Evolution of SARS-CoV-2 in Spain during the First Two Years of the Pandemic: Circulating Variants, Amino Acid Conservation, and Genetic Variability in Structural, Non-Structural, and Accessory Proteins
Abstract
:1. Introduction
2. Results
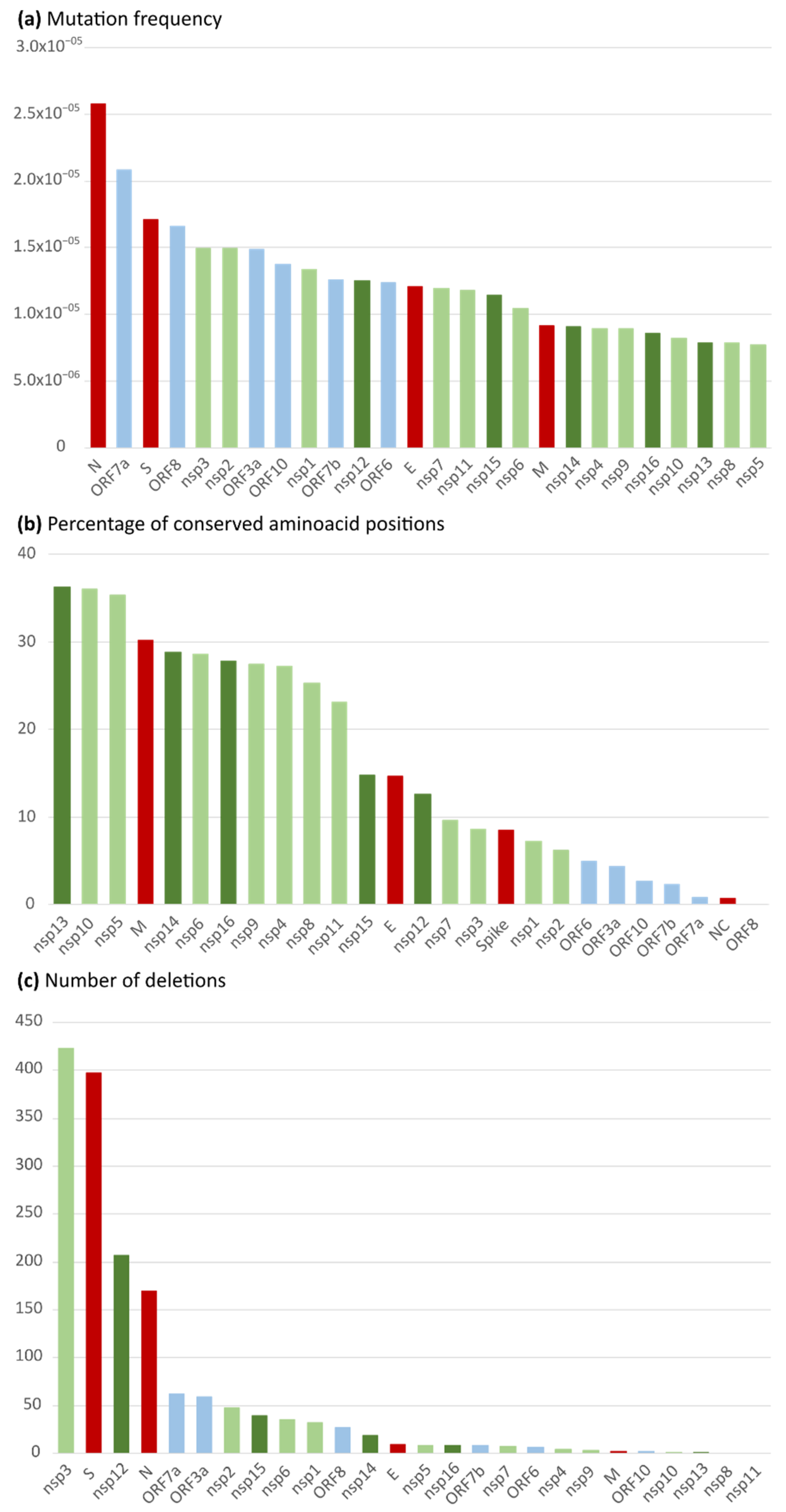
2.1. Nucleotide and Amino Acid Variability in the 26 Spanish SARS-CoV-2 Studied Proteins
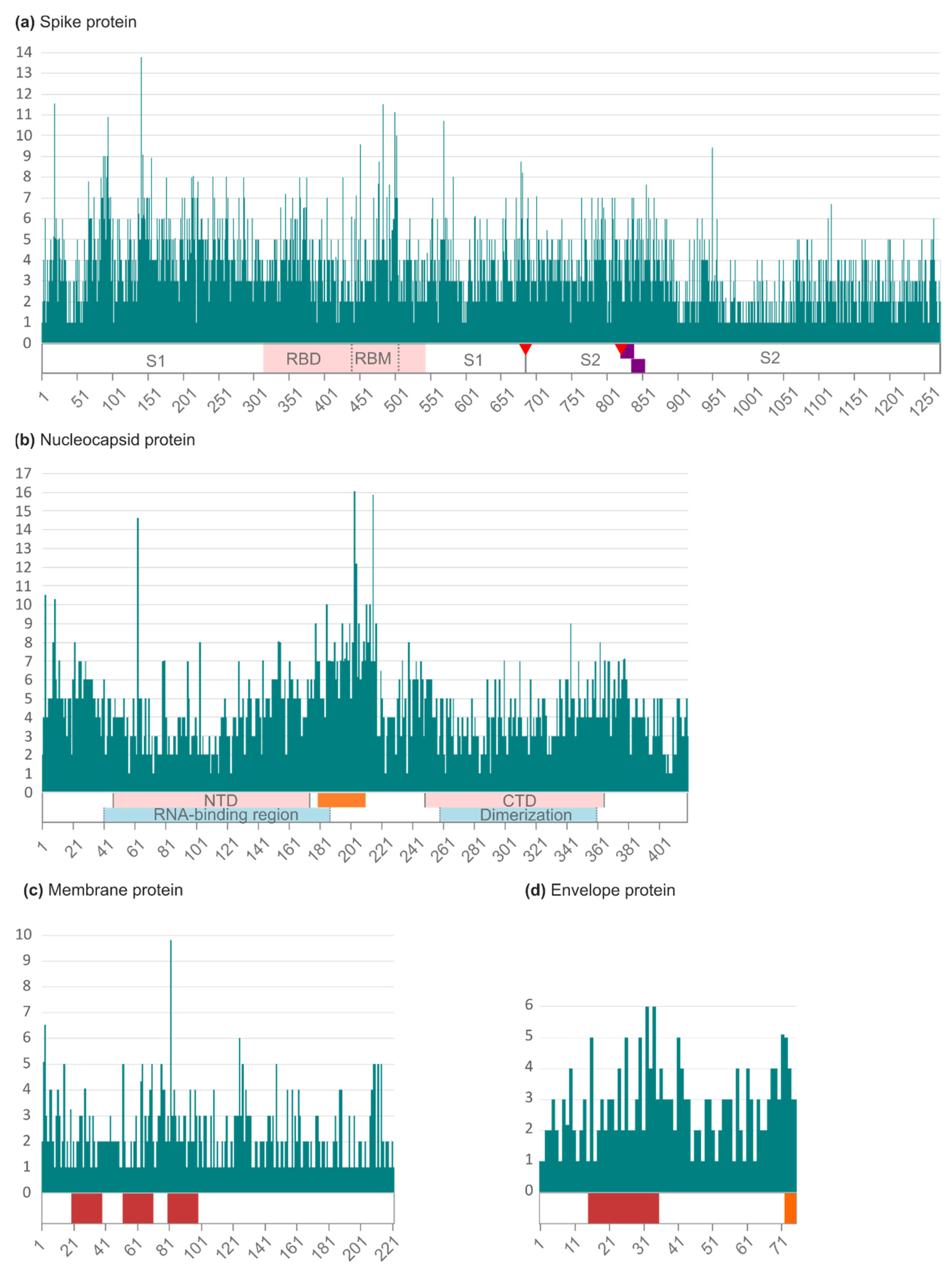
2.2. Amino Acid Variability in Spanish SARS-CoV-2 Structural Proteins
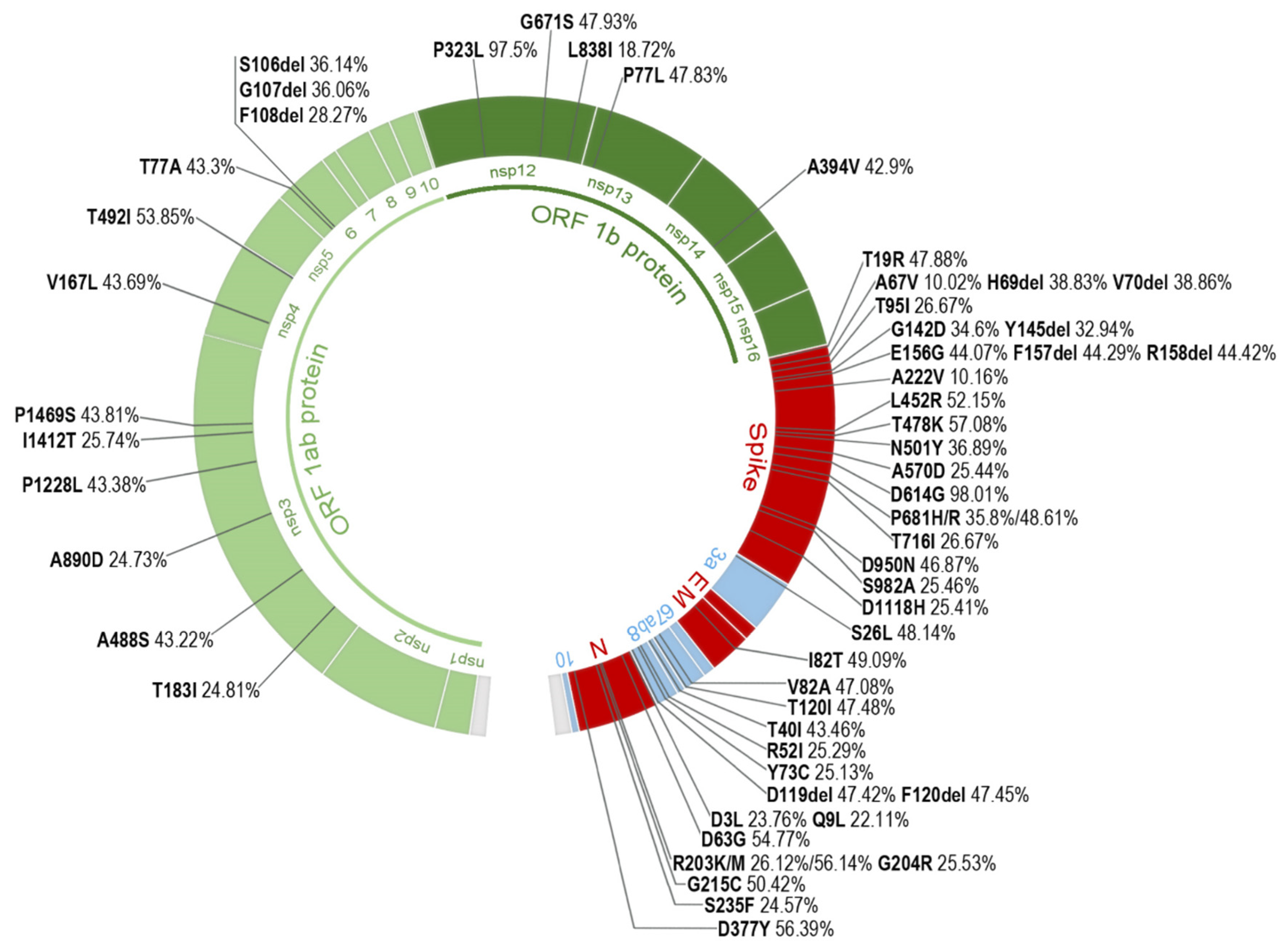
2.3. Most Prevalent aa Changes and Deletions in the Spanish Sequences
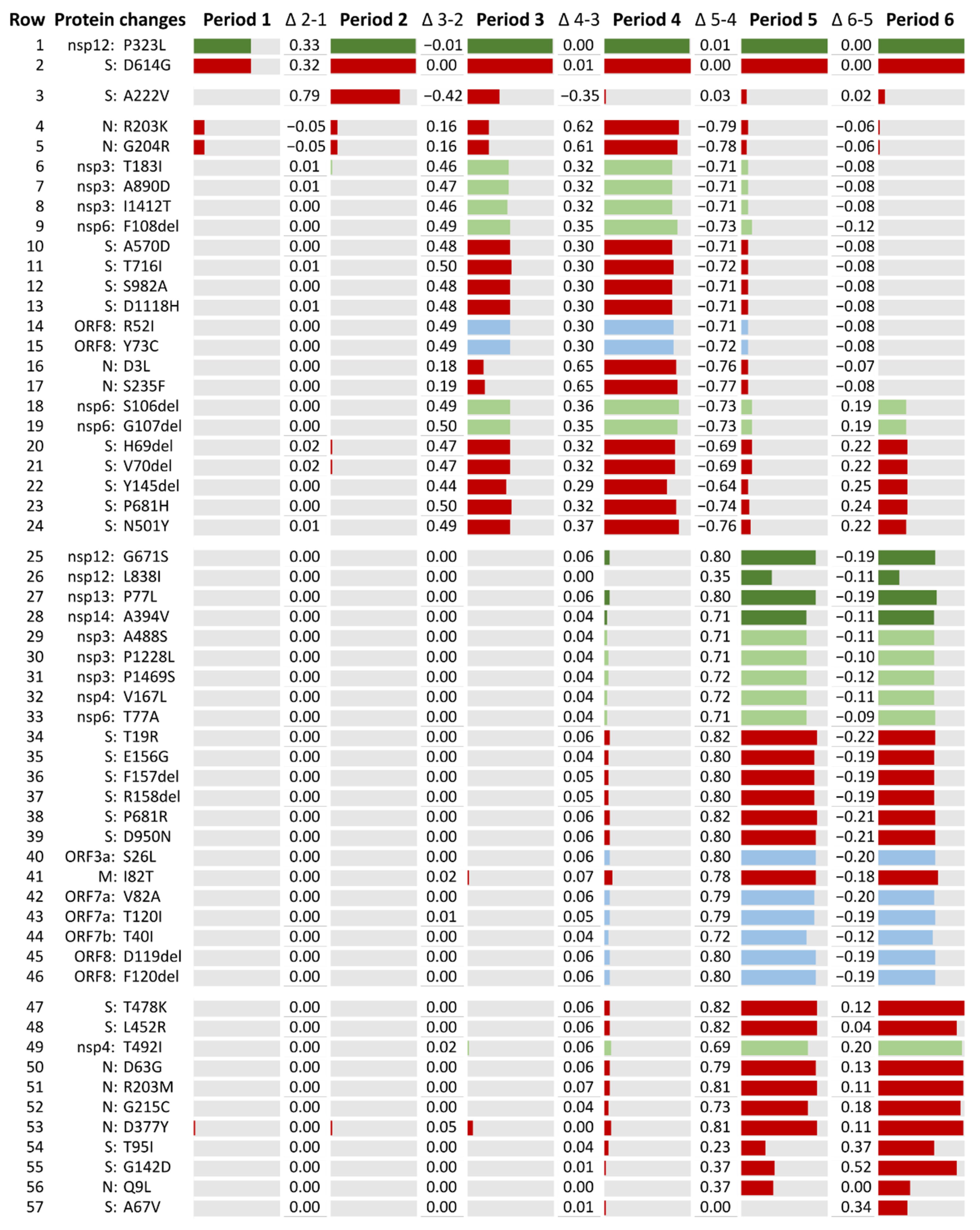
2.4. SARS-CoV-2 Lineages Circulating in Spain during the First Year of the Pandemic per Study Period
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Red Nacional de Vigilancia Epidemiológica CNE CNM (ISCIII) Informe no 116. Situación de COVID-19 en España. Available online: https://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/COVID19_Estrategia_vigilancia_y_control_e_indica (accessed on 8 April 2022).
- Comas, I.; Chiner-Oms, Á.; López, M.G.; González-Candelas, F. INFORME Proyecto COV20/00140 Una Perspectiva Genómica De La Pandemia: Lecciones en Salud Pública; Consejo Superior de Investigaciones Científicas: Valencia, Spain, 2020.
- Gómez-Carballa, A.; Bello, X.; Pardo-Seco, J.; Del Molino, M.L.P.; Martinón-Torres, F.; Salas, A. Phylogeography of SARS-CoV-2 pandemic in Spain: A story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders. Zool. Res. 2020, 41, 605–620. [Google Scholar] [CrossRef]
- España. Ministerio de la Presidencia Real Decreto 463/2020, de 14 de marzo, por el que se declara el estado de alarma para la gestión de la situación de crisis sanitaria ocasionada por el COVID-19. Boletín Of. Del Estado 3692 2020, 67, 25390–25400. [Google Scholar]
- España. Ministerio de la Presidencia Real Decreto 555/2020, de 5 de junio, por el que se prorroga el estado de alarma declarado por el Real Decreto 463/2020, de 14 de marzo, por el que se declara el estado de alarma para la gestión de la situación de crisis sanitaria ocasionada por el COVID-19. Boletín Of. Del Estado 5767 2020, 159, 61561–61567. [Google Scholar]
- España. Ministerio de la Presidencia Real Decreto 900/2020, de 9 de octubre, por el que se declara el estado de alarma para responder ante situaciones de especial riesgo por transmisión no controlada de infecciones causadas por el SARS-CoV-2. Boletín Of. Del Estado 12109 2020, 268, 18987–19106. [Google Scholar]
- España. Ministerio de la Presidencia Real Decreto 926/2020, de 25 de octubre, por el que se declara el estado de alarma para contener la propagación de infecciones causadas por el SARS-CoV-2. Boletín Oficial Del Estado 12898 2020, 282, 61561–61567. [Google Scholar]
- España. Ministerio de la Presidencia Real Decreto 956/2020, de 3 de noviembre, por el que se prorroga el estado de alarma declarado por el Real Decreto 926/2020, de 25 de octubre, por el que se declara el estado de alarma para contener la propagación de infecciones. Boletín Of. Del Estado 13494 2020, 291, 95841–95845. [Google Scholar]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1282, pp. 1–23. [Google Scholar] [CrossRef] [Green Version]
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2005; Volume 287, pp. 1–30. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta. Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Ahmadpour, D.; Ahmadpoor, P. How the COVID-19 Overcomes the Battle? An Approach to Virus Structure. Iran. J. Kidney Dis. 2020, 14, 167–172. [Google Scholar]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Zmasek, C.M.; Lefkowitz, E.J.; Niewiadomska, A.; Scheuermann, R.H. Genomic evolution of the Coronaviridae family. Virology 2022, 570, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.M.; Rottier, P.J.M. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [Green Version]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Fischer, F.; Stegen, C.F.; Masters, P.S.; Samsonoff, W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998, 72, 7885–7894. [Google Scholar] [CrossRef] [Green Version]
- Bos, E.C.; Luytjes, W.; van der Meulen, H.V.; Koerten, H.K.; Spaan, W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 1996, 218, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Maio, F.; Lo Cascio, E.; Babini, G.; Sali, M.; Della Longa, S.; Tilocca, B.; Roncada, P.; Arcovito, A.; Sanguinetti, M.; Scambia, G.; et al. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020, 22, 592–597. [Google Scholar] [CrossRef]
- Toto, A.; Ma, S.; Malagrinò, F.; Visconti, L.; Pagano, L.; Stromgaard, K.; Gianni, S. Comparing the binding properties of peptides mimicking the Envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020, 29, 2038–2042. [Google Scholar] [CrossRef]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef]
- de Haan, C.A.; Vennema, H.; Rottier, P.J. Assembly of the coronavirus envelope: Homotypic interactions between the M proteins. J. Virol. 2000, 74, 4967–4978. [Google Scholar] [CrossRef]
- Mahtarin, R.; Islam, S.; Islam, M.J.; Ullah, M.O.; Ali, M.A.; Halim, M.A. Structure and dynamics of membrane protein in SARS-CoV-2. J. Biomol. Struct. Dyn. 2020; Epub ahead of print. [Google Scholar] [CrossRef]
- Chang, C.; Sue, S.-C.; Yu, T.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.; Hsiao, H.; Wu, W.-J.; Chang, W.-L.; et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006, 13, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.-A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Wang, L.; Fontana, P.; Vora, S.; Zhang, Y.; Fu, T.-M.; Lieberman, J.; Wu, H. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv, 2020; preprint. [Google Scholar] [CrossRef]
- Vankadari, N.; Jeyasankar, N.N.; Lopes, W.J. Structure of the SARS-CoV-2 Nsp1/5’-Untranslated Region Complex and Implications for Potential Therapeutic Targets, a Vaccine, and Virulence. J. Phys. Chem. Lett. 2020, 11, 9659–9668. [Google Scholar] [CrossRef]
- Min, Y.-Q.; Mo, Q.; Wang, J.; Deng, F.; Wang, H.; Ning, Y.-J. SARS-CoV-2 nsp1: Bioinformatics, Potential Structural and Functional Features, and Implications for Drug/Vaccine Designs. Front. Microbiol. 2020, 11, 587317. [Google Scholar] [CrossRef]
- Vann, K.R.; Tencer, A.H.; Kutateladze, T.G. Inhibition of translation and immune responses by the virulence factor Nsp1 of SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 234. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Cornillez-Ty, C.T.; Liao, L.; Yates, J.R., 3rd; Kuhn, P.; Buchmeier, M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009, 83, 10314–10318. [Google Scholar] [CrossRef] [Green Version]
- Freitas, B.T.; Durie, I.A.; Murray, J.; Longo, J.E.; Miller, H.C.; Crich, D.; Hogan, R.J.; Tripp, R.A.; Pegan, S.D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020, 6, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Kusov, Y.; Hilgenfeld, R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018, 149, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 2013, 4, e00524-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagemeijer, M.C.; Monastyrska, I.; Griffith, J.; van der Sluijs, P.; Voortman, J.; van Bergen en Henegouwen, P.M.; Vonk, A.M.; Rottier, P.J.M.; Reggiori, F.; de Haan, C.A.M. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology 2014, 458–459, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Palm, G.J.; Mesters, J.R.; Siddell, S.G.; Ziebuhr, J.; Hilgenfeld, R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002, 21, 3213–3224. [Google Scholar] [CrossRef]
- Xia, B.; Kang, X. Activation and maturation of SARS-CoV main protease. Protein Cell 2011, 2, 282–290. [Google Scholar] [CrossRef] [Green Version]
- Cottam, E.M.; Whelband, M.C.; Wileman, T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 2014, 10, 1426–1441. [Google Scholar] [CrossRef] [Green Version]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [CrossRef] [PubMed]
- Egloff, M.-P.; Ferron, F.; Campanacci, V.; Longhi, S.; Rancurel, C.; Dutartre, H.; Snijder, E.J.; Gorbalenya, A.E.; Cambillau, C.; Canard, B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 2004, 101, 3792–3796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, G.; Fry, E.; Carter, L.; Sainsbury, S.; Walter, T.; Nettleship, J.; Berrow, N.; Owens, R.; Gilbert, R.; Davidson, A.; et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 2004, 12, 341–353. [Google Scholar] [CrossRef]
- Yan, L.; Ge, J.; Zheng, L.; Zhang, Y.; Gao, Y.; Wang, T.; Huang, Y.; Yang, Y.; Gao, S.; Li, M.; et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell 2021, 184, 184–193.e10. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Lugari, A.; Posthuma, C.C.; Zevenhoven, J.C.; Bernard, S.; Betzi, S.; Imbert, I.; Canard, B.; Guillemot, J.-C.; Lécine, P.; et al. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014, 289, 25783–25796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2’-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef] [Green Version]
- Vithani, N.; Ward, M.D.; Zimmerman, M.I.; Novak, B.; Borowsky, J.H.; Singh, S.; Bowman, G.R. SARS-CoV-2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophys. J. 2021, 120, 2880–2889. [Google Scholar] [CrossRef]
- Cheng, A.; Zhang, W.; Xie, Y.; Jiang, W.; Arnold, E.; Sarafianos, S.G.; Ding, J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 2005, 335, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Ahn, D.-G.; Choi, J.-K.; Taylor, D.R.; Oh, J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z.; et al. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, W.; Wojdyla, J.A.; Zhao, R.; Han, R.; Das, R.; Zlatev, I.; Manoharan, M.; Wang, M.; Cui, S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017, 13, e1006474. [Google Scholar] [CrossRef] [PubMed]
- Adedeji, A.O.; Marchand, B.; Te Velthuis, A.J.W.; Snijder, E.J.; Weiss, S.; Eoff, R.L.; Singh, K.; Sarafianos, S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE 2012, 7, e36521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, K.A.; Ziebuhr, J. Human coronavirus 229E nonstructural protein 13: Characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5’-triphosphatase activities. J. Virol. 2004, 78, 7833–7838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Moeller, N.H.; Shi, K.; Demir, Ö.; Banerjee, S.; Yin, L.; Belica, C.; Durfee, C.; Amaro, R.E.; Aihara, H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. bioRxiv, 2021; preprint. [Google Scholar] [CrossRef]
- Ogando, N.S.; Ferron, F.; Decroly, E.; Canard, B.; Posthuma, C.C.; Snijder, E.J. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front. Microbiol. 2019, 10, 1813. [Google Scholar] [CrossRef]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA 2012, 109, 9372–9377. [Google Scholar] [CrossRef] [Green Version]
- Ferron, F.; Subissi, L.; Silveira De Morais, A.T.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA 2018, 115, E162–E171. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Wilamowski, M.; Endres, M.; Godzik, A.; Michalska, K.; Joachimiak, A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020, 29, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Hackbart, M.; Mettelman, R.C.; O’Brien, A.; Mielech, A.M.; Yi, G.; Kao, C.C.; Baker, S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA 2017, 114, E4251–E4260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Su, C.; Ke, M.; Jin, X.; Xu, L.; Zhang, Z.; Wu, A.; Sun, Y.; Yang, Z.; Tien, P.; et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2’-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011, 7, e1002294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswanathan, T.; Arya, S.; Chan, S.-H.; Qi, S.; Dai, N.; Misra, A.; Park, J.-G.; Oladunni, F.; Kovalskyy, D.; Hromas, R.A.; et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020, 11, 3718. [Google Scholar] [CrossRef] [PubMed]
- Silvas, J.A.; Vasquez, D.M.; Park, J.-G.; Chiem, K.; Allué-Guardia, A.; Garcia-Vilanova, A.; Platt, R.N.; Miorin, L.; Kehrer, T.; Cupic, A.; et al. Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice. J. Virol. 2021, 95, e0040221. [Google Scholar] [CrossRef]
- Kanzawa, N.; Nishigaki, K.; Hayashi, T.; Ishii, Y.; Furukawa, S.; Niiro, A.; Yasui, F.; Kohara, M.; Morita, K.; Matsushima, K.; et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006, 580, 6807–6812. [Google Scholar] [CrossRef] [Green Version]
- Issa, E.; Merhi, G.; Panossian, B.; Salloum, T.; Tokajian, S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis. mSystems 2020, 5, e00266-20. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.-Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef]
- Yue, Y.; Nabar, N.R.; Shi, C.-S.; Kamenyeva, O.; Xiao, X.; Hwang, I.-Y.; Wang, M.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018, 9, 904. [Google Scholar] [CrossRef]
- Zhao, J.; Falcón, A.; Zhou, H.; Netland, J.; Enjuanes, L.; Pérez Breña, P.; Perlman, S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 2009, 83, 2368–2373. [Google Scholar] [CrossRef] [Green Version]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Nichols, J.; Snyder, D.T.; Hedges, J.F.; Cicha, C.; Lee, H.; Vanderwood, K.K.; Bimczok, D.; et al. SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression. Cell Rep. 2021, 35, 109197. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.S.; Aljabali, A.A.A.; Panda, P.K.; Ghosh, S.; Attrish, D.; Choudhury, P.P.; Seyran, M.; Pizzol, D.; Adadi, P.; Abd El-Aziz, T.M.; et al. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput. Biol. Med. 2021, 133, 104380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef] [PubMed]
- Flower, T.G.; Buffalo, C.Z.; Hooy, R.M.; Allaire, M.; Ren, X.; Hurley, J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2021785118. [Google Scholar] [CrossRef]
- Lin, X.; Fu, B.; Yin, S.; Li, Z.; Liu, H.; Zhang, H.; Xing, N.; Wang, Y.; Xue, W.; Xiong, Y.; et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience 2021, 24, 102293. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Fahmi, M.; Kitagawa, H.; Yasui, G.; Kubota, Y.; Ito, M. The Functional Classification of ORF8 in SARS-CoV-2 Replication, Immune Evasion, and Viral Pathogenesis Inferred through Phylogenetic Profiling. Evol. Bioinform. Online 2021, 17, 11769343211003080. [Google Scholar] [CrossRef]
- Mena, E.L.; Donahue, C.J.; Vaites, L.P.; Li, J.; Rona, G.; O’Leary, C.; Lignitto, L.; Miwatani-Minter, B.; Paulo, J.A.; Dhabaria, A.; et al. ORF10-Cullin-2-ZYG11B complex is not required for SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2023157118. [Google Scholar] [CrossRef]
- Pancer, K.; Milewska, A.; Owczarek, K.; Dabrowska, A.; Kowalski, M.; Łabaj, P.P.; Branicki, W.; Sanak, M.; Pyrc, K. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020, 16, e1008959. [Google Scholar] [CrossRef]
- Hassan, S.S.; Attrish, D.; Ghosh, S.; Choudhury, P.P.; Uversky, V.N.; Aljabali, A.A.A.; Lundstrom, K.; Uhal, B.D.; Rezaei, N.; Seyran, M.; et al. Notable sequence homology of the ORF10 protein introspects the architecture of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 181, 801–809. [Google Scholar] [CrossRef]
- Li, X.; Hou, P.; Ma, W.; Wang, X.; Wang, H.; Yu, Z.; Chang, H.; Wang, T.; Jin, S.; Wang, X.; et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell. Mol. Immunol. 2022, 19, 67–78. [Google Scholar] [CrossRef] [PubMed]
- van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.S.; Boshier, F.A.T.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 83, 104351. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevajol, M.; Subissi, L.; Decroly, E.; Canard, B.; Imbert, I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014, 194, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Keck, J.G.; Makino, S.; Soe, L.H.; Fleming, J.O.; Stohlman, S.A.; Lai, M.M. RNA recombination of coronavirus. Adv. Exp. Med. Biol. 1987, 218, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- WHO Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 20 September 2021).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Cov-Lineages. Available online: https://cov-lineages.org/lineage_list.html (accessed on 9 May 2022).
- Pango Lineage Nomenclature Pango Network—Helping Track the Transmission and Spread of SARS-CoV-2. Available online: https://www.pango.network/ (accessed on 17 May 2022).
- WHO Statement on Omicron Sublineage BA.2. Available online: https://www.who.int/news/item/22-02-2022-statement-on-omicron-sublineage-ba.2 (accessed on 17 May 2022).
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Kang, T.; Ali, H.; Lai, D. Remdesivir Strongly Binds to RNA-Dependent RNA Polymerase, Membrane Protein, and Main Protease of SARS-CoV-2: Indication From Molecular Modeling and Simulations. Front. Pharmacol. 2021, 12, 710778. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fang, C.; Zhang, Q.; Zhang, R.; Zhao, X.; Duan, Y.; Wang, H.; Zhu, Y.; Feng, L.; Zhao, J.; et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 2021, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Díez-Fuertes, F.; Iglesias-Caballero, M.; García-Pérez, J.; Monzón, S.; Jiménez, P.; Varona, S.; Cuesta, I.; Zaballos, Á.; Jiménez, M.; Checa, L.; et al. A Founder Effect Led Early SARS-CoV-2 Transmission in Spain. J. Virol. 2021, 95, e01583-20. [Google Scholar] [CrossRef] [PubMed]
- Mira-Iglesias, A.; Mengual-Chuliá, B.; Cano, L.; García-Rubio, J.; Tortajada-Girbés, M.; Carballido-Fernández, M.; Mollar-Maseres, J.; Schwarz-Chavarri, G.; García-Esteban, S.; Puig-Barberà, J.; et al. Retrospective screening for SARS-CoV-2 among influenza-like illness hospitalizations: 2018-2019 and 2019-2020 seasons, Valencia region, Spain. Influenza Other Respir. Viruses 2021, 16, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Trobajo-Sanmartín, C.; Miqueleiz, A.; Portillo, M.E.; Fernández-Huerta, M.; Navascués, A.; Sola Sara, P.; Moreno, P.L.; Ordoñez, G.R.; Castilla, J.; Ezpeleta, C. Emergence of SARS-CoV-2 variant B.1.575.2 containing the E484K mutation in the spike protein in Pamplona (Spain) May–June 2021. J. Clin. Microbiol. 2021, 59, e0173621. [Google Scholar] [CrossRef] [PubMed]
- Alcoba-Florez, J.; Lorenzo-Salazar, J.M.; Gil-Campesino, H.; Íñigo-Campos, A.; Martínez de Artola, D.G.; García-Olivares, V.; Díez-Gil, O.; Valenzuela-Fernández, A.; Ciuffreda, L.; González-Montelongo, R.; et al. Monitoring the rise of the SARS-CoV-2 lineage B.1.1.7 in Tenerife (Spain) since mid-December 2020. J. Infect. 2021, 82, e1–e3. [Google Scholar] [CrossRef]
- Viedma, E.; Dahdouh, E.; González-Alba, J.M.; González-Bodi, S.; Martínez-García, L.; Lázaro-Perona, F.; Recio, R.; Rodríguez-Tejedor, M.; Folgueira, M.D.; Cantón, R.; et al. Genomic Epidemiology of SARS-CoV-2 in Madrid, Spain, during the First Wave of the Pandemic: Fast Spread and Early Dominance by D614G Variants. Microorganisms 2021, 9, 454. [Google Scholar] [CrossRef]
- Andrés, C.; Piñana, M.; Borràs-Bermejo, B.; González-Sánchez, A.; García-Cehic, D.; Esperalba, J.; Rando, A.; Zules-Oña, R.-G.; Campos, C.; Codina, M.G.; et al. A year living with SARS-CoV-2: An epidemiological overview of viral lineage circulation by whole-genome sequencing in Barcelona city (Catalonia, Spain). Emerg. Microbes Infect. 2022, 11, 172–181. [Google Scholar] [CrossRef]
- Alves-Cabratosa, L.; Comas-Cufí, M.; Blanch, J.; Martí-Lluch, R.; Ponjoan, A.; Castro-Guardiola, A.; Hurtado-Ganoza, A.; Pérez-Jaén, A.; Rexach-Fumaña, M.; Faixedas-Brunsoms, D.; et al. Individuals With SARS-CoV-2 Infection During the First and Second Waves in Catalonia, Spain: Retrospective Observational Study Using Daily Updated Data. JMIR Public Health Surveill. 2022, 8, e30006. [Google Scholar] [CrossRef]
- Del Águila-Mejía, J.; Wallmann, R.; Calvo-Montes, J.; Rodríguez-Lozano, J.; Valle-Madrazo, T.; Aginagalde-Llorente, A. Secondary Attack Rate, Transmission and Incubation Periods, and Serial Interval of SARS-CoV-2 Omicron Variant, Spain. Emerg. Infect. Dis. 2022, 28, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Sola Campoy, P.J.; Buenestado-Serrano, S.; Pérez-Lago, L.; Rodriguez-Grande, C.; Catalán, P.; Andrés-Zayas, C.; Alcalá, L.; Losada, C.; Rico-Luna, C.; Muñoz, P.; et al. First importations of SARS-CoV-2 P.1 and P.2 variants from Brazil to Spain and early community transmission. Enferm. Infecc. Microbiol. Clin. 2022, 40, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.; Mandal, S.M.; Mondal, S.K.; Mukherjee, S.; Mapder, T.; Ghosh, W.; Chakraborty, R. Trends of mutation accumulation across global SARS-CoV-2 genomes: Implications for the evolution of the novel coronavirus. Genomics 2020, 112, 5331–5342. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Mattiuzzi, C.; Henry, B.M. Updated picture of SARS-CoV-2 variants and mutations. Diagnosis 2021, 9, 11–17. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Zhou, J.; Ye, Q. The Global Epidemic of the SARS-CoV-2 Delta Variant, Key Spike Mutations and Immune Escape. Front. Immunol. 2021, 12, 751778. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef]
- Majumdar, P.; Niyogi, S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol. Infect. 2021, 149, e110. [Google Scholar] [CrossRef]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. Evolution of SARS-CoV-2 Envelope, Membrane, Nucleocapsid, and Spike Structural Proteins from the Beginning of the Pandemic to September 2020: A Global and Regional Approach by Epidemiological Week. Viruses 2021, 13, 243. [Google Scholar] [CrossRef]
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Islam, I.; Mishu, I.D.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 2021, 22, 100997. [Google Scholar] [CrossRef]
- Emam, M.; Oweda, M.; Antunes, A.; El-Hadidi, M. Positive selection as a key player for SARS-CoV-2 pathogenicity: Insights into ORF1ab, S and E genes. Virus Res. 2021, 302, 198472. [Google Scholar] [CrossRef]
- Brown, K.A.; Gubbay, J.; Hopkins, J.; Patel, S.; Buchan, S.A.; Daneman, N.; Goneau, L.W. S-Gene Target Failure as a Marker of Variant, B.1.1.7 Among SARS-CoV-2 Isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA 2021, 325, 2115–2116. [Google Scholar] [CrossRef] [PubMed]
- Buenestado-Serrano, S.; Recio, R.; Sola Campoy, P.J.; Catalán, P.; Folgueira, M.D.; Villa, J.; Muñoz Gallego, I.; de la Cueva, V.M.; Meléndez, M.A.; Andrés Zayas, C.; et al. First confirmation of importation and transmission in Spain of the newly identified SARS-CoV-2 B.1.1.7 variant. Enferm. Infecc. Microbiol. Clin. 2021, S0213-005X(21)00046-X. [Google Scholar] [CrossRef] [PubMed]
- Gobierno de España. Ministerio de Sanidad Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 de Importancia en Salud Pública en España 18 de Marzo de 2021. Available online: https://www.sanidad.gob.es/en/home.htm (accessed on 18 March 2022).
- Metzger, C.M.J.A.; Lienhard, R.; Seth-Smith, H.M.B.; Roloff, T.; Wegner, F.; Sieber, J.; Bel, M.; Greub, G.; Egli, A. PCR performance in the SARS-CoV-2 Omicron variant of concern? Swiss Med. Wkly. 2021, 151, w30120. [Google Scholar] [CrossRef] [PubMed]
- WHO Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 17 May 2022).
- Rahman, M.S.; Islam, M.R.; Hoque, M.N.; Alam, A.S.M.R.U.; Akther, M.; Puspo, J.A.; Akter, S.; Anwar, A.; Sultana, M.; Hossain, M.A. Comprehensive annotations of the mutational spectra of SARS-CoV-2 spike protein: A fast and accurate pipeline. Transbound. Emerg. Dis. 2021, 68, 1625–1638. [Google Scholar] [CrossRef]
- Piccoli, L.; Park, Y.-J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e21. [Google Scholar] [CrossRef]
- Nabel, K.G.; Clark, S.A.; Shankar, S.; Pan, J.; Clark, L.E.; Yang, P.; Coscia, A.; McKay, L.G.A.; Varnum, H.H.; Brusic, V.; et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science 2022, 375, eabl6251. [Google Scholar] [CrossRef]
- Rahman, M.S.; Islam, M.R.; Alam, A.S.M.R.U.; Islam, I.; Hoque, M.N.; Akter, S.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021, 93, 2177–2195. [Google Scholar] [CrossRef]
- Tung, H.Y.L.; Limtung, P. Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein and structure model of sequestration by protein 14-3-3. Biochem. Biophys. Res. Commun. 2020, 532, 134–138. [Google Scholar] [CrossRef]
- Alm, E.; Broberg, E.K.; Connor, T.; Hodcroft, E.B.; Komissarov, A.B.; Maurer-Stroh, S.; Melidou, A.; Neher, R.A.; O’Toole, Á.; Pereyaslov, D. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Euro Surveill. 2020, 25, 2001410. [Google Scholar] [CrossRef]
- Di Giallonardo, F.; Duchene, S.; Puglia, I.; Curini, V.; Profeta, F.; Cammà, C.; Marcacci, M.; Calistri, P.; Holmes, E.C.; Lorusso, A. Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy. Viruses 2020, 12, 1438. [Google Scholar] [CrossRef]
- Hyafil, A.; Moriña, D. Analysis of the impact of lockdown on the reproduction number of the SARS-Cov-2 in Spain. Gac. Sanit. 2021, 35, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.M.; Lombardi Junior, L.P.; Castro, F.F.M.; Yang, A.C. Mathematical modeling of the transmission of SARS-CoV-2-Evaluating the impact of isolation in São Paulo State (Brazil) and lockdown in Spain associated with protective measures on the epidemic of COVID-19. PLoS ONE 2021, 16, e0252271. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.K.; Mudi, S.R. Spike protein D614G and RdRp P323L: The SARS-CoV-2 mutations associated with severity of COVID-19. Genom. Inform. 2020, 18, e44. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Vaughan, T.G.; Crawford, K.H.D.; Althaus, C.L.; Reichmuth, M.L.; Bowen, J.E.; Walls, A.C.; Corti, D.; et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021, 595, 707–712. [Google Scholar] [CrossRef]
- Vilar, S.; Isom, D.G. One Year of SARS-CoV-2: How Much Has the Virus Changed? Biology 2021, 10, 91. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Giles, B.; Meredith, P.; Robson, S.; Smith, G.; Chauhan, A. The SARS-CoV-2 B.1.1.7 variant and increased clinical severity-the jury is out. Lancet Infect. Dis. 2021, 21, 1213–1214. [Google Scholar] [CrossRef]
- Ong, S.W.X.; Chiew, C.J.; Ang, L.W.; Mak, T.-M.; Cui, L.; Toh, M.P.H.S.; Lim, Y.D.; Lee, P.H.; Lee, T.H.; Chia, P.Y.; et al. Clinical and virological features of SARS-CoV-2 variants of concern: A retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin. Infect. Dis. 2021, Aug 23:ciab721. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Sidarovich, A.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Moldenhauer, A.-S.; Winkler, M.S.; Schulz, S.; Jäck, H.-M.; et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 37, 109825. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef] [PubMed]
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Ministerio de Sanidad. Marzo 2021. Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 de Importancia en Salud Pública en España 26 de Marzo de 2021. Available online: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/COVID19_Actualizacion_variantes_20210326.pdf (accessed on 20 September 2021).
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Ministerio de Sanidad. Junio 2021. Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 de Mayor Impacto e Interés en Salud Pública en España 21 de Junio de 2021. Available online: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/COVID19_Actualizacion_variantes_20210621.pdf (accessed on 20 September 2021).
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Ministerio de Sanidad. Agosto 2021. Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 de Importancia en Salud Pública en España 9 de agosto 2021. Available online: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/COVID19_Actualizacion_variantes_20210809.pdf (accessed on 20 September 2021).
- Mishra, T.; Dalavi, R.; Joshi, G.; Kumar, A.; Pandey, P.; Shukla, S.; Mishra, R.K.; Chande, A. SARS-CoV-2 spike E156G/Δ157-158 mutations contribute to increased infectivity and immune escape. Life Sci. Alliance 2022, 5, e202201415. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Woo, H.G. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. 2021, 12, 830527. [Google Scholar] [CrossRef]
- Plante, J.A.; Mitchell, B.M.; Plante, K.S.; Debbink, K.; Weaver, S.C.; Menachery, V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe 2021, 29, 508–515. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv, 2021; preprint. [Google Scholar]
- Ostrov, D.A. Structural Consequences of Variation in SARS-CoV-2 B.1.1.7. J. Cell. Immunol. 2021, 3, 103–108. [Google Scholar] [CrossRef]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.A.T.M.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef]
- Johnson, B.A.; Xie, X.; Bailey, A.L.; Kalveram, B.; Lokugamage, K.G.; Muruato, A.; Zou, J.; Zhang, X.; Juelich, T.; Smith, J.K.; et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021, 591, 293–299. [Google Scholar] [CrossRef]
- Lubinski, B.; Tang, T.; Daniel, S.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B.1.1.7: Role of the P681H mutation. bioRxiv, 2021; preprint. [Google Scholar] [CrossRef]
- Lubinski, B.; Frazier, L.; Phan, M.; Bugumbe, D.; Cunningham, J.L.; Tang, T.; Daniel, S.; Cotten, M.; Jaimes, J.A.; Whittaker, G. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: A case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Zhao, L.P.; Lybrand, T.P.; Gilbert, P.B.; Hawn, T.R.; Schiffer, J.T.; Stamatatos, L.; Payne, T.H.; Carpp, L.N.; Geraghty, D.E.; Jerome, K.R. Tracking SARS-CoV-2 Spike Protein Mutations in the United States (2020/01–2021/03) Using a Statistical Learning Strategy. bioRxiv, 2021; preprint. [Google Scholar] [CrossRef]
- Leary, S.; Gaudieri, S.; Parker, M.D.; Chopra, A.; James, I.; Pakala, S.; Alves, E.; John, M.; Lindsey, B.B.; Keeley, A.J.; et al. Generation of a Novel SARS-CoV-2 Sub-genomic RNA Due to the R203K/G204R Variant in Nucleocapsid: Homologous Recombination has Potential to Change SARS-CoV-2 at Both Protein and RNA Level. Pathog. Immun. 2021, 6, 27–49. [Google Scholar] [CrossRef]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef]
- Ai, J.; Zhang, H.; Zhang, Y.; Lin, K.; Zhang, Y.; Wu, J.; Wan, Y.; Huang, Y.; Song, J.; Fu, Z.; et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2022, 11, 337–343. [Google Scholar] [CrossRef]
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Ministerio de Sanidad. Enero 2022. Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 en España 17 de Enero de 2022. Available online: https://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Integ (accessed on 9 May 2022).
- Centro de Coordinación de Alertas y Emergencias Sanitarias. Ministerio de Sanidad. Febrero 2022. Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 en España 14 de Febrero de 2022. Available online: https://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Integ (accessed on 9 May 2022).
- Centro de Coordinación de Alertas y Emergencias Sanitarias Mayo 2022 Actualización de la Situación Epidemiológica de las Variantes de SARS-CoV-2 en España. 2022. Available online: https://www.sanidad.gob.es/en/home.htm (accessed on 18 March 2022).
- Samrat, S.K.; Tharappel, A.M.; Li, Z.; Li, H. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 2020, 288, 198141. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef]
- Ita, K. Coronavirus Disease (COVID-19): Current Status and Prospects for Drug and Vaccine Development. Arch. Med. Res. 2021, 52, 15–24. [Google Scholar] [CrossRef]
- Grupo de Trabajo Técnico de Vacunación COVID-19 de la Ponencia de Programa y Registro de Vacunaciones Estrategia de Vacunación Frente a COVID-19 en España. Available online: https://www.sanidad.gob.es/profesionales/saludPublica/prevPromocion/vacunaciones/covid19/Actualizaciones_Estrategia_Vacunacion/docs/COVID-19_Actualizacion1_EstrategiaVacunacion.pdf (accessed on 8 May 2022).
- Premkumar, L.; Segovia-Chumbez, B.; Jadi, R.; Martinez, D.R.; Raut, R.; Markmann, A.; Cornaby, C.; Bartelt, L.; Weiss, S.; Park, Y.; et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020, 5, eabc8413. [Google Scholar] [CrossRef]
- Yuan, M.; Liu, H.; Wu, N.C.; Wilson, I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021, 538, 192–203. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Wei, G.-W. Mechanisms of SARS-CoV-2 Evolution Revealing Vaccine-Resistant Mutations in Europe and America. J. Phys. Chem. Lett. 2021, 12, 11850–11857. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Wei, G.-W. Review of the mechanisms of SARS-CoV-2 evolution and transmission. arXiv 2021, arXiv:2109.08148v1. [Google Scholar]
- Niesen, M.J.M.; Anand, P.; Silvert, E.; Suratekar, R.; Pawlowski, C.; Ghosh, P.; Lenehan, P.; Hughes, T.; Zemmour, D.; O’Horo, J.C.; et al. COVID-19 vaccines dampen genomic diversity of SARS-CoV-2: Unvaccinated patients exhibit more antigenic mutational variance. medRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Yeh, T.-Y.; Contreras, G.P. Full vaccination is imperative to suppress SARS-CoV-2 delta variant mutation frequency. medRxiv 2021. preprint. [Google Scholar] [CrossRef]
- Barandalla, I.; Alvarez, C.; Barreiro, P.; de Mendoza, C.; González-Crespo, R.; Soriano, V. Impact of scaling up SARS-CoV-2 vaccination on COVID-19 hospitalizations in Spain. Int. J. Infect. Dis. 2021, 112, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Mazagatos, C.; Monge, S.; Olmedo, C.; Vega, L.; Gallego, P.; Martín-Merino, E.; Sierra, M.J.; Limia, A.; Larrauri, A. Effectiveness of mRNA COVID-19 vaccines in preventing SARS-CoV-2 infections and COVID-19 hospitalisations and deaths in elderly long-term care facility residents, Spain, weeks 53 2020 to 13 2021. Euro Surveill. 2021, 26, 2100452. [Google Scholar] [CrossRef]
- Harder, T.; Külper-Schiek, W.; Reda, S.; Treskova-Schwarzbach, M.; Koch, J.; Vygen-Bonnet, S.; Wichmann, O. Effectiveness of COVID-19 vaccines against SARS-CoV-2 infection with the Delta (B.1.617.2) variant: Second interim results of a living systematic review and meta-analysis, 1 January to 25 August 2021. Euro Surveill. 2021, 26, 2100920. [Google Scholar] [CrossRef]
- Bansal, N.; Raturi, M.; Bansal, Y. SARS-CoV-2 variants in immunocompromised COVID-19 patients: The underlying causes and the way forward. Transfus. Clin. Biol. 2022, 29, 161–163. [Google Scholar] [CrossRef]
- Weigang, S.; Fuchs, J.; Zimmer, G.; Schnepf, D.; Kern, L.; Beer, J.; Luxenburger, H.; Ankerhold, J.; Falcone, V.; Kemming, J.; et al. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat. Commun. 2021, 12, 6405. [Google Scholar] [CrossRef]
- España. Ministerio de la Presidencia Real Decreto 286/2022, de 19 de abril, por el que se modifica la obligatoriedad del uso de mascarillas durante la situación de crisis sanitaria ocasionada por la COVID-19. Boletín Of. Del Estado 2022-6449 2022, 94, 53729–53732. [Google Scholar]
- Pfizer Pfizer and BioNTech Initiate Study to Evaluate Omicron-Based COVID-19 Vaccine in Adults 18 to 55 Years of Age|Pfizer. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-initiate-study-evaluate-omicron-based# (accessed on 18 May 2022).
- Moderna mRNA Medicines We Are Currently Developing. Available online: https://www.modernatx.com/research/product-pipeline (accessed on 18 May 2022).
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, X.; Du, L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin. Ther. Targets 2021, 25, 415–421. [Google Scholar] [CrossRef]
- Ravichandran, S.; Coyle, E.M.; Klenow, L.; Tang, J.; Grubbs, G.; Liu, S.; Wang, T.; Golding, H.; Khurana, S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020, 12, eabc3539. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Deng, Y.; Huang, B.; Han, D.; Wang, W.; Huang, M.; Zhai, C.; Zhao, Z.; Yang, R.; Zhao, Y.; et al. DNA Vaccines Expressing the Envelope and Membrane Proteins Provide Partial Protection Against SARS-CoV-2 in Mice. Front. Immunol. 2022, 13, 827605. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-M. Universal COVID-19 Vaccine Targeting SARS-CoV-2 Envelope Protein. World, J. Vaccines 2021, 11, 19–27. [Google Scholar] [CrossRef]
- Sun, J.; Zhuang, Z.; Zheng, J.; Li, K.; Wong, R.L.-Y.; Liu, D.; Huang, J.; He, J.; Zhu, A.; Zhao, J.; et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020, 182, 734–743.e5. [Google Scholar] [CrossRef]
- Liu, W.J.; Zhao, M.; Liu, K.; Xu, K.; Wong, G.; Tan, W.; Gao, G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017, 137, 82–92. [Google Scholar] [CrossRef]
- WHO. COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 4 June 2022).
- Shih, H.-I.; Wu, C.-J.; Tu, Y.-F.; Chi, C.-Y. Fighting COVID-19: A quick review of diagnoses, therapies, and vaccines. Biomed. J. 2020, 43, 341–354. [Google Scholar] [CrossRef]
- Uzunova, K.; Filipova, E.; Pavlova, V.; Vekov, T. Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS-CoV-2. Biomed. Pharmacother. 2020, 131, 110668. [Google Scholar] [CrossRef]
- WHO. Therapeutics and COVID-19: Living Guideline. Available online: https://app.magicapp.org/#/guideline/nBkO1E/rec/LwrMyv (accessed on 4 June 2022).
- National Institutes of Health. COVID-19 Therapeutics Prioritized for Testing in Clinical Trials|National Institutes of Health (NIH). Available online: https://www.nih.gov/research-training/medical-research-initiatives/activ/covid-19-therapeutics-prioritized-testing-clinical-trials (accessed on 4 June 2022).
- FDA. Coronavirus (COVID-19)|Drugs|FDA. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 4 June 2022).
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Dampalla, C.S.; Zheng, J.; Perera, K.D.; Wong, L.-Y.R.; Meyerholz, D.K.; Nguyen, H.N.; Kashipathy, M.M.; Battaile, K.P.; Lovell, S.; Kim, Y.; et al. Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2101555118. [Google Scholar] [CrossRef]
- Vandyck, K.; Deval, J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr. Opin. Virol. 2021, 49, 36–40. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Recommends Highly Successful COVID-19 Therapy and Calls for Wide Geographical Distribution and transparency from Originator. Available online: https://www.who.int/news/item/22-04-2022-who-recommends-highly-Successful-covid-19-therapy-and-calls-for-wide-geographical-distribution-and-transparency-from-originator (accessed on 4 June 2022).
- Mahase, E. COVID-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef] [PubMed]
- CNE. Gobierno de España. Evolución Pandemia COVID-19. Available online: https://cnecovid.isciii.es/covid19/#ccaa (accessed on 20 September 2021).
- Burgos, M.; Llácer, T.; Reinosa, R.; Rubio-Garrido, M.; González, A.; Holguín, A. Impaired genotypic resistance interpretation due to HIV-1 variant specific Markers. In Proceedings of the 10th IAS Conference on HIV Science, Mexico City, México, 21–24 July 2019. [Google Scholar]
- Troyano-Hernáez, P.; Reinosa, R.; Burgos, M.C.; Holguín, Á. Short Communication: Update in Natural Antiretroviral Resistance-Associated Mutations Among HIV Type 2 Variants and Discrepancies Across HIV Type 2 Resistance Interpretation Tools. AIDS Res. Hum. Retrovir. 2021, 37, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. Marcadores genéticos en la proteína de la Cápside p24 en los grupos, subtipos, sub-subtipos y recombinantes del VIH-1. In Proceedings of the XI CONGRESO NACIONAL GeSIDA, Toledo, Spain, 10–13 December 2019; pp. 124–125. [Google Scholar]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. Mutaciones en la proteína Spike de SARS-CoV-2 por Comunidades Autónomas en secuencias españolas recogidas hasta junio 2020. In Proceedings of the I Congreso Nacional COVID-19, Virtual Congress, Spain, 13 September 2020; p. 76. [Google Scholar]
- Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. HIV Capsid Protein Genetic Diversity Across HIV-1 Variants and Impact on New Capsid-Inhibitor Lenacapavir. Front. Microbiol. 2022, 13, 854974. [Google Scholar] [CrossRef]
- Kabat, E.A.; Wu, T.T.; Bilofsky, H. Unusual distributions of amino acids in complementarity-determining (hypervariable) segments of heavy and light chains of immunoglobulins and their possible roles in specificity of antibody-combining sites. J. Biol. Chem. 1977, 252, 6609–6616. [Google Scholar] [CrossRef]
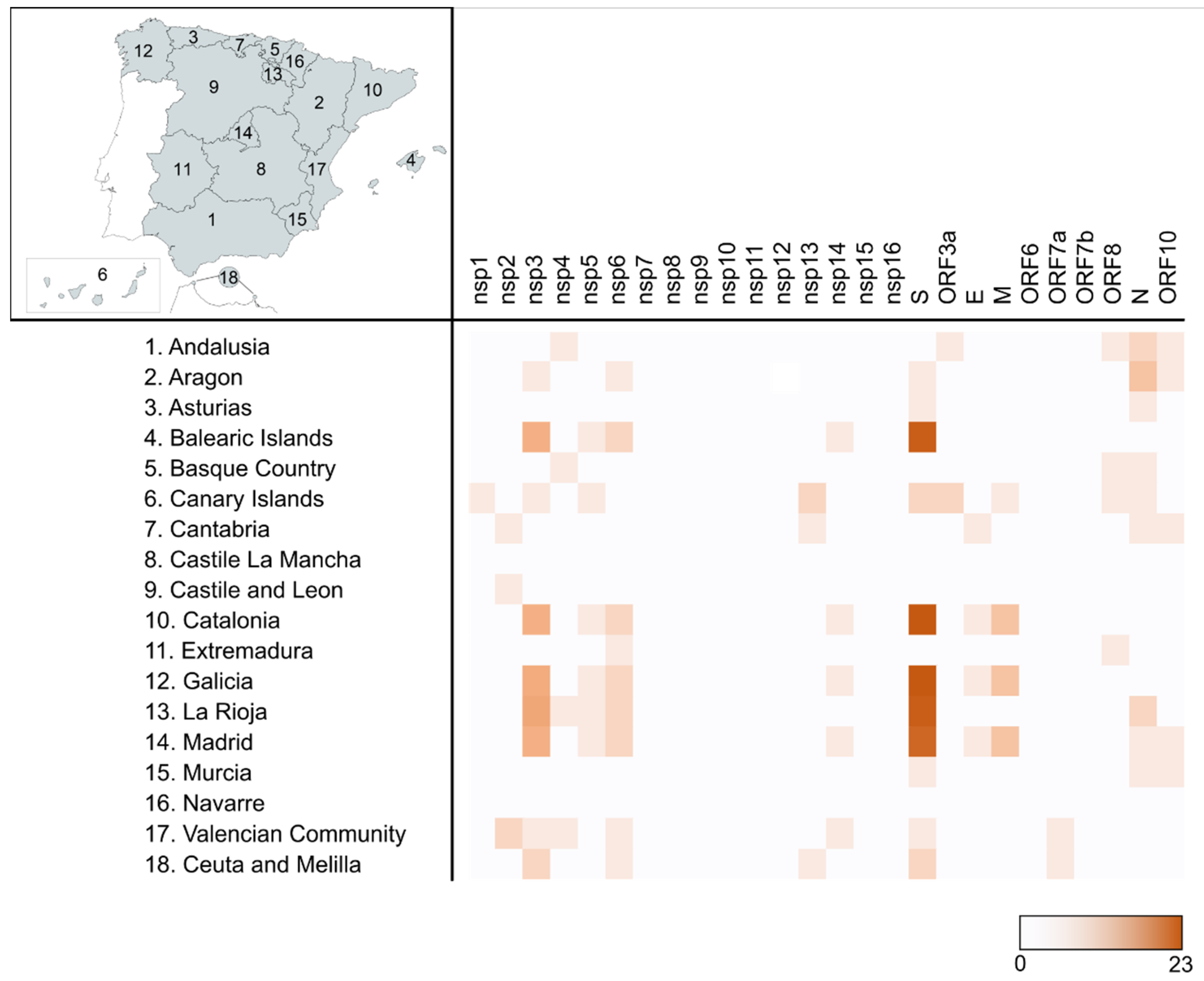

| Protein | Proposed Molecular Function |
|---|---|
| I. Structural proteins | |
| Spike (S) | Class I fusion protein that mediates attachment to the host cell’s receptor angiotensin-converting enzyme 2 (ACE2) through the receptor-binding domain (RBD), and fusion of viral and cellular membranes [21,22,23] |
| Envelope (E) | Viral assembly and release through interaction with M protein [24,25,26,27], epithelial cells’ tight junctions’ disruption by interaction with PALS1 [28,29]. |
| Membrane (M) | Virion shape, participates in E assembly and N attachment to the viral genome, interacts with S [30,31,32]. |
| Nucleocapsid (N) | Nucleocapsid protein, binding to RNA genome, participates in transcription and replication, interaction with M during viral assembly [27,30,33,34], type I IFN inhibition [35,36]. |
| II. Nonstructural proteins | |
| nsp1 | Leader protein, suppresses host gene expression by ribosome association, mediates RNA replication [37,38,39,40,41], type I IFN inhibition [35,37,42,43]. |
| nsp2 | Related to the disruption of intracellular host signaling in SARS-CoV infections [44]. |
| nsp3 | Papain-like protease [45,46], polyprotein processing [47]. Type I IFN inhibition [35,46], implicated in membrane structure formation that is induced upon CoV infection and with which the RTC is thought to be associated [48,49,50]. |
| nsp4 | Implicated in membrane structure formation that is induced upon CoV infection and with which the RTC is thought to be associated [48,49]. |
| nsp5 | Chymotrypsin-like protease (3CLpro) (main protease), polyprotein processing [51,52]. |
| nsp6 | Induction of autophagosomes and limit of autophagosome expansion [53]. INF inhibition [43], implicated in membrane structure formation that is induced upon CoV infection and with which the RTC is thought to be associated [48]. |
| nsp7 | Processivity cofactor for RdRp [54,55]. |
| nsp8 | Processivity cofactor for RdRp [54,55]. |
| nsp9 | Single-strand nucleic acid-binding protein [56,57]. Possibly involved in the capping process: nsp9 may inhibit nsp12 NiRAN GTase activity in an intermediate state of RTC for further cap structure synthesis [58]. |
| nsp10 | Increases nsp14 exoribonuclease and nsp16 2′-O-methyltransferase activities [54,59,60,61]. |
| nsp11 | Unknown |
| nsp12 | RNA-dependent RNA polymerase (RdRp), replication and transcription of the viral RNA genome [62,63,64], type I IFN inhibition [35]. |
| nsp13 | Superfamily 1 helicase with a zinc-binding domain involved in RTC: participates in capping [58], unwinds RNA duplexes with 5′ to 3′ direction [65,66,67], and has 5’ triphosphophatase activity [68]. Type I INF inhibition [35,43,69]. |
| nsp14 | Proofreading exoribonuclease and N7 guanine-methyl transferase activity involved in the viral mRNA cap synthesis [70,71,72,73,74]. |
| nsp15 | Uridylate-specific endoribonuclease activity [75], may counteract double-strand RNA sensing [76]. Type I INF inhibition [69]. |
| nsp16 | 2′-O-Methyltransferase: mRNAs cap 2′-O-ribose methylation to the 5′-cap structure [60,77,78]. |
| III. Accessory proteins | |
| 3a | Type I INF inhibition [43], virulence [79], NF-κB activation [80,81], JNK and IL-8 activation [80], ion-channel activity [81], enhanced production of inflammatory chemokines [80], apoptosis induction, and necrosis [82,83]. |
| 6 | Type I INF inhibition [35,36,43,69], enhances viral replication [84], virulence [79]. |
| 7a | Type I INF inhibition [43], NF-κB activation [80], JNK and IL-8 activation [80], modulation of the inflammatory response [85]. |
| 7b | Unknown |
| 8 | Type I INF inhibition [36], mediates immune evasion [86,87,88] and inflammation [89], interacts with proteins involved in ER protein quality control and ubiquitin-dependent endoplasmic reticulum-associated degradation pathways [90,91]. |
| 10 | There is controversy regarding its expression and whether it is a coding protein [92,93]. May affect the immune response [94,95]. |
| Locus | Number of Sequences | Location | Length (bp) | Number of Polymorphisms | Ts:Tv Ratio | Mean Mutation Frequency |
|---|---|---|---|---|---|---|
| nsp1 | 86,080 | 266–805 | 540 | 621 | 1:0.64 | 1.34 × 10−5 |
| nsp2 | 85,659 | 806–2719 | 1914 | 2446 | 1:0.87 | 1.49 × 10−5 |
| nsp3 | 83,819 | 2720–8554 | 5835 | 7310 | 1:0.98 | 1.49 × 10−5 |
| nsp4 | 84,434 | 8555–10,054 | 1500 | 1130 | 1:0.49 | 8.92 × 10−6 |
| nsp5 | 85,208 | 10,055–10,972 | 918 | 605 | 1:0.44 | 7.73 × 10−6 |
| nsp6 | 85,511 | 10,973–11,842 | 870 | 777 | 1:0.73 | 1.04 × 10−5 |
| nsp7 | 86,668 | 11,843–12,091 | 249 | 257 | 1:0.78 | 1.19 × 10−5 |
| nsp8 | 86,849 | 12,092–12,685 | 594 | 405 | 1:0.43 | 7.85 × 10−6 |
| nsp9 | 86,713 | 12,686–13,024 | 339 | 262 | 1:0.45 | 8.91 × 10−6 |
| nsp10 | 84,592 | 13,025–13,441 | 417 | 290 | 1:0.51 | 8.22 × 10−6 |
| nsp11 | 84,593 | 13,442–13,480 | 39 | 39 | 1:1.29 | 1.18 × 10−5 |
| nsp12 | 84,069 | 13,442–16,236 | 2796 | 2934 | 1:1 | 1.25 × 10−5 |
| nsp13 | 85,477 | 16,237–18,039 | 1803 | 1212 | 1:0.49 | 7.86 × 10−6 |
| nsp14 | 84,666 | 18,040–19,620 | 1581 | 1210 | 1:0.50 | 9.04 × 10−6 |
| nsp15 | 85,788 | 19,621–20,658 | 1038 | 1021 | 1:0.78 | 1.15 × 10−5 |
| nsp16 | 85,050 | 20,659–21,552 | 894 | 651 | 1:0.65 | 8.56 × 10−6 |
| gene S | 83,928 | 21,563–25,384 | 3819 | 5486 | 1:1.28 | 1.71 × 10−5 |
| ORF3a | 86,034 | 25,393–26,220 | 825 | 1055 | 1:0.90 | 1.49 × 10−5 |
| gene E | 85,937 | 26,245–26,472 | 225 | 234 | 1:0.92 | 1.21 × 10−5 |
| gene M | 85,720 | 26,523–27,191 | 666 | 522 | 1:0.65 | 9.14 × 10−6 |
| ORF6 | 85,701 | 27,202–27,387 | 183 | 194 | 1:0.81 | 1.24 × 10−5 |
| ORF7a | 82,217 | 27,394–27,759 | 363 | 621 | 1:1.16 | 2.08 × 10−5 |
| ORF7b | 82,083 | 27,756–27,887 | 129 | 133 | 1:0.82 | 1.26 × 10−5 |
| ORF8 | 84,992 | 27,894–28,259 | 363 | 513 | 1:0.92 | 1.66 × 10−5 |
| gene N | 70,124 | 28,274–29,533 | 1257 | 2277 | 1:1.49 | 2.58 × 10−5 |
| ORF10 | 82,312 | 29,558–29,674 | 114 | 129 | 1:0.55 | 1.37 × 10−5 |
| Complete Genome | 32,334 | 1:0.90 | 1.24 × 10−5 | |||
| Non-structural proteins | 21,170 | 1:0.78 | 1.05 × 10−5 | |||
| Structural proteins | 8519 | 1:2.26 | 1.60 × 10−5 | |||
| Accessory proteins | 2645 | 1:0.93 | 1.52 × 10−5 | |||
| Protein | Number of Sequences | Length (aa) | Number of Changes (aa; Deletions; Stops) | Mean Changes per Sequence * | Variable Positions (%) | aa Conservation (%) |
|---|---|---|---|---|---|---|
| nsp1 | 86,080 | 180 | 438 (404; 32; 2) | 0.10 | 92.78 | 99.95 |
| nsp2 | 85,659 | 638 | 1614 (1545; 48; 21) | 0.41 | 93.73 | 99.94 |
| nsp3 | 83,819 | 1945 | 4921 (4364; 423; 134) | 3.22 | 91.36 | 99.83 |
| nsp4 | 84,434 | 500 | 671 (661; 4; 6) | 1.15 | 72.80 | 99.77 |
| nsp5 | 85,208 | 306 | 334 (322; 9; 3) | 0.16 | 64.71 | 99.95 |
| nsp6 | 85,511 | 290 | 514 (471; 35; 8) | 1.79 | 81.03 | 99.38 |
| nsp7 | 86,668 | 83 | 144 (129; 7; 8) | 0.02 | 90.36 | 99.98 |
| nsp8 | 86,849 | 198 | 237 (236; 0; 1) | 0.03 | 74.75 | 99.98 |
| nsp9 | 86,713 | 113 | 146 (139; 3; 4) | 0.03 | 72.57 | 99.97 |
| nsp10 | 84,592 | 139 | 154 (152; 1; 1) | 0.02 | 64.03 | 99.98 |
| nsp11 | 84,593 | 13 | 20 (20; 0; 0) | 0.00 | 76.92 | 99.97 |
| nsp12 | 84,069 | 932 | 1832 (1526; 207; 99) | 1.88 | 87.34 | 99.80 |
| nsp13 | 85,477 | 601 | 648 (638; 1; 9) | 0.69 | 63.73 | 99.89 |
| nsp14 | 84,666 | 527 | 734 (704; 19; 11) | 0.72 | 71.16 | 99.86 |
| nsp15 | 85,788 | 346 | 659 (600; 39; 20) | 0.09 | 85.26 | 99.98 |
| nsp16 | 85,050 | 298 | 385 (371; 8; 6) | 0.07 | 72.15 | 99.98 |
| S | 83,928 | 1273 | 3838 (3318; 397; 123) | 10.80 | 91.52 | 99.13 |
| ORF3a | 86,034 | 275 | 811 (736; 59; 16) | 0.87 | 95.64 | 99.68 |
| E | 85,937 | 75 | 150 (133; 10; 7) | 0.12 | 85.33 | 99.84 |
| M | 85,720 | 222 | 288 (281; 2; 5) | 0.78 | 69.82 | 99.64 |
| ORF6 | 85,701 | 61 | 138 (123; 6; 9) | 0.02 | 95.08 | 99.97 |
| ORF7a | 82,217 | 121 | 499 (408; 62; 29) | 1.08 | 99.17 | 99.10 |
| ORF7b | 82,083 | 43 | 105 (92; 8; 5) | 0.45 | 97.67 | 98.96 |
| ORF8 | 84,992 | 121 | 396 (338; 27; 31) | 1.60 | 100.00 | 98.67 |
| N | 70,124 | 419 | 1661 (1459; 170; 32) | 3.79 | 99.28 | 99.09 |
| ORF10 | 82,312 | 38 | 96 (91; 2; 3) | 0.09 | 97.37 | 99.77 |
| Complete genome | 9757 | 21,433 (19,261; 1579; 593) | 1.15 | 84.06 | 99.69 | |
| Non-structural proteins | 7109 | 13,451 (12,282; 836; 333) | 1.25 | 79.19 | 99.84 | |
| Structural proteins | 1989 | 5937 (5191; 579; 167) | 3.87 | 86.49 | 99.42 | |
| Accessory proteins | 659 | 2045 (1788; 164; 93) | 0.68 | 97.49 | 99.36 | |
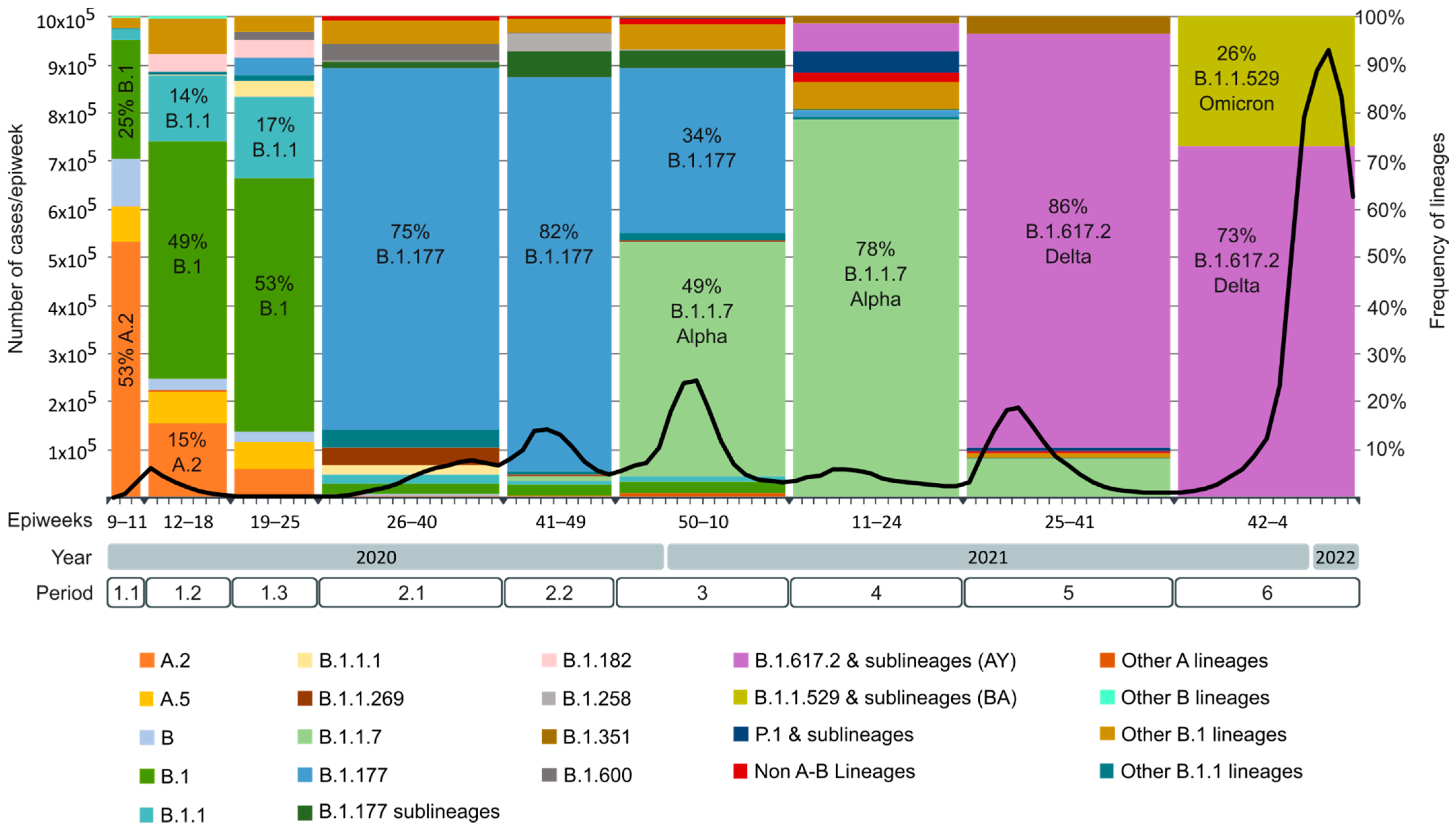
| Periods | Epiweeks | Dates | Relevant Events |
|---|---|---|---|
| Period 1 | 09.2020 to 25.2020 | 24 February 2020 to 20 June 2020 | First Spanish COVID-19 wave. First state of emergency. |
| 1.1 | 09.2020 to 11.2020 | 24 February 2020 to 14 March 2020 | From the beginning of the pandemic until the national lockdown (15 March 2020). |
| 1.2 | 12.2020 to 18.2020 | 15 March 2020 to 02 May 2020 | From the national lockdown until the beginning of the national deconfinement plan. |
| 1.3 | 19.2020 to 25.2020 | 03 May 2020 to 20 June 2020 | End of the first epidemic wave. |
| Period 2 | 26.2020 to 49.2020 | 21 June 2020 to 05 December 2020 | Second COVID-19 Spanish wave. |
| 2.1 | 26.2020 to 40.2020 | 21 June 2020 to 03 October 2020 | First peak of incidence after 2020 summer with a rise in the Rt* on early July. |
| 2.2 | 41.2020 to 49.2020 | 04 October 2020 to 05 December 2020 | Second peak of incidence before 2020 winter with another rise in the Rt in mid-October. Second state of emergency and beginning of the third state of emergency. |
| Period 3 | 50.2020 to 10.2021 | 06 December 2020 to 13 March 2021 | Third Spanish epidemic wave. Introduction of B.1.1.7 or Alpha variant. Start of the COVID-19 vaccination campaign. |
| Period 4 | 11.2021 to 24.2021 | 14 March 2021 to 19 June 2021 | Fourth Spanish epidemic wave. Alpha became the main circulating variant in Spain. Introduction of Delta variant during the last half of the period. End of the third state of emergency in May. |
| Period 5 | 25.2021 to 41.2021 | 20 June 2021 to 16 October 2021 | Fifth Spanish epidemic wave. Delta became the main circulating variant in Spain. |
| Period 6 | 42.2021 to 04.2022 | 17 October 2021 to 29 January 2022 | Sixth Spanish epidemic wave. Introduction of the Omicron variant, which quickly became the main circulating variant in Spain. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troyano-Hernáez, P.; Reinosa, R.; Holguín, Á. Evolution of SARS-CoV-2 in Spain during the First Two Years of the Pandemic: Circulating Variants, Amino Acid Conservation, and Genetic Variability in Structural, Non-Structural, and Accessory Proteins. Int. J. Mol. Sci. 2022, 23, 6394. https://doi.org/10.3390/ijms23126394
Troyano-Hernáez P, Reinosa R, Holguín Á. Evolution of SARS-CoV-2 in Spain during the First Two Years of the Pandemic: Circulating Variants, Amino Acid Conservation, and Genetic Variability in Structural, Non-Structural, and Accessory Proteins. International Journal of Molecular Sciences. 2022; 23(12):6394. https://doi.org/10.3390/ijms23126394
Chicago/Turabian StyleTroyano-Hernáez, Paloma, Roberto Reinosa, and África Holguín. 2022. "Evolution of SARS-CoV-2 in Spain during the First Two Years of the Pandemic: Circulating Variants, Amino Acid Conservation, and Genetic Variability in Structural, Non-Structural, and Accessory Proteins" International Journal of Molecular Sciences 23, no. 12: 6394. https://doi.org/10.3390/ijms23126394
APA StyleTroyano-Hernáez, P., Reinosa, R., & Holguín, Á. (2022). Evolution of SARS-CoV-2 in Spain during the First Two Years of the Pandemic: Circulating Variants, Amino Acid Conservation, and Genetic Variability in Structural, Non-Structural, and Accessory Proteins. International Journal of Molecular Sciences, 23(12), 6394. https://doi.org/10.3390/ijms23126394








