Transcriptome Analysis Reveals the Regulatory Networks of Cytokinin in Promoting Floral Feminization in Castanea henryi
Abstract
1. Introduction
2. Results
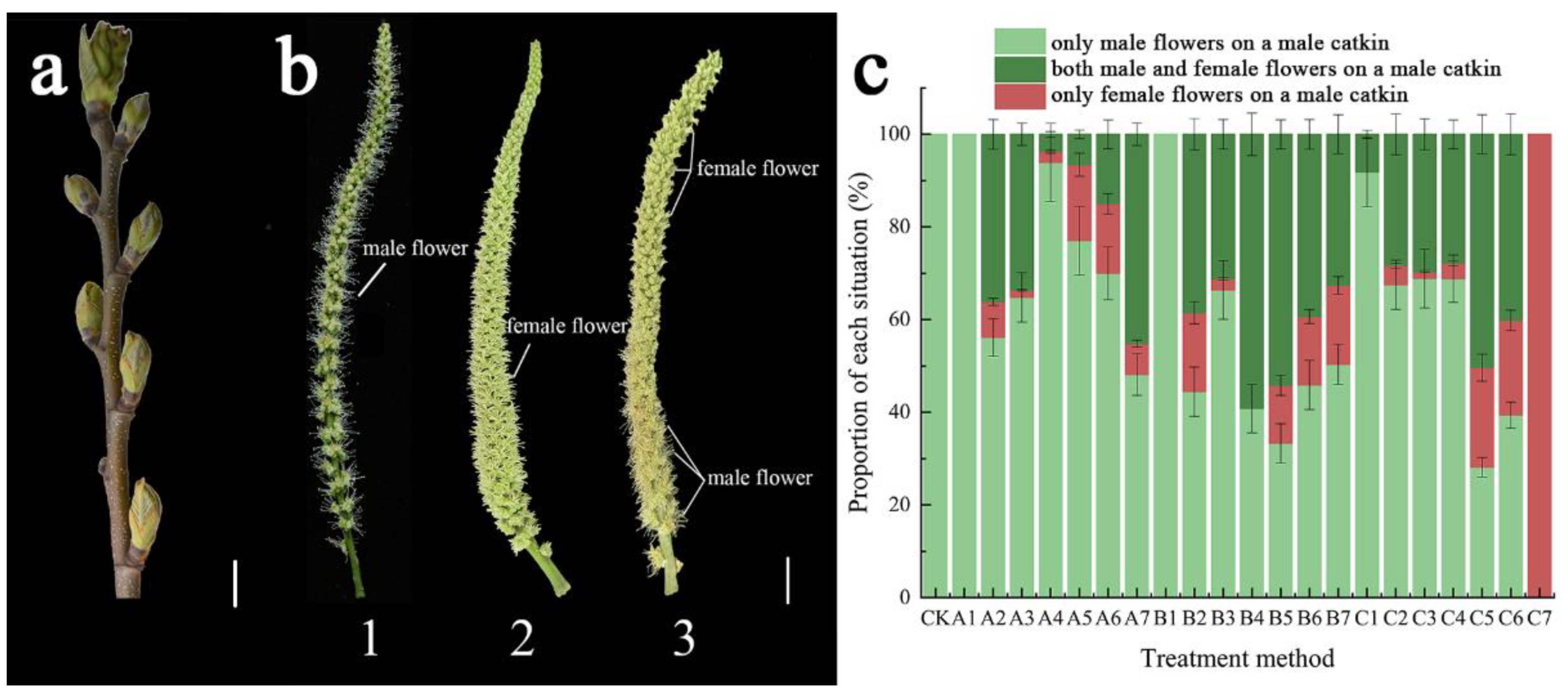
2.1. Feminization Effects of CPPU Treatment on Floral Development in C. henryi
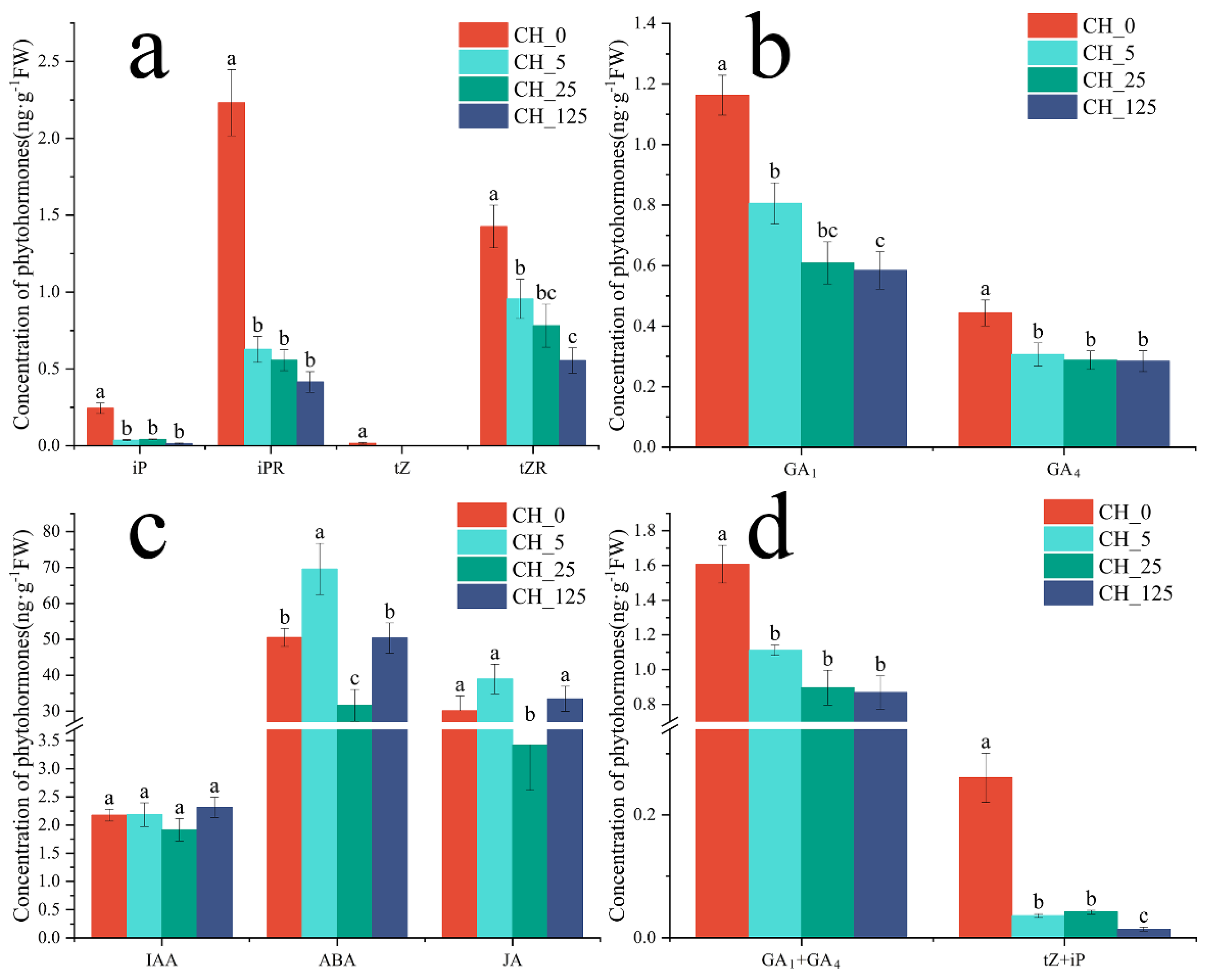
2.2. Effect of CPPU Treatment on the Content of Endogenous Hormones in C. henryi Male Catkins
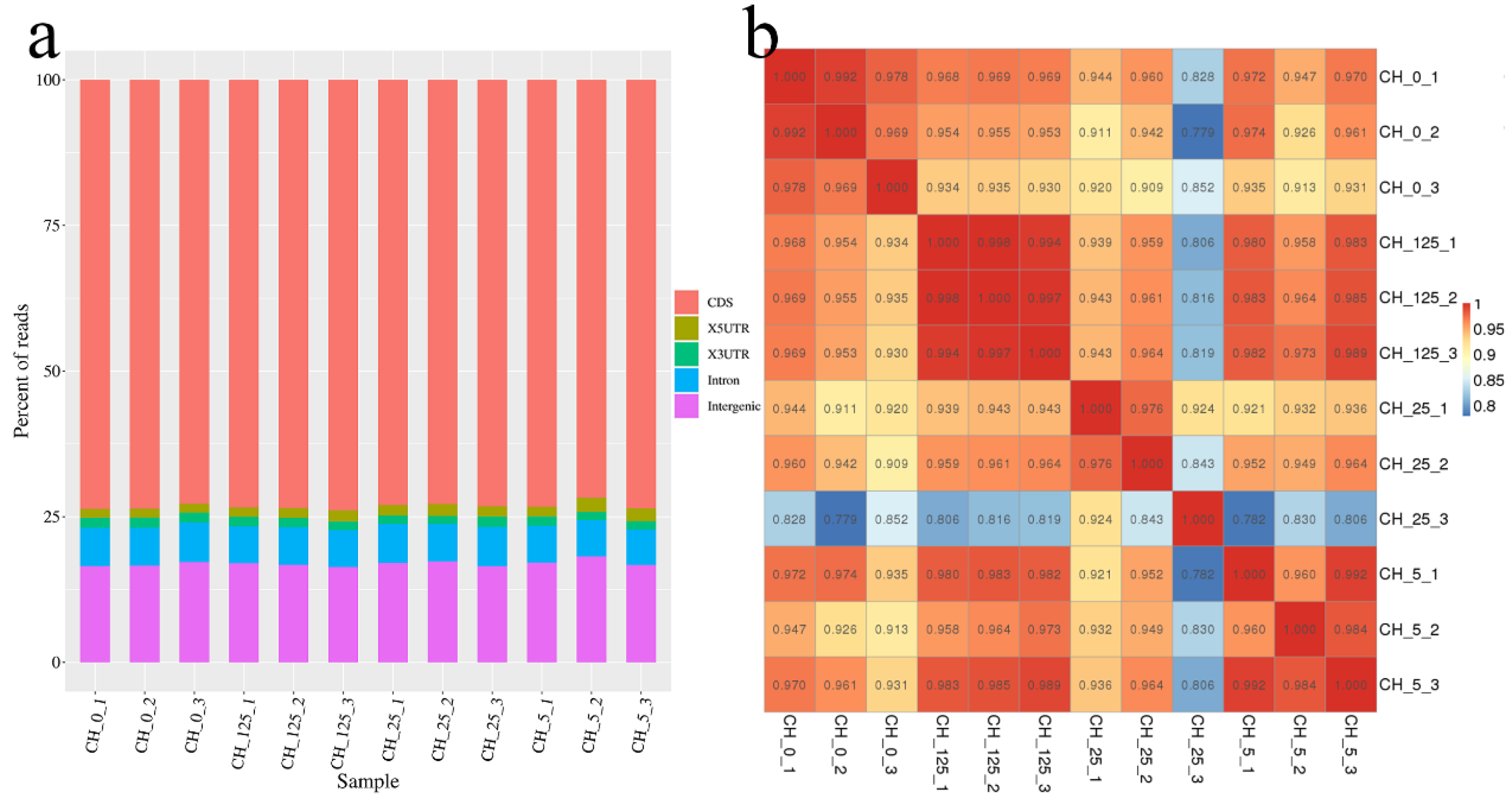
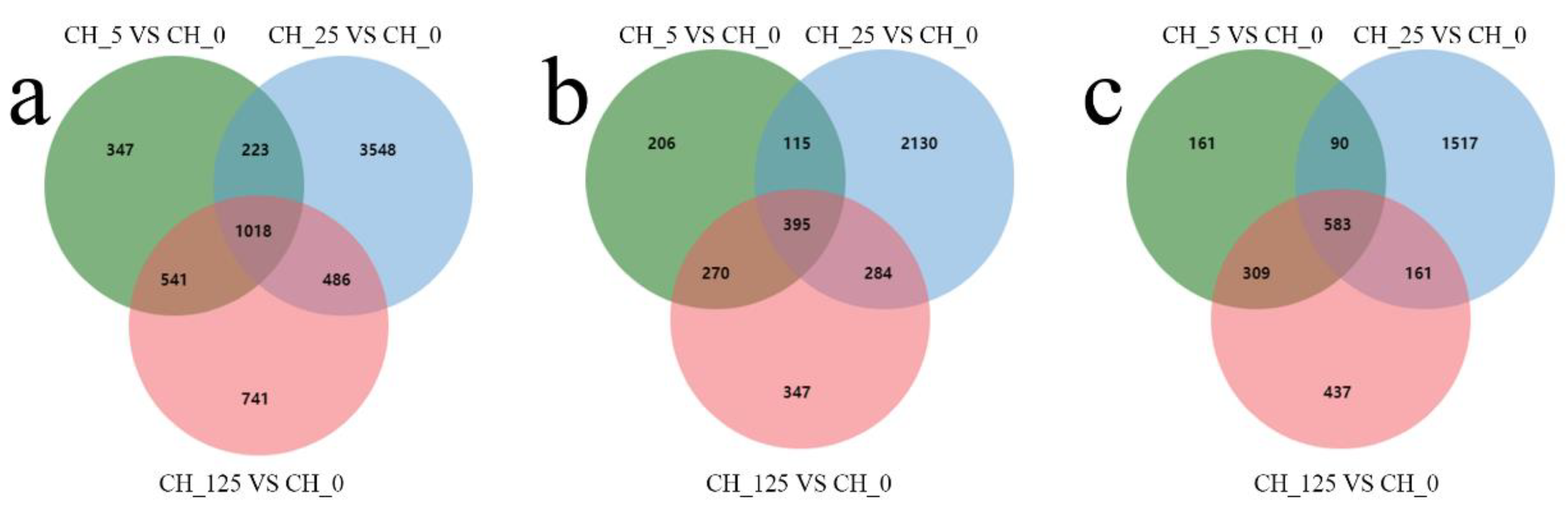
2.3. Transcriptome Sequencing of the Male Catkins of C. henryi in Response to CPPU Treatment
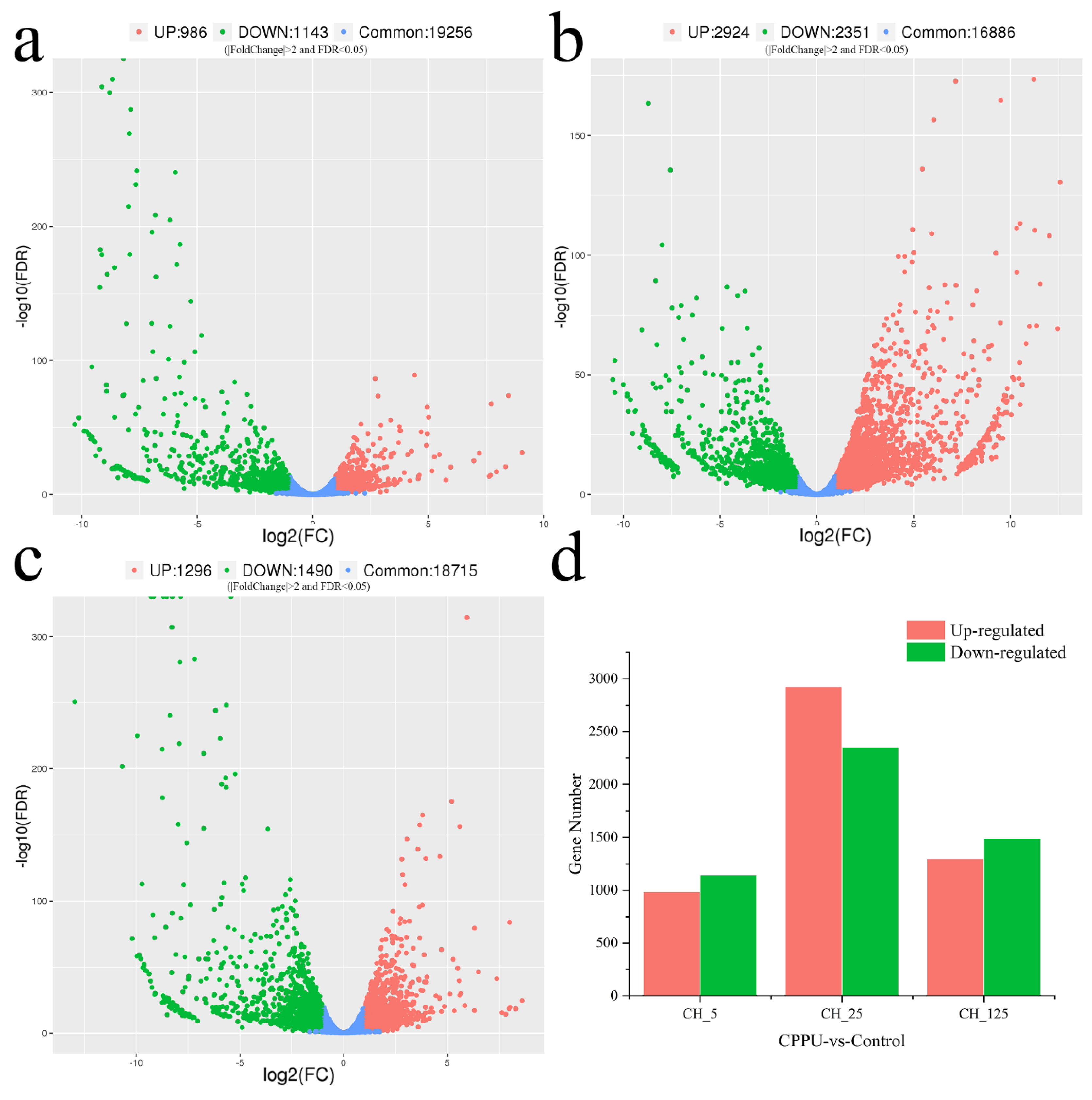
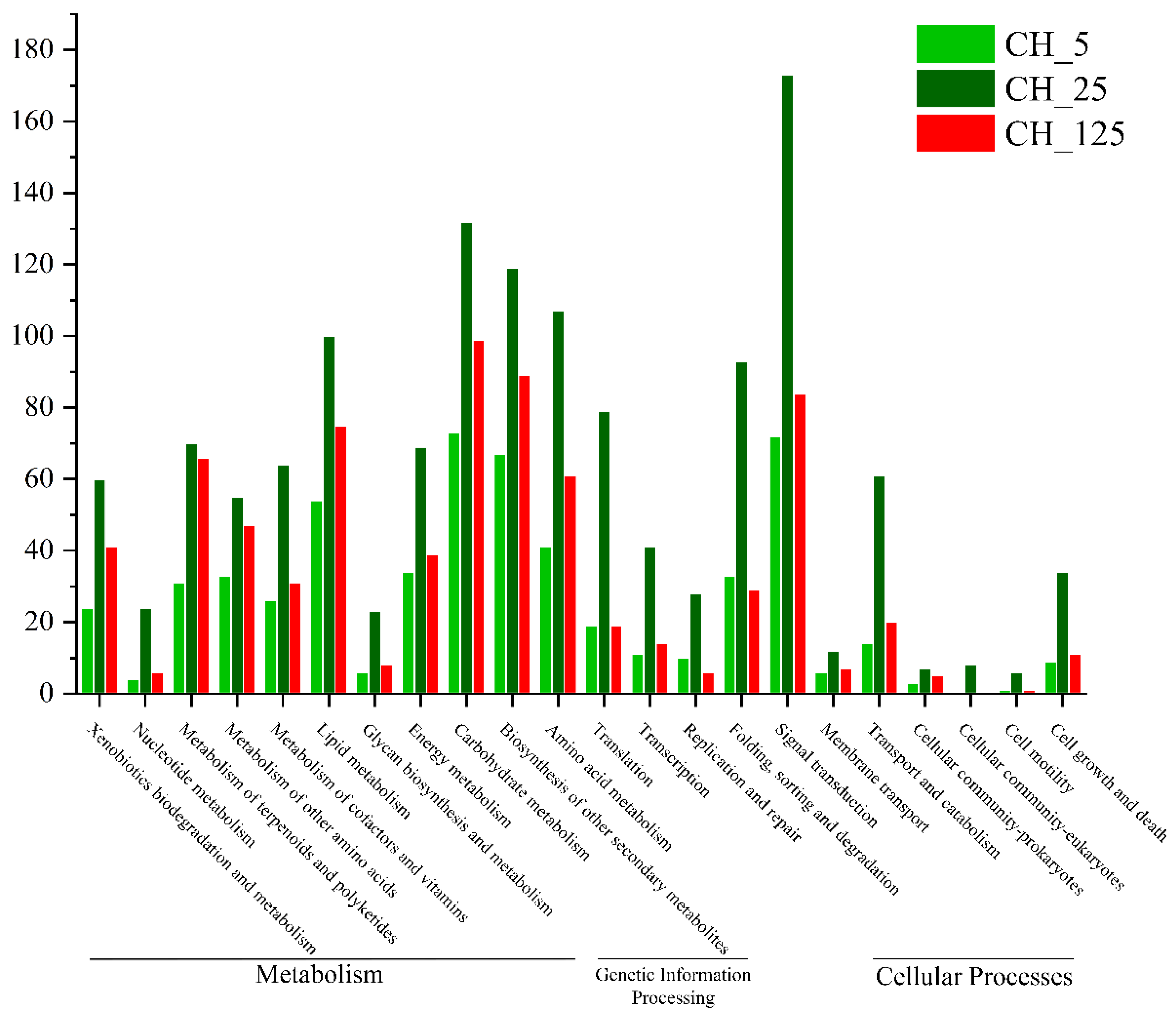
2.4. Functional Annotation and Classification of Differentially Expressed Genes (DEGs)
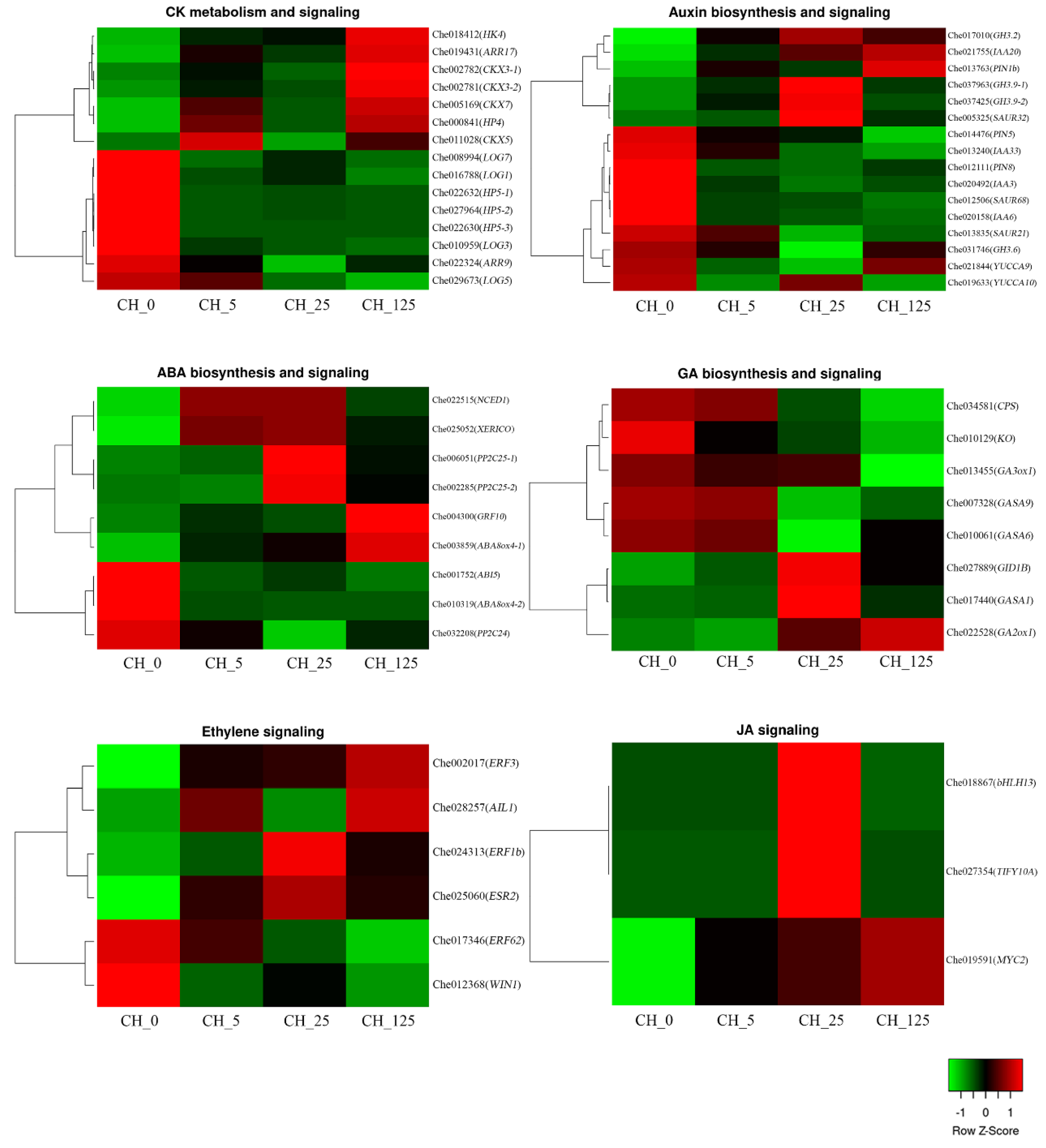
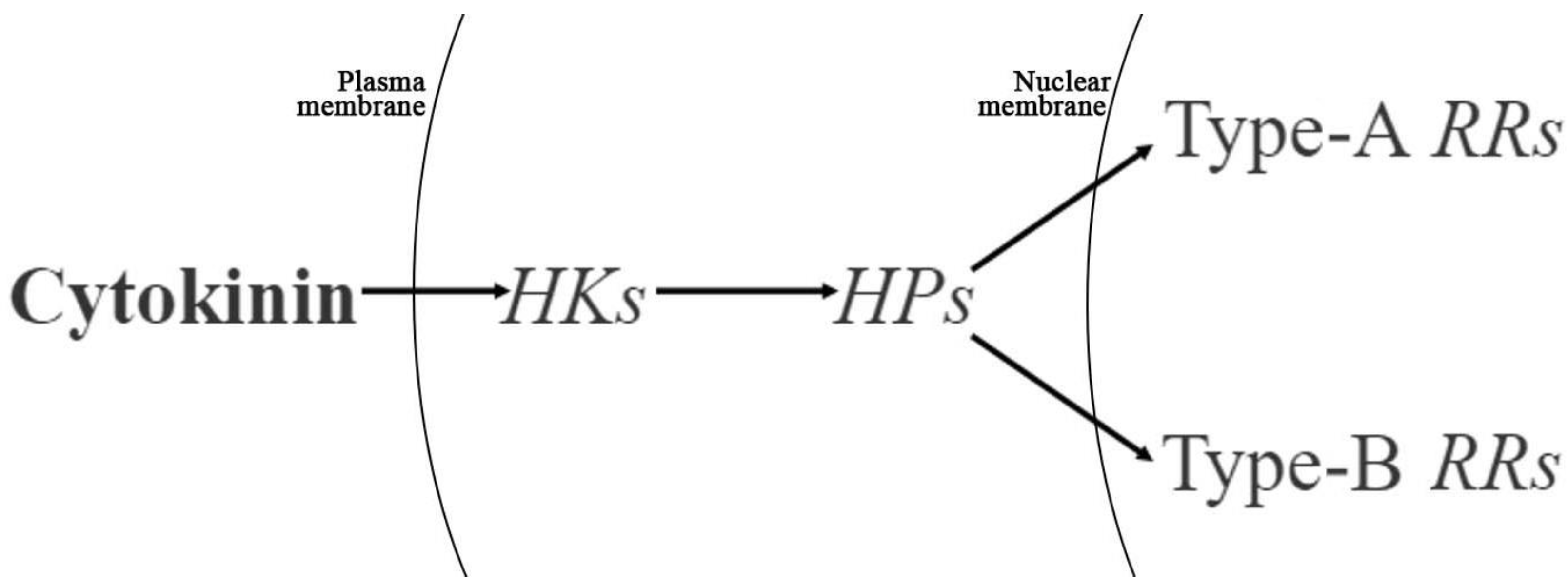
2.5. DEGs Related to Endogenous CK Biosynthesis and Signaling
2.6. DEGs Related to Endogenous Hormonal Biosynthesis and Signaling
2.7. Screening of Key Genes in the Hormone Biosynthesis Pathway
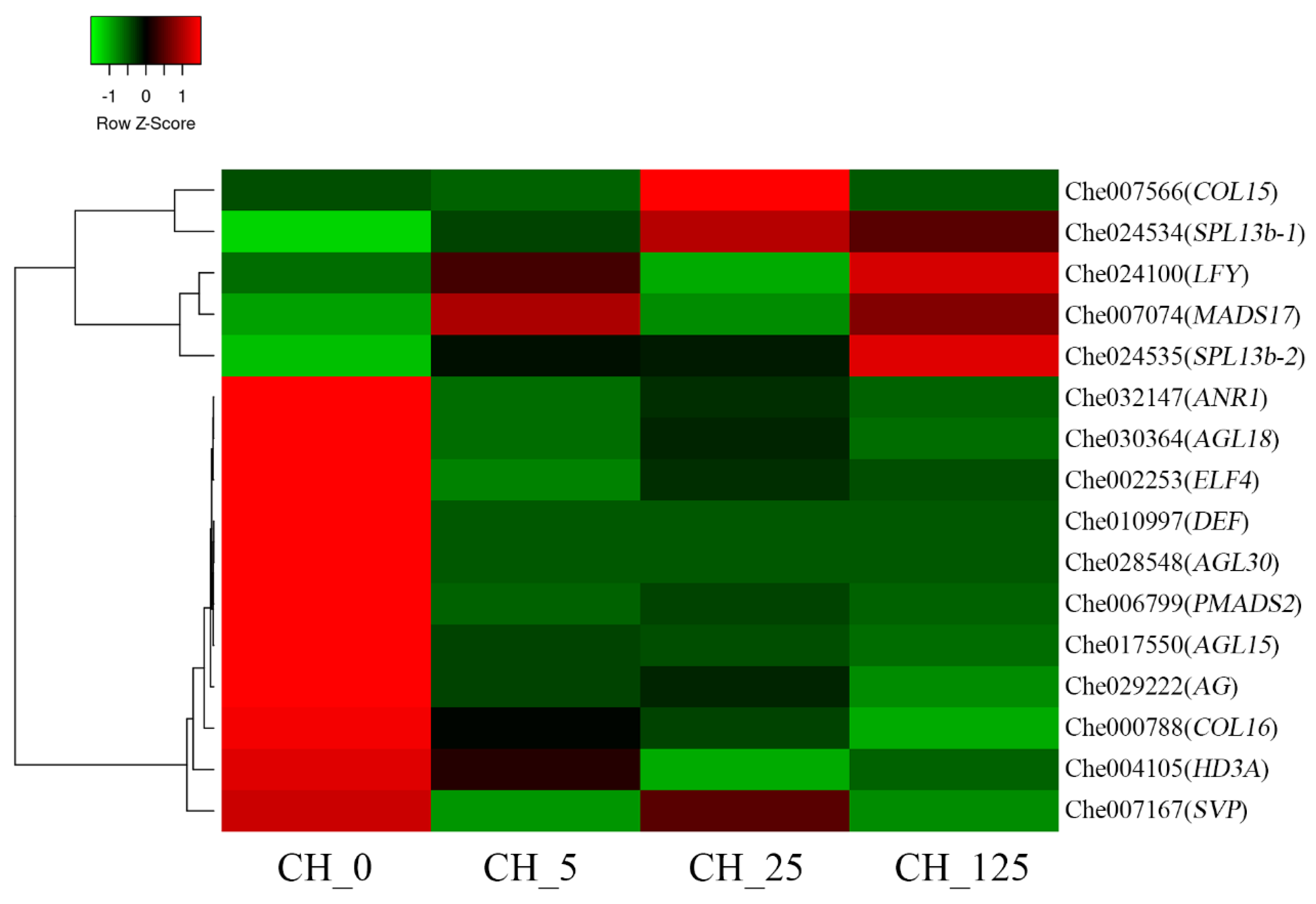
2.8. Identification of the DEGs Related to Floral Development
3. Discussion
3.1. CPPU Has Strong Feminization Effects on the Floral Development in C. henryi
3.2. Exogenous Cytokinin Treatment Alters Endogenous Cytokinin Levels and the Expression of Cytokinin Biosynthesis and Signaling-Related Genes
3.3. Exogenous CK Treatment Alters Endogenous GA Levels and the Expression of GA Biosynthesis and Signaling-Related Genes
3.4. Exogenous CK Treatment Alters the Levels of Other Endogenous Phytohormones
3.5. CK Regulated the Expression of Genes Related to Floral Organ Development
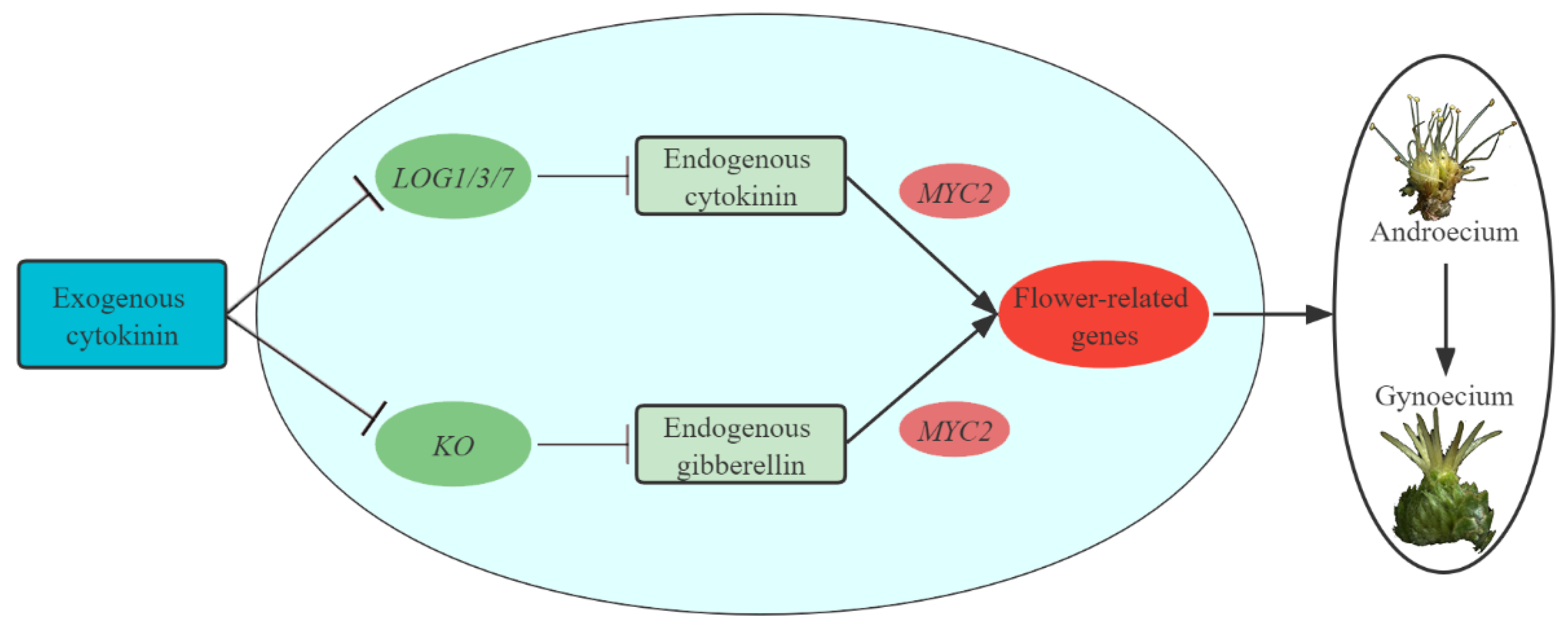
3.6. Regulatory Networks Participate in Promoting Floral Feminization in C. henryi
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Treatment
4.2. Sample Collection
4.3. Determination of Endogenous Hormones
4.4. Illumina Transcriptome Sequencing and Assembly of Clean Reads
4.5. Transcript Annotation and Gene Expression Analysis
4.6. Validation of DEGs by RT-qPCR
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ertürk, Ü.; Mert, C.; Soylu, A. Chemical composition of fruits of some important chestnut cultivars. Braz. Arch. Biol. Technol. 2006, 49, 183–188. [Google Scholar] [CrossRef]
- Fan, X.; Yuan, D.; Tang, J.; Tian, X.; Zhang, X.; Wang, B.; Tan, X. Biological characteristics of flowering and pollination in Castanea henryi. Sci. Silvae Sin. 2014, 50, 42–48. [Google Scholar] [CrossRef]
- Chen, M.S.; Pan, B.Z.; Fu, Q.; Tao, Y.B.; Herrera, J.M.; Niu, L.; Ni, J.; Dong, Y.; Zhao, M.L.; Xu, Z.F. Comparative Transcriptome Analysis between Gynoecious and Monoecious Plants Identifies Regulatory Networks Controlling Sex Determination in Jatropha curcas. Front. Plant Sci. 2017, 7, 1953. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, O.-A.; Sonia, V.-S.; Aldebaran, C.; Dubrovsky, J.G.; Felipe, C.-G. Inception of maleness: Auxin contribution to flower masculinization in the dioecious cactus Opuntia stenopetala. Planta 2012, 236, 225–238. [Google Scholar] [CrossRef]
- Yin, T.; Quinn, J.A. Tests of a Mechanistic Model of One Hormone Regulating Both Sexes in Cucumis sativus (Cucurbitaceae). Am. J. Bot. 1995, 82, 1537–1546. [Google Scholar] [CrossRef]
- Bang-Zhen, P.; Mao-Sheng, C.; Jun, N.; Zeng-Fu, X. Transcriptome of the inflorescence meristems of the biofuel plant Jatropha curcas treated with cytokinin. BMC Genom. 2014, 15, 974. [Google Scholar] [CrossRef]
- Irish, E.E.; Nelson, T. Sex Determination in Monoecious and Dioecious Plants. Plant Cell 1989, 1, 737. [Google Scholar] [CrossRef]
- Pan, B.-Z.; Xu, Z.-F. Benzyladenine Treatment Significantly Increases the Seed Yield of the Biofuel Plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef]
- Ming, X.; Tao, Y.-B.; Fu, Q.; Tang, M.; He, H.; Chen, M.-S.; Pan, B.-Z.; Xu, Z.-F. Flower-Specific Overproduction of Cytokinins Altered Flower Development and Sex Expression in the Perennial Woody Plant Jatropha curcas L. Int. J. Mol. Sci. 2020, 21, 640. [Google Scholar] [CrossRef]
- Bruce, M.; Zwar, J. Cytokinin activity of some substituted ureas and thioureas. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 1966, 165, 245–265. [Google Scholar] [CrossRef]
- Jun, A.; Aimin, L.; Changyu, L.; Jun, W.; Yujie, S. The effect of cytokinin on sex conversion of male plants of Vitis amurensis. Acta Hortic. Sin. 2002, 29, 163. [Google Scholar] [CrossRef]
- Hu, X.; Hu, F.; Fan, H.; Wang, X.; Han, B.; Lin, Y. Effects of five plant growth regulators on blooming and fruit-setting of “Feizixiao’ litchi. Southwest China J. Agric. Sci. 2016, 29, 915–919. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, N.; Sun, T.-p.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef] [PubMed]
- Ivo, F.; Marta, K.; Tomás, H.; Jitka, F.; Petr, G. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef]
- Michael, A.; Wensheng, Q.; François, M.; Barbara, M. Adenine phosphoribosyltransferase isoforms of Arabidopsis and their potential contributions to adenine and cytokinin metabolism. Physiol. Plant. 2002, 115, 56–68. [Google Scholar] [CrossRef]
- Šmehilová, M.; Dobrůšková, J.; Novák, O.; Takáč, T.; Galuszka, P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 2016, 7, 1264. [Google Scholar] [CrossRef]
- Ferreira, F.J.; Kieber, J.J. Cytokinin signaling. Curr. Opin. Plant Biol. 2005, 8, 518–525. [Google Scholar] [CrossRef]
- Xiaoming, F.; Deyi, Y.; Xiaoming, T.; Zhoujun, Z.; Meilan, L.; Heping, C. Comprehensive Transcriptome Analysis of Phytohormone Biosynthesis and Signaling Genes in the Flowers of Chinese Chinquapin (Castanea henryi). J. Agric. Food Chem. 2017, 65, 1429–1450. [Google Scholar] [CrossRef]
- Weigel, D.; Meyerowitz, E.M. The ABCs of floral homeotic genes. Cell 1994, 78, 203–209. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Zürcher, E.; Müller, B. Cytokinin Synthesis, Signaling, and Function—Advances and New Insights. Int. Rev. Cell Mol. Biol. 2016, 324, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.; Calderon-Urrea, A. The sex determination process in maize. Science 1994, 266, 1501–1505. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J. Auxin and Sexuality in Cannabis sativa. Physiol. Plant. 1956, 9, 153–162. [Google Scholar] [CrossRef]
- Ando, S.; Sato, Y.; Kamachi, S.I.; Sakai, S. Isolation of a MADS-box gene (ERAF17) and correlation of its expression with the induction of formation of female flowers by ethylene in cucumber plants (Cucumis sativus L.). Planta 2001, 213, 943–952. [Google Scholar] [CrossRef]
- Takashi, A.; Henry, I.M.; Haruka, O.; Takuya, M.; Kenji, B.; Ikuo, K.; Ryutaro, T. A Y-Encoded Suppressor of Feminization Arose via Lineage-Specific Duplication of a Cytokinin Response Regulator in Kiwifruit. Plant Cell 2018, 30, 780–795. [Google Scholar] [CrossRef]
- Moncaleán, P.; Alonso, P.; Centeno, M.; Cortizo, M.; Rodríguez, A.; Fernández, B.; Ordás, R. Organogenic responses of Pinus pinea cotyledons to hormonal treatments: BA metabolism and cytokinin content. Tree Physiol. 2005, 25, 1–9. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Jones, R.J.; Schreiber, B. Role and function of cytokinin oxidase in plants. Plant Growth Regul. 1997, 23, 123–134. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin Signaling Networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef]
- Muller, B.; Sheen, J. Advances in cytokinin signaling. Science 2007, 318, 68–69. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.; Kieber, J.J. Cytokinin signaling: Two-components and more. Trends Plant Sci. 2008, 13, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Morrone, D.; Chen, X.; Coates, R.M.; Peters, R.J. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. 2010, 431, 337–347. [Google Scholar] [CrossRef]
- Xu, M.; Lu, Y.; Yang, H.; He, J.; Hu, Z.; Hu, X.; Luan, M.; Zhang, L.; Fan, Y.; Wang, L. ZmGRF, a GA regulatory factor from maize, promotes flowering and plant growth in Arabidopsis. Plant Mol. Biol. 2015, 87, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Sheldon, C.C.; Olive, M.R.; Walker, A.R.; Zeevaart, J.A.D.; Peacock, W.J.; Dennis, E.S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 1998, 95, 9019–9024. [Google Scholar] [CrossRef] [PubMed]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Kotilainen, M.; Helariutta, Y.; Mehto, M.; Pöllänen, E.; Albert, V.A.; Elomaa, P.; Teeri, T.H. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 1999, 11, 1093–1104. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.-G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Golenberg, E.M.; West, N.W. Hormonal interactions and gene regulation can link monoecy and environmental plasticity to the evolution of dioecy in plants. Am. J. Bot. 2013, 100, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Astot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044. [Google Scholar] [CrossRef] [PubMed]
- Ponnala, L.; Wang, Y.; Sun, Q.; van Wijk, K.J. Correlation of mRNA and protein abundance in the developing maize leaf. The Plant J. 2014, 78, 424–440. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.A.; Bowman, J.L.; Kempin, S.A.; Ma, H.; Meyerowitz, E.M.; Yanofsky, M.F. Manipulation of flower structure in transgenic tobacco. Cell 1992, 71, 133–143. [Google Scholar] [CrossRef]
- Meyerowitz, E.M.; Weigel, D. Activation of Floral Homeotic Genes in Arabidopsis. Science 1993, 261, 1723–1726. [Google Scholar] [CrossRef]
- Jack, T. Molecular and genetic mechanisms of floral control. Plant Cell 2004, 16, S1–S17. [Google Scholar] [CrossRef]
- Jeon, J.-S.; Jang, S.; Lee, S.; Nam, J.; Kim, C.; Lee, S.-H.; Chung, Y.-Y.; Kim, S.-R.; Lee, Y.H.; Cho, Y.-G. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 2000, 12, 871–884. [Google Scholar] [CrossRef]
- Hu, Y.; Liang, W.; Yin, C.; Yang, X.; Ping, B.; Li, A.; Jia, R.; Chen, M.; Luo, Z.; Cai, Q.; et al. Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 2015, 8, 1366–1384. [Google Scholar] [CrossRef]
- Rijpkema, A.; Gerats, T.; Vandenbussche, M. Genetics of floral development in Petunia. Adv. Bot. Res. 2006, 44, 237–278. [Google Scholar] [CrossRef]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Caruso, L.V.; Downie, A.B.; Perry, S.E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 2004, 16, 1206–1219. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Yamashino, T.; Matsushika, A.; Fujimori, T.; Sato, S.; Kato, T.; Tabata, S.; Mizuno, T. A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 619–629. [Google Scholar] [CrossRef]
- Jang, G.; Chang, S.H.; Um, T.Y.; Lee, S.; Kim, J.-K.; Choi, Y.D. Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Sci. Rep. 2017, 7, 10212. [Google Scholar] [CrossRef]
- Hong, G.-J.; Xue, X.-Y.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Fan, X.; Yuan, D.; Tang, J.; Tian, X.; Zhang, L.; Zou, F.; Tan, X. Sporogenesis and gametogenesis in Chinese chinquapin (Castanea henryi (Skam) Rehder & Wilson) and their systematic implications. Trees 2015, 29, 1713–1723. [Google Scholar] [CrossRef]
- Bian, T.; Ma, Y.; Guo, J.; Wu, Y.; Shi, D.; Guo, X. Herbaceous peony (Paeonia lactiflora Pall.) PlDELLA gene negatively regulates dormancy release and plant growth. Plant Sci. 2020, 297, 110539. [Google Scholar] [CrossRef]
- Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Daehwan, K.; Ben, L.; Steven, L.S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Mihaela, P.; Geo, M.P.; Corina, M.A.; Tsung-Cheng, C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Yang, L.; Smyth, G.K.; Wei, S. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Kenneth, M.; Lorian, S.; Barbara, W. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Consortium, G.O. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Guangchuang, Y.; Li-Gen, W.; Yanyan, H.; Qing-Yu, H. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Minoru, K.; Susumu, G.; Yoko, S.; Masayuki, K.; Miho, F.; Mao, T. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Cole, T.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

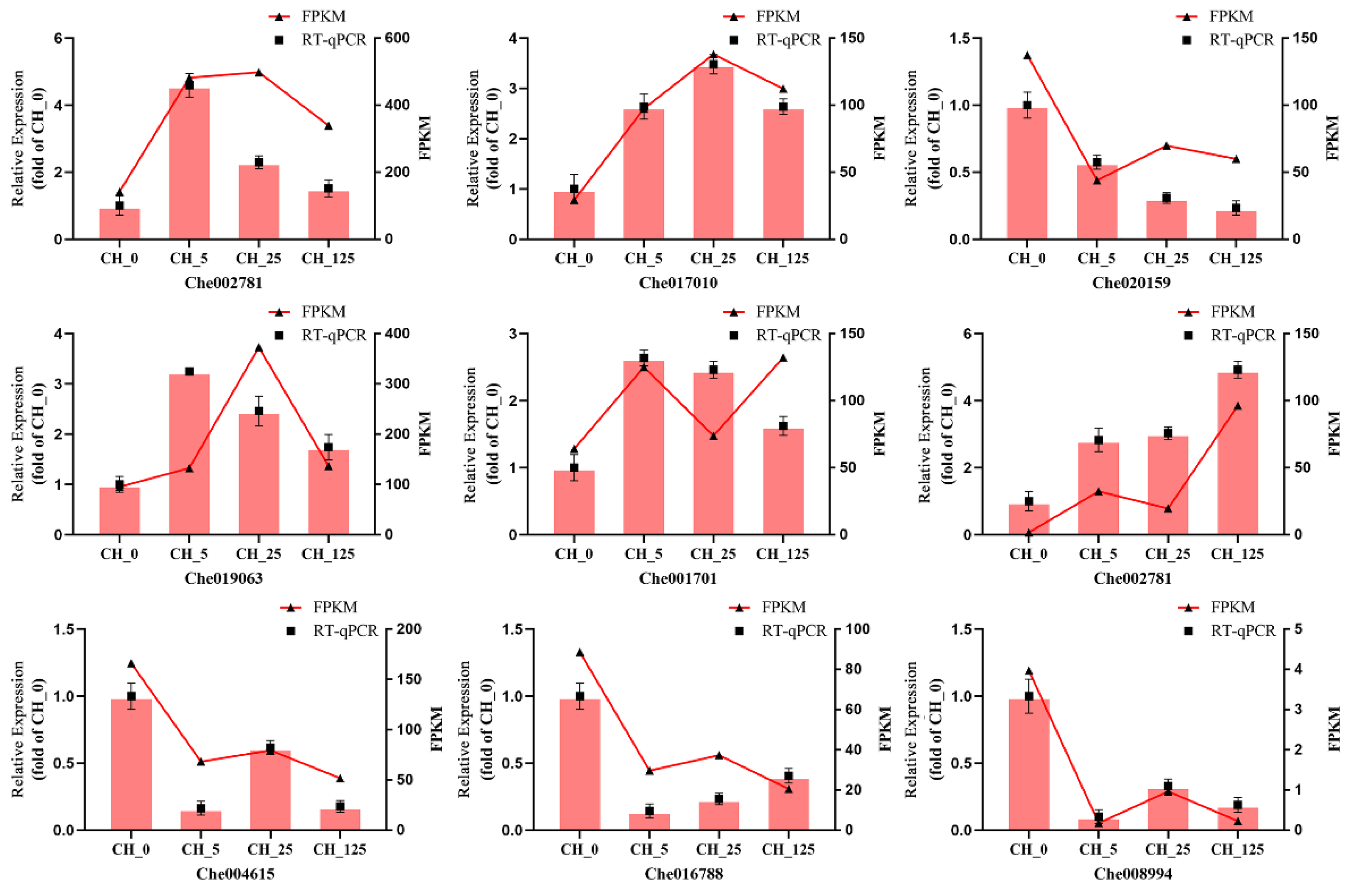
| Sample | RawReads | RawBases | CleanReads | CleanBases | CleanRatio | Q20 | Q30 | GC |
|---|---|---|---|---|---|---|---|---|
| CH_0_1 | 66,611,830 | 9.99 × 109 | 66,065,838 | 9.67 × 109 | 99.18% | 97.81% | 93.75% | 45.35% |
| CH_0_2 | 66,288,344 | 9.94 × 109 | 65,736,958 | 9.63 × 109 | 99.17% | 97.65% | 93.33% | 45.51% |
| CH_0_3 | 60,326,280 | 9.05 × 109 | 59,880,596 | 8.67 × 109 | 99.26% | 97.84% | 93.78% | 45.05% |
| CH_5_1 | 57,234,618 | 8.59 × 109 | 56,636,914 | 8.32 × 109 | 98.96% | 97.41% | 92.83% | 46.13% |
| CH_5_2 | 56,737,570 | 8.51 × 109 | 56,058,944 | 8.31 × 109 | 98.80% | 97.23% | 92.51% | 45.81% |
| CH_5_3 | 60,847,540 | 9.13 × 109 | 60,135,430 | 8.83 × 109 | 98.83% | 97.35% | 92.75% | 45.96% |
| CH_25_1 | 65,534,342 | 9.83 × 109 | 64,933,306 | 9.49 × 109 | 99.08% | 97.63% | 93.35% | 45.33% |
| CH_25_2 | 64,267,422 | 9.64 × 109 | 63,506,822 | 9.29 × 109 | 98.82% | 97.50% | 93.10% | 45.98% |
| CH_25_3 | 48,063,476 | 7.21 × 109 | 47,646,260 | 6.97 × 109 | 99.13% | 97.76% | 93.52% | 43.58% |
| CH_125_1 | 61,642,840 | 9.25 × 109 | 60,986,388 | 8.94 × 109 | 98.94% | 97.57% | 93.21% | 46.12% |
| CH_125_2 | 61,918,812 | 9.29 × 109 | 61,382,232 | 9.03 × 109 | 99.13% | 97.68% | 93.43% | 45.97% |
| CH_125_3 | 64,129,072 | 9.62 × 109 | 63,554,386 | 9.37 × 109 | 99.10% | 97.65% | 93.41% | 45.73% |
| Gene ID | Direction | Primer sequence (5′–3′) | Annealing Temperature (°C) | Amplicon (bp) |
|---|---|---|---|---|
| Che006645 | Forward | GGAGTTCAAGAACCGGACACCATC | 54.8 | 85 |
| Reverse | CAAGCAAACCGAGCATTCATGTTCC | |||
| Che017010 | Forward | CGAAGACGATTCCTGGTCACTACG | 54.7 | 81 |
| Reverse | GTACTTCCTCACTTGGCGAGTTAGC | |||
| Che020159 | Forward | CGGTGAGAGCATCAAGGAAGAACG | 55.9 | 81 |
| Reverse | CCCCATTTGCAGGGTCCATAAGC | |||
| Che019063 | Forward | AACAACGCCTCCAAGCTCTAATCG | 54.9 | 142 |
| Reverse | GCCTTATCGTCCTCGCCTTTGTAG | |||
| Che001701 | Forward | CAGTGGCAGGAGCTTGAACTACAG | 55.2 | 131 |
| Reverse | AGAGGGTGATGGAGGAAGTAAGGTG | |||
| Che002781 | Forward | ATGGCTCGTAATGGGGTTGTTGTG | 55.9 | 87 |
| Reverse | TGACGGGGTTCCAGAGACAGTG | |||
| Che004615 | Forward | CGTCATTGGGGTTCATGGGTATCC | 55.4 | 89 |
| Reverse | GCCTCTTCAGCAGTCTCGAATGTG | |||
| Che008994 | Forward | TTTATCTCGCCAACCGCACGTC | 55.8 | 126 |
| Reverse | CACTCCTTCCCAAACCAGCTTCG | |||
| Che016788 | Forward | GAGGTTGCCATGTTCTCGGAGTG | 55.1 | 118 |
| Reverse | GCCATTTCTGCCTTCCTTTGATGC | |||
| GAPDH | Forward | AGCAAGGACTGGAGAGGTGGAAG | 56.1 | 136 |
| Reverse | CGGTAGGAACACGGAAAGCCATC |
| Variable | CKX3-1 | CKX3-2 | CKX7 | CKX5 | LOG1 | LOG3 | LOG7 | LOG5 |
|---|---|---|---|---|---|---|---|---|
| tZ + iP | −0.58 | −0.66 | −0.80 | −0.47 | 0.991 ** | 0.996 ** | 0.987 * | 0.79 |
| Variable | KO | CPS | GA2ox1 | GA3ox1 |
|---|---|---|---|---|
| GA1 + GA4 | 0.973 * | 0.81 | −0.74 | 0.62 |
| A (5 mg·L−1) | B (25 mg·L−1) | C (125 mg·L−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3.22 | 3.29 | 4.5 | 3.22 | 3.29 | 4.5 | 3.22 | 3.29 | 4.5 | |||
| 1 | √ | 1 | √ | 1 | √ | ||||||
| 2 | √ | 2 | √ | 2 | √ | ||||||
| 3 | √ | 3 | √ | 3 | √ | ||||||
| 4 | √ | √ | 4 | √ | √ | 4 | √ | √ | |||
| 5 | √ | √ | 5 | √ | √ | 5 | √ | √ | |||
| 6 | √ | √ | 6 | √ | √ | 6 | √ | √ | |||
| 7 | √ | √ | √ | 7 | √ | √ | √ | 7 | √ | √ | √ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.-L.; Zhu, Z.-J.; Qiu, Q.; Fan, X.-M.; Yuan, D.-Y. Transcriptome Analysis Reveals the Regulatory Networks of Cytokinin in Promoting Floral Feminization in Castanea henryi. Int. J. Mol. Sci. 2022, 23, 6389. https://doi.org/10.3390/ijms23126389
Wu G-L, Zhu Z-J, Qiu Q, Fan X-M, Yuan D-Y. Transcriptome Analysis Reveals the Regulatory Networks of Cytokinin in Promoting Floral Feminization in Castanea henryi. International Journal of Molecular Sciences. 2022; 23(12):6389. https://doi.org/10.3390/ijms23126389
Chicago/Turabian StyleWu, Guo-Long, Zhou-Jun Zhu, Qi Qiu, Xiao-Ming Fan, and De-Yi Yuan. 2022. "Transcriptome Analysis Reveals the Regulatory Networks of Cytokinin in Promoting Floral Feminization in Castanea henryi" International Journal of Molecular Sciences 23, no. 12: 6389. https://doi.org/10.3390/ijms23126389
APA StyleWu, G.-L., Zhu, Z.-J., Qiu, Q., Fan, X.-M., & Yuan, D.-Y. (2022). Transcriptome Analysis Reveals the Regulatory Networks of Cytokinin in Promoting Floral Feminization in Castanea henryi. International Journal of Molecular Sciences, 23(12), 6389. https://doi.org/10.3390/ijms23126389





