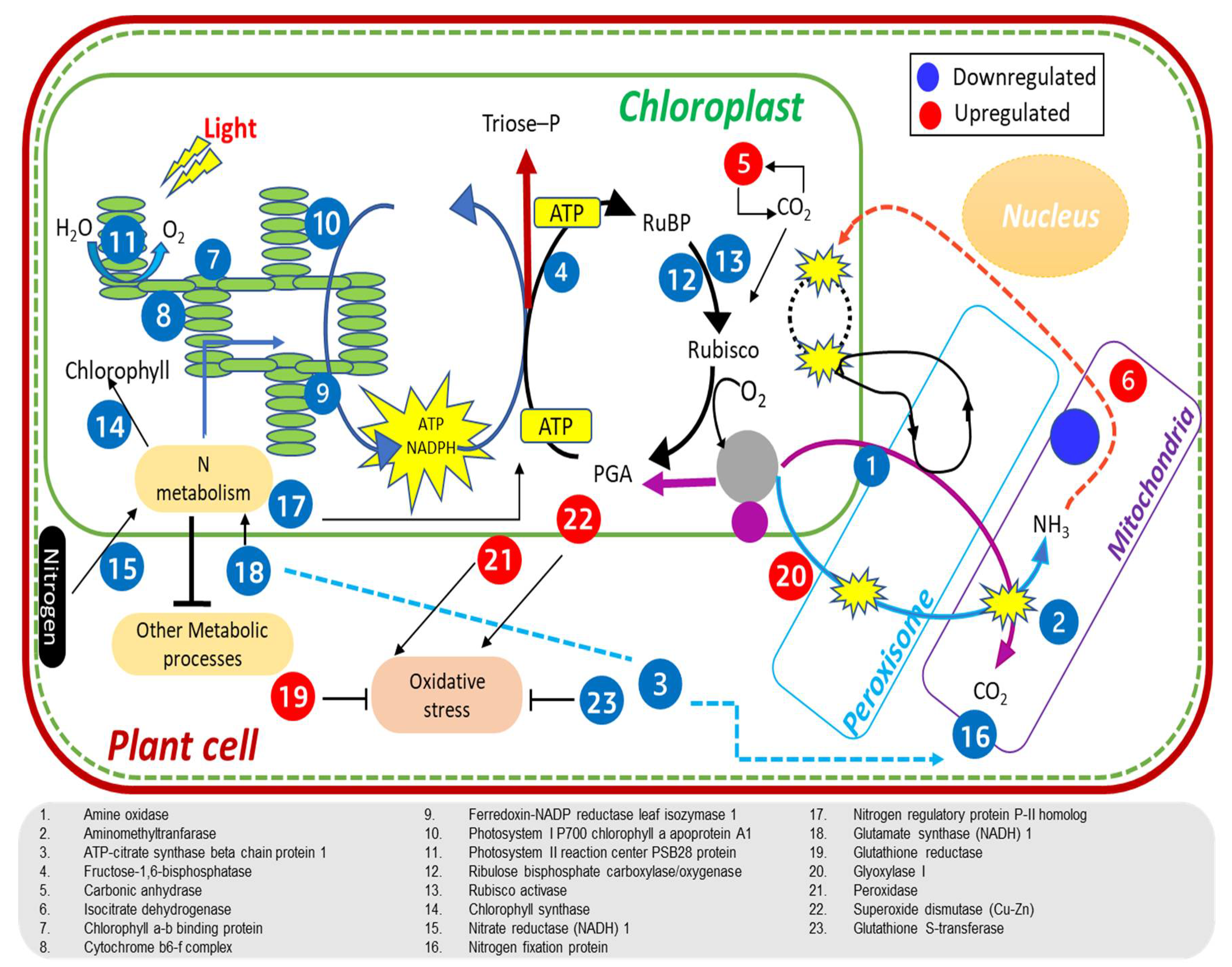
2.9. Proteins Involved in Photosynthesis and Carbon Assimilation Are Downregulated in Nagina 22
2.10. Variation in Differential Expression of Protein Turnover, N Metabolism, and Stress Related Proteins
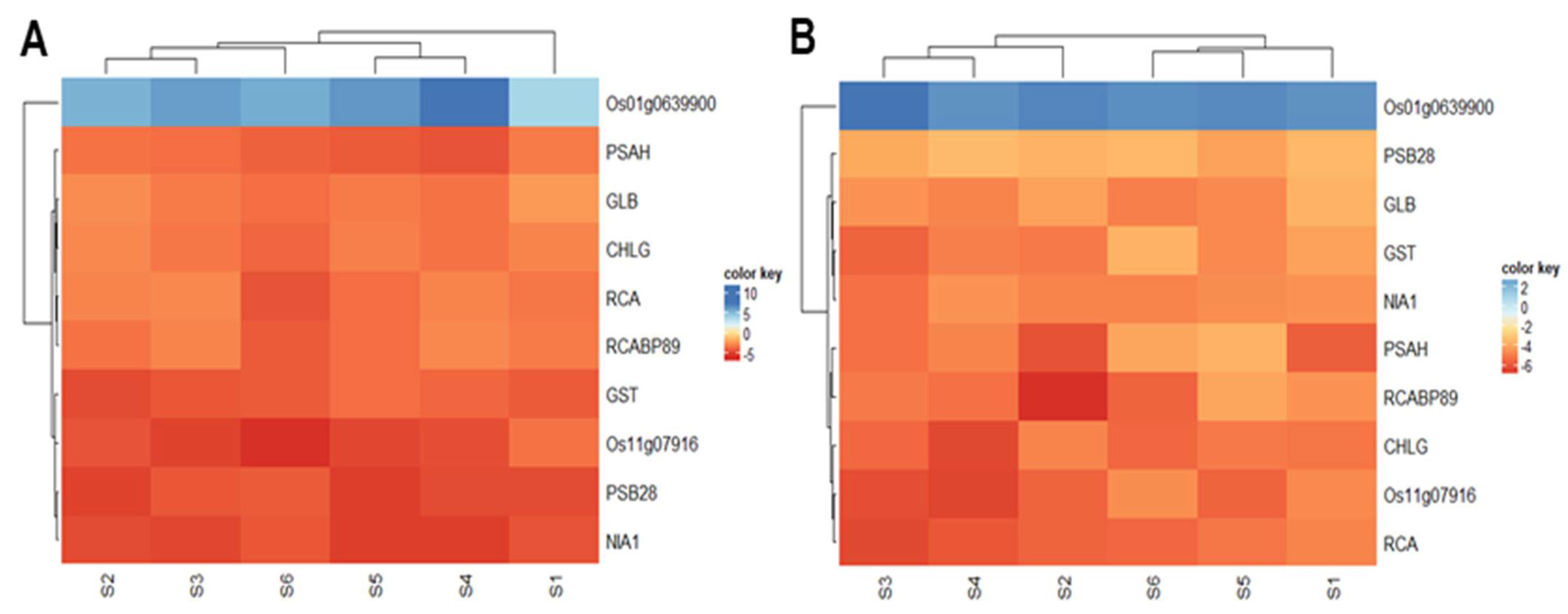
2.11. Validation of Gene Expression of Some Proteins by Quantitative Real Time-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of Photosynthesis and Chlorophyll Fluorescence of PSII
4.3. Estimation of Rubisco Activity and Soluble Protein Content of Leaf
4.4. Analysis of Growth and Yield Parameters
4.5. Analysis of Leaf NPS Contents and N Use Efficiency
4.6. Proteome Analysis
4.7. Quantitative Real-Time PCR Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Bouain, N.; Krouk, G.; Lacombe, B.; Rouached, H. Getting to the root of plant mineral nutrition: Combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci. 2019, 24, 542–552. [Google Scholar] [CrossRef]
- Swarbreck, S.M.; Wang, M.; Wang, Y.; Kindred, D.; Sylvester-Bradley, R.; Shi, W.; Bentley, A.R.; Griffiths, H. A Roadmap for Lowering Crop Nitrogen Requirement. Trends Plant Sci. 2019, 24, 892–904. [Google Scholar] [CrossRef]
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating targets for sustainable intensification. Bioscience 2017, 67, 386–391. [Google Scholar] [CrossRef]
- Wang, X. Managing Land Carrying Capacity: Key to Achieving Sustainable Production Systems for Food Security. Land 2022, 11, 484. [Google Scholar] [CrossRef]
- Glass, A.D. Nitrogen use efficiency of crop plants: Physiological constraints upon nitrogen absorption. CRC Crit. Rev. Plant Sci. 2003, 22, 453–470. [Google Scholar] [CrossRef]
- Gutiérrez, R.A. Systems biology for enhanced plant nitrogen nutrition. Science 2012, 336, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Thangarajan, R.; Bolan, N.S.; Kunhikrishnan, A.; Wijesekara, H.; Xu, Y.; Tsang, D.C.W.; Song, H.; Ok, Y.S.; Hou, D. The potential value of biochar in the mitigation of gaseous emission of nitrogen. Sci. Total Environ. 2018, 612, 257–268. [Google Scholar] [CrossRef]
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004, 9, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Hawkesford, M.J. Genetic variation in traits for nitrogen use efficiency in wheat. J. Exp. Bot. 2017, 68, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron. J. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Cañas, R.A.; Quilleré, I.; Gallais, A.; Hirel, B. Can genetic variability for nitrogen metabolism in the developing ear of maize be exploited to improve yield? New Phytol. 2012, 194, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E. Storage proteins of vegetative plant tissues. Annu. Rev. Plant Physiol. Plant Mol. 1994, 45, 303–322. [Google Scholar] [CrossRef]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Diaz, C.; Lemaître, T.; Christ, A.; Azzopardi, M.; Kato, Y.; Sato, F.; MorotGaudry, J.F.; Le Dily, F.; Masclaux-Daubresse, C. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 2008, 147, 1437–1449. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Chardon, F. Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 2131–2142. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.H.; Yang, J.J.; Liu, Y.D.; Shen, F.F. Protein degradation and nitrogen remobilization during leaf senescence. J. Plant Biol. 2008, 51, 11–19. [Google Scholar] [CrossRef]
- Kong, L.; Wang, F.; Zhang, R.; Feng, B.; Si, J.; Li, S.; Zhang, B. High nitrogen rate inhibits proteolysis and decreases the export of leaf pre-stored proteins to grains in wheat (Triticum aestivum). Int. J. Agric. Biol. 2012, 14, 1009–1013. [Google Scholar]
- Carmo-Silva, E.; Andralojc, P.J.; Scales, J.C.; Driever, S.M.; Mead, A.; Lawson, T.; Raines, C.A.; Parry, M.A.J. Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J. Exp. Bot. 2017, 68, 3473–3486. [Google Scholar] [CrossRef] [PubMed]
- Barbottin, A.; Lecomte, C.; Bouchard, C.; Jeuffroy, M.H. Nitrogen remobilization during grain filling in wheat: Genotypic and environmental effects. Crop Sci. 2005, 45, 1141–1150. [Google Scholar] [CrossRef]
- Gaju, O.; Allard, V.; Martre, P.; Le Gouis, J.; Moreau, D.; Bogard, M.; Hubbart, S.; Foulkes, M.J. Nitrogen partitioning and remobilization in relation to leaf senescence, grain yield and grain nitrogen concentration in wheat cultivars. Field Crops Res. 2014, 155, 213–223. [Google Scholar] [CrossRef]
- Idowu, O.; Wang, Y.; Homma, K.; Nakazaki, T.; Xu, Z.; Shiraiwa, T. Interaction of erect panicle genotype and nitrogen fertilizer application on the source-sink ratio and nitrogen use efficiency in rice. Field Crops Res. 2022, 278, 108430. [Google Scholar] [CrossRef]
- Wang, L.; Czedik-Eysenberg, A.; Mertz, R.A.; Si, Y.; Tohge, T.; Nunes-Nesi, A.; Arrivault, S.; Dedow, L.K.; Bryant, D.W.; Zhou, W.; et al. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 2014, 32, 1158–1165. [Google Scholar] [CrossRef]
- Bassi, D.; Menossi, M.; Mattiello, L. Nitrogen supply influences photosynthesis establishment along the sugarcane leaf. Sci. Rep. 2018, 8, 2327. [Google Scholar] [CrossRef]
- Evans, J.R.; Clarke, V.C. The nitrogen cost of photosynthesis. J. Exp. Bot. 2018, 70, 7–15. [Google Scholar] [CrossRef]
- Allison, J.; Williams, H.; Pammenter, N. Effect of specific leaf nitrogen content on photosynthesis of sugarcane. Ann. Appl. Biol. 1997, 131, 339–350. [Google Scholar] [CrossRef]
- Tendler, A.; Wolf, B.C.; Tiwari, V.; Alon, U.; Danon, A. Fold-change response of photosynthesis to step increases of light level. iScience 2018, 8, 126–137. [Google Scholar] [CrossRef]
- Yin, X.; Schapendonk, A.H.; Struik, P.C. Exploring the optimum nitrogen partitioning to predict the acclimation of C3 leaf photosynthesis to varying growth conditions. J. Exp. Bot. 2018, 70, 2435–2447. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 1–11. [Google Scholar] [CrossRef]
- Tantray, A.Y.; Bashir, S.S.; Ahmad, A. Low nitrogen stress regulates chlorophyll fluorescence in coordination with photosynthesis and Rubisco efficiency of rice. Physiol. Mol. Biol. Plants 2020, 26, 83–94. [Google Scholar] [CrossRef]
- Hubbart, S.; Peng, S.; Horton, P.; Chen, Y.; Murchie, E.H. Trends in leaf photosynthesis in historical rice varieties developed in the Philippines since 1966. J. Exp. Bot. 2007, 58, 3429–3438. [Google Scholar] [CrossRef]
- Meng, X.; Wang, X.; Zhang, Z.; Xiong, S.; Wei, Y.; Guo, J.; Zhang, J.; Wang, L.; Ma, X.; Tegeder, M. Transcriptomic, proteomic, and physiological studies reveal key players in wheat nitrogen use efficiency under both high and low nitrogen supply. J. Exp. Bot. 2021, 72, 4435–4456. [Google Scholar] [CrossRef]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef]
- Caputo, C.; Barneix, A.J. Export of amino acids to the phloem in relation to N supply in wheat. Physiol. Plant. 1997, 101, 853–860. [Google Scholar] [CrossRef]
- Shi, W.M.; Xu, W.F.; Li, S.M.; Zhao, X.Q.; Dong, G.Q. Responses of two rice cultivars differing in seedling-stage nitrogen use efficiency to growth under low-nitrogen conditions. Plant Soil. 2010, 326, 291–302. [Google Scholar] [CrossRef]
- Zörb, C.; Ludewig, U.; Hawkesford, M.J. Perspective on wheat yield and quality with reduced nitrogen supply. Trends Plant Sci. 2018, 23, 1029–1037. [Google Scholar] [CrossRef]
- Simpson, R.J.; Lambers, H.; Dalling, M.J. Nitrogen redistribution during grain growth in wheat (Triticum aestivum L.): IV. Development of a quantitative model of the translocation of nitrogen to the grain. Plant Physiol. 1983, 71, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; McCaig, T.N.; DePauw, R.M.; Clarke, J.M. Flag leaf physiological traits in two high-yielding Canada Western Red Spring wheat cultivars. Can. J. Plant Sci. 2008, 88, 35–42. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, M.J.; Slafer, G.A.; Berry, P.; Parry, M.A.; Snape, J.W.; Angus, W.J. Raising yield potential in wheat. J. Exp. Bot. 2009, 60, 1899–1918. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Wang, F.; Yuan, J.; Zhang, Y.; Zhao, Z.; Abulizi, B.; Wen, X.; Kang, M.; Tang, F. Screening and comprehensive evaluation of rice (Oryza sativa L. subsp. japonica Kato) germplasm resources for nitrogen efficiency in Xinjiang, China. Plant Genet. Resour. 2020, 18, 179–189. [Google Scholar] [CrossRef]
- Zhu, K.; Zhou, Q.; Shen, Y.; Yan, J.; Xu, Y.; Wang, Z.; Yang, J. Agronomic and physiological performance of an indica–japonica rice variety with a high yield and high nitrogen use efficiency. Crop Sci. 2020, 60, 1556–1568. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Chu, C. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- Hao, Q.N.; Zhou, X.A.; Sha, A.H.; Wang, C.; Zhou, R.; Chen, S.L. Identification of genes associated with nitrogen-use efficiency by genome wide transcriptional analysis of two soybean genotypes. BMC Genom. 2011, 12, 525. [Google Scholar] [CrossRef]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genom. 2014, 15, 179. [Google Scholar] [CrossRef]
- Zamboni, A.; Astolfi, S.; Zuchi, S.; Pii, Y.; Guardini, K.; Tononi, P.; Varanini, Z. Nitrate induction triggers different transcriptional changes in a high and a low nitrogen use efficiency maize inbred line. J. Integr. Plant Biol. 2014, 56, 1080–1094. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016, 16, 30. [Google Scholar] [CrossRef]
- Sinha, S.K.; Sevanthi, A.M.; Chaudhary, S.; Tyagi, P.; Venkadesan, S.; Rani, M.; Mandal, P.K. Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic N starvation reveals differences in chloroplast and starch metabolism-related genes. Genes 2018, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, G.; Yang, N.; White, P.J.; Ye, X.; Cai, H.; Lu, J.; Shi, L.; Xu, F. Comparative genome and transcriptome analysis unravels key factors of nitrogen use efficiency in Brassica napus L. Plant Cell Environ. 2020, 43, 712–731. [Google Scholar] [CrossRef] [PubMed]
- Passioura, J.B. Scaling up: The essence of effective agricultural research. Funct. Plant Biol. 2010, 37, 585–591. [Google Scholar] [CrossRef]
- Bacilio, M.; Moreno, M.; Lopez-Aguilar, D.R.; Bashan, Y. Scaling from the growth chamber to the greenhouse to the field: Demonstration of diminishing effects of mitigation of salinity in peppers inoculated with plant growth-promoting bacterium and humic acids. Appl. Soil Ecol. 2017, 119, 327–338. [Google Scholar] [CrossRef]
- Bennett, E.; Roberts, J.A.; Wagstaff, C. Manipulating resource allocation in plants. J. Exp. Bot. 2012, 63, 3391–3400. [Google Scholar] [CrossRef] [PubMed]
- Perchlik, M.; Tegeder, M. Leaf amino acid supply affects photosynthetic and plant nitrogen use efficiency under nitrogen stress. Plant Physiol. 2018, 178, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.I.; Allen, D.J.; Ortiz-Lopez, A.; Hernández, G. Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 2001, 52, 1071–1081. [Google Scholar] [CrossRef]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef]
- Pan, S.; Liu, H.; Mo, Z.; Patterson, B.; Duan, M.; Tian, H.; Hu, S.; Tang, X. Effects of nitrogen and shading on root morphologies, nutrient accumulation, and photosynthetic parameters in different rice genotypes. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Peng, S.; Buresh, R.J.; Huang, J.; Zhong, X.; Zou, Y.; Yang, J.; Wang, G.; Liu, Y.; Hu, R.; Tang, Q.; et al. Improving nitrogen fertilization in rice by site specific N management. A review. Agron. Sustain. Dev. 2010, 30, 649–656. [Google Scholar] [CrossRef]
- Yang, H.; Ge, C.S.; Ying, W.; Yang, J.P.; Li, J.W.; He, J.J. Effect of shading on leaf SPAD values and the characteristics of photosynthesis and morphology of rice canopy. Plant Nutr. Fertil. Sci. 2014, 20, 580–587. [Google Scholar]
- Brooks, M.D.; Niyogi, K.K. Use of a pulse-amplitude modulated chlorophyll fluorometer to study the efficiency of photosynthesis in Arabidopsis plants. In Chloroplast Research in Arabidopsis; Humana Press: Totowa, NJ, USA, 2011; pp. 299–310. [Google Scholar]
- Müller, P.; Li, X.P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 2016, 354, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.A.; Ceretta, C.A.; Soriani, H.H.; Tiecher, T.L.; Soares, C.R.; Rossato, L.V.; Nicoloso, F.T.; Brunetto, G.; Paranhos, J.T.; Cornejo, P. Rhizophagus clarus and phosphate alter the physiological responses of Crotalaria juncea cultivated in soil with a high Cu level. Appl. Soil Ecol. 2015, 91, 37–47. [Google Scholar] [CrossRef]
- Govindjee, U. Non-Photochemical Quenching and Energy Dissipation in Plants, Algae and Cyanobacteria; Demmig-Adams, B., Garab, G., Adams, W., III, Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 40, pp. 297–314. [Google Scholar]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Ann. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Driever, S.M.; Lawson, T.; Andralojc, P.J.; Raines, C.A.; Parry, M.A. Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J. Exp. Bot. 2014, 65, 4959–4973. [Google Scholar] [CrossRef]
- Murata, Y. Dependence of the potential productivity and efficiency in solar energy utilization on leaf photosynthetic capacity in crop species. Jpn. J. Crop Sci. 1981, 50, 223–232. [Google Scholar] [CrossRef]
- Evans, J.R. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 1989, 78, 9–19. [Google Scholar] [CrossRef]
- Makino, A.; Sakashita, H.; Hidema, J.; Mae, T.; Ojima, K.; Osmond, B. Distinctive responses of ribulose-1,5-bisphosphate carboxylase and carbonic anhydrase in wheat leaves to nitrogen nutrition and their possible relationships to CO2 transfer resistance. Plant Physiol. 1992, 100, 1737–1743. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Ren, T.; Hussain, S.; Guo, C.; Wang, S.; Cong, R.; Li, X. Effects of nitrogen and tiller type on grain yield and physiological responses in rice. AoB Plants 2017, 9, 1–14. [Google Scholar] [CrossRef]
- Yoshida, H.; Horie, T.; Shiraiwa, T. A model explaining genotypic and environmental variation of rice spikelet number per unit area measured by cross-locational experiments in Asia. Field Crops Res. 2006, 97, 337–343. [Google Scholar] [CrossRef]
- Fageria, N.K.; Moreira, A.; Coelho, A.M. Yield and yield components of upland rice as influenced by nitrogen sources. J. Plant Nutr. 2011, 34, 361–370. [Google Scholar] [CrossRef]
- Muurinen, S.; Slafer, G.A.; Peltonen-Sainio, P. Breeding effects on nitrogen use efficiency of spring cereals under northern conditions. Crop Sci. 2006, 46, 561–568. [Google Scholar] [CrossRef]
- Liang, Z.; Bronson, K.F.; Thorp, K.R.; Mon, J.; Badaruddin, M.; Wang, G. Cultivar and N fertilizer rate affect yield and N use efficiency in irrigated durum wheat. Crop Sci. 2014, 54, 1175–1183. [Google Scholar] [CrossRef]
- Barraclough, P.B.; Lopez-Bellido, R.; Hawkesford, M.J. Genotypic variation in the uptake, partitioning and remobilisation of nitrogen during grain-filling in wheat. Field Crops Res. 2014, 156, 242–248. [Google Scholar] [CrossRef]
- Tegeder, M.; Perchlik, M. The importance of organic nitrogen transport processes for plant productivity and nitrogen use efficiency. In Engineering Nitrogen Utilization in Crop Plants; Shrawat, A.K., Zayed, A., Lightfoot, D.A., Eds.; Springer: New York, NY, USA, 2018; pp. 233–253. [Google Scholar]
- Song, C.; Zeng, F.; Feibo, W.; Ma, W.; Zhang, G. Proteomic analysis of nitrogen stress-responsive proteins in two rice cultivars differing in N utilization efficiency. J. Integr. Omics 2011, 1, 78–87. [Google Scholar]
- Ding, C.; Chang, Z.; Wang, Y.; You, S.; Wang, S.; Ding, Y. Proteomic analysis reveals that developing leaves are more sensitive to nitrogen fertilizer than mature leaves. J. Plant Growth Regul. 2018, 37, 426–437. [Google Scholar] [CrossRef]
- Carmo-Silva, E.; Scales, J.C.; Madgwick, P.J.; Parry, M.A. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015, 38, 1817–1832. [Google Scholar] [CrossRef]
- Ermakova, M.; Lopez-Calcagno, P.E.; Raines, C.A.; Furbank, R.T.; von Caemmerer, S. Overexpression of the Rieske FeS protein of the cytochrome b6f complex increases C4 photosynthesis in Setaria viridis. Commun. Biol. 2019, 2, 314. [Google Scholar] [CrossRef]
- Kim, D.H.; Shibato, J.; Kim, D.W.; Oh, M.K.; Kim, M.K.; Shim, I.S.; Rakwal, R. Gel-based proteomics approach for detecting low nitrogen responsive proteins in cultivated rice species. Physiol. Mol. Biol. Plants 2009, 15, 31–41. [Google Scholar] [CrossRef][Green Version]
- Hakeem, K.R.; Chandna, R.; Ahmad, A.; Qureshi, M.I.; Iqbal, M. Proteomic analysis for low and high nitrogen-responsive proteins in the leaves of rice genotypes grown at three nitrogen levels. Appl. Biochem. Biotechnol. 2012, 168, 834–850. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Li, W.; Hu, D.; Shi, X.; Zhang, X.; Zhang, F.; Fu, Z.; Ding, D.; Liu, Z.; Tang, J. Biological responses and proteomic changes in maize seedlings under nitrogen deficiency. Plant Mol. Biol. Rep. 2015, 33, 490–504. [Google Scholar] [CrossRef]
- Jiang, J.; Gai, Z.; Wang, Y.; Fan, K.; Sun, L.; Wang, H.; Ding, Z. Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lea, P.J.; Miflin, B.J. Nitrogen assimilation and its relevance to crop improvement. In Nitrogen Metabolism in Plants in the Post-genomic Era; Foyer, C., Zhang, H., Eds.; Blackwell Publishing: Oxford, UK, 2011; pp. 1–40. [Google Scholar]
- Uhrig, R.G.; Ng, K.K.; Moorhead, G.B. PII in higher plants: A modern role for an ancient protein. Trends Plant Sci. 2009, 14, 505–511. [Google Scholar] [CrossRef]
- Ohashi, M.; Ishiyama, K.; Kojima, S.; Konishi, N.; Sasaki, K.; Miyao, M.; Hayakawa, T.; Yamaya, T. Outgrowth of rice tillers requires availability of glutamine in the basal portions of shoots. Rice 2018, 11, 1–11. [Google Scholar] [CrossRef]
- Martin, A.; Lee, J.; Kichey, T.; Gerentes, D.; Zivy, M.; Tatout, C.; Dubois, F.; Balliau, T.; Valot, B.; Davanture, M.; et al. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006, 18, 3252–3274. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Popova, A.A.; Shestibratov, K.A. Genetic engineering and genome editing for improving nitrogen use efficiency in plants. Cells 2021, 10, 3303. [Google Scholar] [CrossRef]
- Gao, Y.; de Bang, T.C.; Schjoerring, J.K. Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 2019, 17, 1209–1221. [Google Scholar] [CrossRef]
- Kudlicki, W.; Coffman, A.; Kramer, G.; Hardesty, B. Renaturation of rhodanese by translational elongation factor (EF) Tu: Protein refolding by EFTu flexing. J. Biol. Chem. 1997, 272, 32206–32210. [Google Scholar] [CrossRef]
- Hummel, M.; Cordewener, J.H.; de Groot, J.C.; Smeekens, S.; America, A.H.; Hanson, J. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MS E proteomics. Proteomics 2012, 12, 1024–1038. [Google Scholar] [CrossRef]
- Graifer, D.; Karpova, G. Roles of ribosomal proteins in the functioning of translational machinery of eukaryotes. Biochimie 2015, 109, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Higuchi, M.; Kondou, Y.; Ichikawa, T.; Iwabuchi, M.; Hirochika, H.; Matsui, M.; Oda, K. A novel chloroplast protein, CEST induces tolerance to multiple environmental stresses and reduces photooxidative damage in transgenic Arabidopsis. J. Exp. Bot. 2011, 62, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Challabathula, D.; Analin, B.; Mohanan, A.; Bakka, K. Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and-tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 2022, 268, 153583. [Google Scholar] [CrossRef] [PubMed]
- Zakari, S.A.; Asad, M.A.U.; Han, Z.; Zhao, Q.; Cheng, F. Relationship of nitrogen deficiency-induced leaf senescence with ROS generation and ABA concentration in rice flag leaves. J. Plant Growth Regul. 2020, 39, 1503–1517. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Wang, Y.; Wu, J.; Kang, K.Y.; Kim, S.T. Physiological and proteomic analysis of young rice leaves grown under nitrogen-starvation conditions. Plant Biotechnol. Rep. 2011, 5, 309–315. [Google Scholar] [CrossRef]
- Singh Shivay, Y.; Prasad, R.; Pal, M. Effect of levels and sources of sulfur on yield, sulfur and nitrogen concentration and uptake and S-use efficiency in basmati rice. Commun. Soil Sci. Plant Anal. 2014, 45, 2468–2479. [Google Scholar] [CrossRef]
- Yadav, J.; Verma, J.P.; Jaiswal, D.K.; Kumar, A. Evaluation of PGPR and different concentration of phosphorus level on plant growth, yield and nutrient content of rice (Oryza sativa). Ecol. Eng. 2014, 62, 123–128. [Google Scholar]
- Usuda, H. The activation state of ribulose 1, 5-bisphosphate carboxylase in maize leaves in dark and light. Plant Cell Physiol. 1985, 26, 1455–1463. [Google Scholar]
- Carvalho, L.C.; Esquível, M.G.; Martins, I.; Ricardo, C.P.; Amâncio, S. Monitoring the stability of Rubisco in micropropagated grapevine (Vitis vinifera L.) by two-dimensional electrophoresis. J. Plant Physiol. 2005, 162, 365–374. [Google Scholar] [CrossRef]
- Yoshida, S. Fundamentals of Rice Crop Science; International Rice Research Institute: Los Baños, Laguna, 1981. [Google Scholar]
- Mahajan, G.R.; Pandey, R.N.; Sahoo, R.N.; Gupta, V.K.; Datta, S.C.; Kumar, D. Monitoring nitrogen, phosphorus and sulphur in hybrid rice (Oryza sativa L.) using hyperspectral remote sensing. Precis. Agric. 2017, 18, 736–761. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [CrossRef]
- Tantray, A.Y.; Ali, H.M.; Ahmad, A. Analysis of proteomic profile of contrasting phosphorus responsive rice cultivars grown under phosphorus deficiency. Agronomy 2020, 10, 1028. [Google Scholar] [CrossRef]
- Koenig, T.; Menze, B.H.; Kirchner, M.; Monigatti, F.; Parker, K.C.; Patterson, T.; Steen, J.J.; Hamprecht, F.A.; Steen, H. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J. Proteome Res. 2008, 7, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2006, 34, D173–D180. [Google Scholar] [CrossRef]
- Wu, C.H.; Apweiler, R.; Bairoch, A.; Natale, D.A.; Barker, W.C.; Boeckmann, B.; Suzek, B. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J.; et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 1–12. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.J.; Kim, T.H.; Lim, P.O.; Masclaux-Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]

| Traits | Growth Stages | Panvel | Nagina 22 | ||
|---|---|---|---|---|---|
| N-100% | N-50% | N-100% | N-50% | ||
| Fv/Fm | 3rd tiller | 0.796 ± 0.036 | 0.781 ± 0.032 | 0.794 ± 0.043 | 0.735 ± 0.034 * |
| 6th tiller | 0.809 ± 0.041 | 0.787 ± 0.028 | 0.801 ± 0.029 | 0.723 ± 0.042 * | |
| Flag leaf | 0.825 ± 0.037 | 0.799 ± 0.035 | 0.807 ± 0.033 | 0.729 ± 0.038 * | |
| Booting | 0.822 ± 0.044 | 0.776 ± 0.042 * | 0.805 ± 0.061 | 0.672 ± 0.028 ** | |
| Panicle | 0.821 ± 0.041 | 0.728 ± 0.056 * | 0.803 ± 0.054 | 0.662 ± 0.035 ** | |
| Milk stage | 0.821 ± 0.035 | 0.716 ± 0.049 ** | 0.806 ± 0.048 | 0.658 ± 0.046 *** | |
| ΦPSII | 3rd tiller | 0.684 ± 0.026 | 0.671 ± 0.031 | 0.677 ± 0.026 | 0.658 ± 0.029 |
| 6th tiller | 0.754 ± 0.033 | 0.722 ± 0.035 | 0.738 ± 0.025 | 0.654 ± 0.021 ** | |
| Flag leaf | 0.762 ± 0.024 | 0.682 ± 0.032 * | 0.743 ± 0.036 | 0.632 ± 0.042 *** | |
| Booting | 0.746 ± 0.038 | 0.668 ± 0.028 * | 0.725 ± 0.031 | 0.605 ± 0.029 *** | |
| Panicle | 0.715 ± 0.041 | 0.637 ± 0.037 * | 0.692 ± 0.035 | 0.578 ± 0.044 *** | |
| Milk stage | 0.672 ± 0.036 | 0.605 ± 0.039 * | 0.664 ± 0.034 | 0.527 ± 0.037 *** | |
| ETR | 3rd tiller | 164.55 ± 11.4 | 161.43 ± 12.1 | 162.87 ± 10.3 | 158.30 ± 10.4 |
| 6th tiller | 181.39 ± 10.3 | 173.70 ± 11.7 | 177.55 ± 12.7 | 157.34 ± 11.5 * | |
| Flag leaf | 183.32 ± 09.7 | 164.09 ± 13.4 * | 178.75 ± 11.6 | 152.04 ± 11.9 ** | |
| Booting | 179.47 ± 13.5 | 160.70 ± 12.6 * | 174.42 ± 12.9 | 145.55 ± 12.1 ** | |
| Panicle | 172.01 ± 12.8 | 153.25 ± 11.4 * | 166.48 ± 13.2 | 139.05 ± 10.6 ** | |
| Milk stage | 161.67 ± 14.2 | 145.55 ± 15.3 * | 159.74 ± 14.5 | 126.78 ± 11.2 *** | |
| qP | 3rd tiller | 0.807 ± 0.024 | 0.798 ± 0.031 | 0.802 ± 0.025 | 0.779 ± 0.032 |
| 6th tiller | 0.825 ± 0.036 | 0.812 ± 0.028 | 0.819 ± 0.029 | 0.736 ± 0.027 * | |
| Flag leaf | 0.867 ± 0.027 | 0.801 ± 0.037 * | 0.855 ± 0.031 | 0.758 ± 0.025 ** | |
| Booting | 0.832 ± 0.041 | 0.763 ± 0.043 * | 0.821 ± 0.036 | 0.724 ± 0.032 ** | |
| Panicle | 0.801 ± 0.038 | 0.714 ± 0.041 ** | 0.793 ± 0.028 | 0.687 ± 0.044 *** | |
| Milk stage | 0.785 ± 0.023 | 0.678 ± 0.036 ** | 0.778 ± 0.042 | 0.583 ± 0.041 *** | |
| NPQ | 3rd tiller | 0.064 ± 0.011 | 0.071 ± 0.016 | 0.068 ± 0.014 | 0.089 ± 0.015 * |
| 6th tiller | 0.042 ± 0.013 | 0.053 ± 0.012 | 0.047 ± 0.011 | 0.073 ± 0.013 * | |
| Flag leaf | 0.023 ± 0.008 | 0.033 ± 0.010 | 0.029 ± 0.010 | 0.056 ± 0.014 * | |
| Booting | 0.046 ± 0.012 | 0.054 ± 0.013 | 0.048 ± 0.013 | 0.073 ± 0.016 ** | |
| Panicle | 0.073 ± 0.019 | 0.091 ± 0.017 * | 0.075 ± 0.020 | 0.115 ± 0.022 *** | |
| Milk stage | 0.096 ± 0.014 | 0.125 ± 0.023 ** | 0.101 ± 0.019 | 0.168 ± 0.034 *** | |
| Traits | Growth Stages | Panvel | Nagina 22 | ||
|---|---|---|---|---|---|
| N-100% | N-50% | N-100% | N-50% | ||
| Rubisco enzyme activity (n mol−1 g−1 FW) | 3rd tiller | 215.9 ± 12.2 | 203.3 ± 14.3 | 208.8 ± 11.6 | 197.5 ± 13.8 |
| 6th tiller | 224.2 ± 11.9 | 207.2 ± 11.5 | 218.3 ± 13.2 | 199.9± 12.4 * | |
| Flag leaf | 234.6 ± 13.5 | 211.2 ± 11.6 * | 229.3 ± 12.3 | 202.3 ± 14.6 * | |
| Booting | 235.4 ± 09.8 | 209.4 ± 11.3 * | 230.8 ± 13.6 | 198.5 ± 12.8 ** | |
| Panicle | 235.7 ± 14.1 | 204.9 ± 11.5 ** | 228.2 ± 12.2 | 186.4 ± 11.6 *** | |
| Milk stage | 234.9 ± 10.2 | 201.3 ± 13.7 ** | 225.7 ± 11.2 | 175.2 ± 12.2 *** | |
| Rubisco protein content (mg m−2 FW) | 3rd tiller | 0.139 ± 0.025 | 0.134 ± 0.031 | 0.132 ± 0.027 | 0.125 ± 0.026 |
| 6th tiller | 0.152 ± 0.032 | 0.141 ± 0.028 | 0.143 ± 0.021 | 0.130 ± 0.023 * | |
| Flag leaf | 0.163 ± 0.029 | 0.148 ± 0.025 * | 0.156 ± 0.027 | 0.138 ± 0.028 * | |
| Booting | 0.167 ± 0.033 | 0.154 ± 0.027 | 0.162 ± 0.031 | 0.141 ± 0.029 ** | |
| Panicle | 0.171 ± 0.022 | 0.155 ± 0.026 * | 0.165 ± 0.024 | 0.140 ± 0.028 ** | |
| Milk stage | 0.174 ± 0.027 | 0.154 ± 0.023 * | 0.169 ± 0.025 | 0.142 ± 0.021 ** | |
| Soluble protein content (mg g−2 FW) | 3rd tiller | 29.31 ± 3.23 | 23.82 ± 2.42 | 24.56 ± 3.31 | 20.13 ± 4.11 |
| 6th tiller | 32.49 ± 4.21 | 25.35 ± 3.26 | 28.78 ± 3.45 | 24.89 ± 3.27 * | |
| Flag leaf | 35.46 ± 2.78 | 30.04 ± 3.16 | 32.16 ± 3.28 | 27.55 ± 3.14 * | |
| Booting | 36.19 ± 4.03 | 32.66 ± 4.15 * | 33.73 ± 3.79 | 27.49 ± 3.26 ** | |
| Panicle | 35.96 ± 3.99 | 31.72 ± 3.48 * | 33.11 ± 4.12 | 26.12 ± 3.65 ** | |
| Milk stage | 36.11 ± 4.09 | 30.37 ± 3.67 * | 33.51 ± 4.55 | 24.47 ± 4.13 *** | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tantray, A.Y.; Hazzazi, Y.; Ahmad, A. Physiological, Agronomical, and Proteomic Studies Reveal Crucial Players in Rice Nitrogen Use Efficiency under Low Nitrogen Supply. Int. J. Mol. Sci. 2022, 23, 6410. https://doi.org/10.3390/ijms23126410
Tantray AY, Hazzazi Y, Ahmad A. Physiological, Agronomical, and Proteomic Studies Reveal Crucial Players in Rice Nitrogen Use Efficiency under Low Nitrogen Supply. International Journal of Molecular Sciences. 2022; 23(12):6410. https://doi.org/10.3390/ijms23126410
Chicago/Turabian StyleTantray, Aadil Yousuf, Yehia Hazzazi, and Altaf Ahmad. 2022. "Physiological, Agronomical, and Proteomic Studies Reveal Crucial Players in Rice Nitrogen Use Efficiency under Low Nitrogen Supply" International Journal of Molecular Sciences 23, no. 12: 6410. https://doi.org/10.3390/ijms23126410
APA StyleTantray, A. Y., Hazzazi, Y., & Ahmad, A. (2022). Physiological, Agronomical, and Proteomic Studies Reveal Crucial Players in Rice Nitrogen Use Efficiency under Low Nitrogen Supply. International Journal of Molecular Sciences, 23(12), 6410. https://doi.org/10.3390/ijms23126410







