Abstract
The recent novel coronavirus (SARS-CoV-2) disease (COVID-19) outbreak created a severe public health burden worldwide. Unfortunately, the SARS-CoV-2 variant is still spreading at an unprecedented speed in many countries and regions. There is still a lack of effective treatment for moderate and severe COVID-19 patients, due to a lack of understanding of the SARS-CoV-2 life cycle. Lysosomes, which act as “garbage disposals” for nearly all types of eukaryotic cells, were shown in numerous studies to support SARS-CoV-2 replication. Lysosome-associated pathways are required for virus entry and exit during replication. In this review, we summarize experimental evidence demonstrating a correlation between lysosomal function and SARS-CoV-2 replication, and the development of lysosomal perturbation drugs as anti-SARS-CoV-2 agents.
1. Introduction
Coronaviruses (CoVs), a highly diverse family of enveloped positive-sense single-stranded RNA viruses, belong to the subfamily Orthocoronavirinae within the family Coronaviridae. Four genera are found within the subfamily Orthocoronavirinae, namely Alphacoronavirus, Betacoronavirus (βCoV), Gammacoronavirus, and Deltacoronavirus. βCoVs exclusively infect mammalian species, leading mainly to respiratory and enteric illnesses that can be deadly [1,2]. Notably, βCoV species that can cause severe, life-threatening respiratory illnesses in humans include Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [3]. To date, no prophylactic or therapeutic treatment is approved to prevent the severe illness and permanent lung damage caused by these βCoV species once the infection progresses beyond the early infection stage [4,5].
SARS-CoV-2, a member of the βCoV SARS phylogenetic cluster, is the pathogen responsible for causing the “coronavirus disease of 2019” (COVID-19) pandemic. This pandemic continues to pose insurmountable challenges to healthcare systems and healthcare professionals globally, while both the disease itself and local and national restrictions to contain the pandemic continue to directly and indirectly unleash social, health, and economic devastation on humankind. To prevent and treat this disease, more effective vaccines, drugs, and other tools are urgently needed; however, whether the use of such tools will finally put an end to COVID-19 is still debated [6,7]. On the one hand, although vaccination is underway in some countries, SARS-CoV-2 mutations and the incomplete protection afforded by vaccines (even against the unmutated virus) led to vaccine hesitancy that allowed the virus to adapt and continue to spread and cause illness. On the other hand, the United States Food and Drug Administration (FDA) approved several neutralizing antibodies, such as bamlanivimab, etesevimab, casirivimab, and imdevimab, for the treatment of hospitalized COVID-19 patients [8]. However, their extensive usage was limited by the complex intravenous administration route required, and high cost [9]. Notably, the FDA authorized Merck’s molnupiravir and Pfizer’s Paxlovid for the treatment of COVID-19 [10,11,12]. Molnupiravir, a biological prodrug of NHC (b-D-N(4)-hydroxycytidine), is a novel nucleoside analogue with a broad-spectrum antiviral activity against SARS-CoV-2 [13]. In clinical trials, molnupiravir showed beneficial effects for mild to moderate COVID-19 patients [13,14]. Paxlovid is a combination of two oral drugs: nirmatrelvir and ritonavir [15]. The FDA authorized the emergency use of Paxlovid for the treatment of mild-to-moderate COVID-19 in adults and children (12 years of age and older weighing at least 88 pounds) with a positive test for the virus that causes COVID-19, and who are at high risk for progression to severe COVID-19, including hospitalization or death [11,15]. However, as with any medication, Paxlovid and molnupiravir still have risks of more serious side effects [10,11,12]. Molnupiravir should not be taken if patients are pregnant, and Paxlovid has important safety risks for patients with transplants or kidney disease [9,11,14,16,17]. In addition, most of the approved therapeutic options, including neutralizing antibodies and oral drugs for COVID-19, are still mainly targeted at patients with mild disease, and prevent mild disease from turning into severe disease; however, there is still a lack of effective treatment for moderate and severe COVID-19 patients, which is an urgent problem to be solved clinically [9,18]. In the meantime, researchers demonstrated that the SARS-CoV-2 infection process comprised a series of steps that could be targeted to inhibit infection [19]. Nevertheless, even though several potential and promising therapeutic targets are identified to date, post-exposure antiviral therapies still need to be repeatedly optimized and upgraded to apply to people of different ages, as well as to make inexpensive, broad-spectrum anti-novel coronavirus drugs with low toxicity available.
Understanding the life cycle of SARS-CoV-2 will greatly facilitate future discoveries to guide the design of new therapeutics to treat COVID-19 [3]. Toward this goal, a series of SARS-CoV-2 studies uncovered mechanistic details of βCoV entry and replication processes in host cells. Briefly, a βCoV particle can enter a cell only after it binds to a specific cell receptor, and the subsequent fusion of viral and host cell membranes of endosomes deliver the viral genome to the cytoplasm, where it directs virus genome replication and protein synthesis prior to virion assembly. During the assembly of new virions, a portion of the endoplasmic reticulum (ER) membrane is appropriated by the virus to serve as a lipid envelope. This lipid membrane is subsequently populated with viral transmembrane proteins to package the viral genome and associated nucleocapsid proteins. However, the process by which newly assembled virions leave host cells is still unclear. The latest research shows that βCoVs exit cells by hijacking a lysosome-based pathway instead of a known biosynthetic secretory pathway commonly used by other enveloped viruses [20]. This finding is critically important, as it potentially opens up new therapeutic avenues to target lysosomotropic molecules toward inhibiting lysosomal function as a strategy to block βCoVs infection and slow virus spread.
2. Role of Lysosomes in SARS-CoV-2 Replication
Lysosomes, which were discovered by de Duve in the 1950s, are subcellular organelles found in nearly all types of eukaryotic cells that perform core degradative and metabolic cellular functions [21,22,23]. Lysosomes form via a budding process that pinches off membrane vesicles from the trans-Golgi network, a region of the Golgi complex responsible for sorting newly synthesized proteins [24,25]. This process generates single membrane-enclosed, spherical, dynamic, and heterogeneous organelles that vary in position, morphology, size, and contents of enzymes and substrates [13]. Importantly, lysosomes contain an acidic environment that is maintained within a low pH range of 4.5–5.5 via proton pump activity [26,27]. This low-pH environment supports activities of various hydrolytic enzymes, such as proteases, nucleases, and phosphatases that catalyze hydrolysis reactions to digest macromolecules such as proteins, nucleic acids, lipids, and carbohydrates. In addition, lysosomal fusion with endosomes or phagosomes leads to lysosomal digestion of endosome-derived small molecules and cell surface proteins or phagosome-derived large particles, such as apoptotic cell corpses and pathogenic bacteria. Furthermore, lysosomes participate in the autophagy of cytoplasmic contents, including damaged mitochondria, ER membranes, and lysosomes [28]. Lysosomes are also able to release their contents extracellularly through lysosomal exocytosis [29]. Lysosomes traffic towards the proximity of the plasma membrane (PM) along microtubules, and the lysosomal membrane and PM fuse together via a Ca2+-dependent process [30]. Lysosome exocytosis was shown to be important for PM wound repair by providing the additional membrane. Lysosomal secretion is commonly observed for lysosome-related organelles which exist in some specialized cell types [26,31]. In addition, the secretion of lysosomal content with regular lysosomes into extracellular space was shown in many physiological and pathological conditions [32,33].
Notably, lysosomes play key roles in host antiviral defenses through virus degradation and modulating the metabolic turnover of proteins related to immune response-associated biological signal pathways [34,35]. For example, core lysosomal proteins of human monocytic leukemia cells, including galactosidase beta 1, cathepsin A, cathepsin B, hexosaminidase subunit alpha, and hexosaminidase subunit beta, were shown to play key functional roles in cell resistance to vesicular stomatitis virus infection, an illness caused by an RNA virus [36]. As another example, Fernández de Castro et al. reported that infection with reovirus, a nonenveloped RNA virus, triggered an increase in lysosomal number and size, while also increasing lysosomal pH from ∼4.5–5 to ∼6.1 [37]. This work revealed that viral proteins are recruited to reovirus inclusions to assist in the assembly of viral components to form virions. However, before the initiation of virion assembly, modified lysosomes move toward the reovirus inclusions. Mature virions are found inside lysosomes as they are assembled and released for egress via lysosomal exocytosis, thus demonstrating that reovirus replication is a lysosome-dependent process [37]. Taken together, these and the results of other studies suggest that lysosomes may have complex (positive or negative) roles in the infection and replication processes associated with numerous viruses.
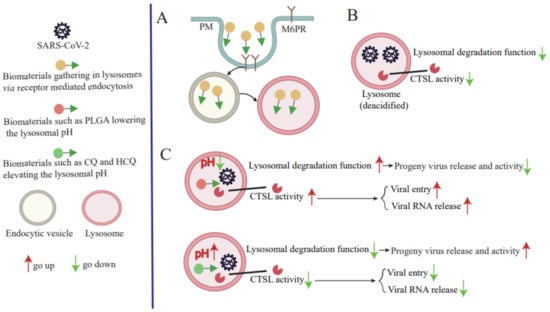
CoVs are enveloped viruses that contain the largest known viral RNA genome, which ranges in size from 26 to 32 kilobases (kb) [38]. The SARS-CoV-2 genome size is ~ 29.9 kb and shares approximately 82% sequence identity with the genome sequences of SARS-CoV and MERS-CoV, and 96.3% sequence identity with the bat CoV RaTG13 genome sequence. Comparisons of predicted protein sequences of essential enzymes and structural proteins of these viruses indicate they possess >90% identity, as determined using whole-genome sequence alignments [38,39]. Importantly, SARS-CoV-2, SARS-CoV, and MERS-CoV genomes resemble one another in that each possesses a 5′ end m7G cap structure, m7GpppA1, and a 3′-end ~ 30–60-nt-long (47-nt median length) poly-A tail that together maintain viral genome stability and prevent cellular exoribonuclease digestion. Moreover, each genome is packaged in association with viral nucleocapsid (N) proteins within a large ribonucleoprotein complex that is enclosed by an envelope membrane containing viral spike (S), envelope (E), and membrane (M) proteins. The initial steps of SARS-CoV-2 infection involve specific binding of S protein to the cellular entry receptor, namely angiotensin-converting enzyme 2 (ACE2) [40]. After initial binding of the virus to cell-surface ACE2, SARS-CoV-2 S protein is cleaved by cell-surface serine protease TMPRSS2 and cathepsin L (CTS L) to form two distinct protein fragments (S1 and S2), which are essential for S1/S2 priming, which triggers virus entry into cells via an endosome-dependent mechanism [41,42,43]. S1 is composed of an N-terminal domain, which is involved in sugar bindings, and a C-terminal domain capable of recognizing human ACE2. S2 contains the putative fusion peptide, the heptad repeats (HR1 and HR2), and is involved in the viral membrane fusion. S1 binds to ACE2 of the host cell during the virus entry, and S2 fuses, allowing the genomes to enter the host cells [44]. During the early stages of SARS-CoV-2 infection, endocytosis is followed by endosome-lysosome fusion that enables the release of SARS-CoV-2 viral RNA into the host cell cytoplasm. Indeed, different lysosomal protease activities associated with different host species were found to determine host species tropisms of various CoVs, since endosomal-lysosomal fusion is required for initiation of a complex program of viral gene expression that is highly regulated in space and time [45]. This program begins with initiation of the SARS-CoV-2 genomic replication that leads to synthesis of full-length negative-sense genomic copies that function as templates for a generation of new positive-sense copies of viral genomic RNA. In turn, these newly synthesized positive-sense genomes either: (1) engage in protein translation that generates viral nonstructural (NSP) and accessory proteins; (2) serve as replication-transcription complexes; or (3) are packaged to form new virions [3]. Open reading frames (ORFs) encoding viral structural proteins are located within the 3′-end one-third of SARS-CoV-2 genomes, while ORFs encoding accessory proteins are interspersed between these ORFs. Importantly, CoVs structural proteins participate in virion assembly and membrane budding of new virions that enable virions to leave the endoplasmic reticulum (ER) and enter the Golgi compartment. After virions enter the Golgi compartment, they can exit the cell via exocytosis to perpetuate virus transmission. Notably, recent evidence shows that SARS-CoV-2 virions preferentially exit infected cells using the lysosomal trafficking pathway after normal lysosomal pathway functions, such as lysosomal acidification, lysosomal enzyme activity, and antigen presentation, were disrupted by viral activities [20]. Taken together, results of these and numerous other studies suggest that SARS-CoV-2 replication steps, especially virus entry, virus RNA release into the cytoplasm of the cell, and progeny virus release from infected cells, are closely tied to lysosomal dysfunction (Figure 1).
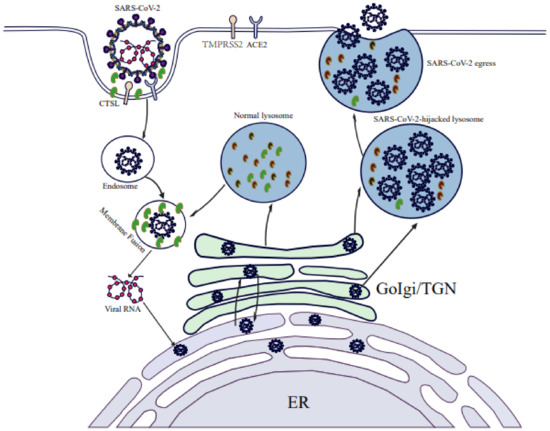
Figure 1.
SARS-CoV-2 replication is dependent on a lysosome-based pathway. SARS-CoV-2 entry into the host cells is mediated by the endocytic pathway. The entry of SARS-CoV-2 and mature virus RNA release into the cytoplasm requires CTSL activity and an acid lysosomal environment. Progeny viruses exit cells by hijacking a lysosome-based pathway.
3. SARS-CoV-2 Infection Leads to Lysosomal Dysfunction
The main function of lysosomes, to degrade or break down macromolecules, is critically important for numerous cellular processes in addition to its function as the cell’s “garbage disposal” [46]. In fact, lysosomes also act as signaling hubs that monitor intracellular levels of nutrients and energy within larger networking platforms that interconnect multiple signaling pathways related to signal transduction and autophagy regulation [47,48]. Moreover, lysosomes interact with other intracellular organelles (e.g., mitochondria, ER) that together maintain cell homeostasis [26,49,50]. Furthermore, these organelles also degrade and recycle intracellular and extracellular wastes generated during cell secretion of substances and PM repair, while also playing important roles in many other physiological and pathological processes, such as immune responses and tumorigenesis [30,50,51,52].
Indeed, for decades CoVs were detected in lysosomes during late stages of infection, although a connection between lysosomes and viral success was only proposed recently [53]. For example, Ghosh et al. recently reported results of a pioneering investigation that revealed a vital role of lysosomes in SARS-CoV-2 release. Briefly, they found that SARS-CoV-2 exploits the small Arf-like Ras family GTPase (ARL8b)-dependent lysosomal exocytosis pathway for release into the extracellular environment, and that inhibition of Rab7 GTPase, an enzyme involved in endosomal-lysosomal transport of materials, prevents egress of SARS-CoV-2 virions from cells that were associated with reduction in lysosomal number and limited endolysosomal maturation [20]. These results, when considered together with the established fact that normally the acidic environment within lysosomes helps destroy viruses and other pathogens before they leave cells, imply that SARS-CoV-2 may hijack the lysosomal acidification process by somehow promoting lysosomal deacidification that significantly weakens activities of lysosomal degradative enzymes and disrupts lysosome-dependent antigen cross-presentation [20]. Taken together, these clues support a possible scenario whereby SARS-CoV-2 infection leads to lysosomal dysfunction that lessens the ability of infected cells to degrade and recycle intracellular and extracellular materials through autophagy and endocytosis (Figure 2). In turn, virus-induced lysosomal dysfunction interferes with other cellular functions, such as PM repair, the immune response, signal transduction, pathogen elimination, and so on (Figure 2). As a result, SARS-CoV-2 virions that exit cells remain intact and ready to infect other cells to begin a new infection cycle. In addition, deacidification of SARS-CoV-2-hijacked lysosomes may alter immune system functions to account for observed COVID-19-associated immune system abnormalities [20,51].
Figure 2.
SARS-CoV-2 infection induces lysosomal dysfunction. SARS-CoV-2 infection leads to lysosome deacidification and promotes lysosomal exocytosis. SARS-CoV-2-hijacked lysosomes participate in the degradation and recycling of intracellular and extracellular material through autophagy and endocytosis to provide energy and source molecules. Exocytosis of SARS-CoV-2-hijacked lysosomes contributes to PM repair, the immune response, pathogen elimination, and progeny virus release in stores. SARS-CoV-2-hijacked lysosomes sense nutrients and activate metabolic signal transduction.
Lysosomal exocytosis results in the PM localization of lysosomal membrane proteins and also the release of lysosomal contents into the extracellular environment [54,55]. ARL8b localizes to late endosomes/lysosomes and regulates their movement to the PM and, ultimately, their exocytosis. During SARS-CoV-2-hijacking lysosome exocytosis, the multisubunit complex (BORC) recruiting ARL8b (BORC-ARL8b) complex drives the anterograde transport of lysosomes from perinuclear regions to the vicinity of the PM [20]. Fusion of lysosomes with the PM is mediated by the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex composed of VAMP7, STX4, and SNAP23, and also requires an increase in the intracellular and/or localized Ca2+ level. Chen et al. showed that SARS-CoV-2 infection promoted lysosomal exocytosis (Figure 2) and revealed a mechanism by which SARS-CoV-2 interacts with host factors to promote its extracellular egress [43]. Briefly, SARS-CoV-2 NSP (ORF3a) facilitates lysosomal targeting of the BORC-ARL8b complex, which mediates the trafficking of lysosomes to the vicinity of the PM, and exocytosis-related SNARE proteins [56]. The Ca2+ channel TRPML3 is required for SARS-CoV-2 ORF3a-mediated lysosomal exocytosis [56]. Ser171 and Trp193 in SARS-CoV-2 ORF3a are critical for promoting lysosomal exocytosis and blocking autophagy [56]. However, further research is needed to shed more light on the role of ORF3a in the lysosomal exocytosis-mediated egress of SARS-CoV-2.
Lysosomal degradation of materials is tightly regulated by pH and hydrolase activities. The basal lysosomal pH range of 4.5–5.0 is associated with a buffering capacity of 19 ± 6 mM/pH unit [57]. Each lysosome consists of an outer membrane that surrounds an interior acidic environment. The low internal pH is maintained via proton pumps acting through multiple types of membrane channels that include chloride channels (e.g., CLC-7, CLC-6, CLC-3), calcium channels (e.g., TRPML1), and vacuolar H+-ATPase (V-ATPase) [58]. Meanwhile, lysosomes contain numerous hydrolases, which are synthesized within the ER followed by entry into lysosomes via at least two main mechanisms: one mechanism mainly involves modification of hydrolytic enzymes through the addition of mannose 6-phosphate receptor (M6PR) residues that are recognized by the Golgi apparatus then transported to the lysosome; the other mechanism involves transport by lysosomal integral membrane protein 2 (LIMP-2) of hydrolases such as glucocerebrosidases into lysosomes [59,60]. Normal lysosomal degradative functions are important for cellular functions, as dysregulated lysosomal degradation can lead to impaired endocytic function, autophagic degradation, and macromolecule biogenesis and transport that are associated with proteinopathic neurodegenerative diseases, metabolic disorders, and immunological diseases [61,62,63]. In addition, lysosomal degradation dysfunction can disrupt functions of other organelles (e.g., mitochondria), leading to increased production of reactive oxygen species and inflammatory cytokines that can contribute to pathogenesis of inflammatory diseases, cancer, and infectious diseases [64,65,66].
Remarkably, host cell fusion with βCoV particles, which occurs after virions bind to their respective host cell receptors, is induced by lysosomal hydrolases (e.g., CTSL) that are sensitive to lysosomal pH [3,43]. After fusion is complete, βCoV viral particles enter the endolysosomal intracellular transport system, where the low-pH environment supports cleavage of viral proteins and subsequent release of viral RNA into the cytoplasm, and viral exit from cells is due to viral hijacking of a lysosome-based pathway [3,7,30]. Although SARS-CoV-2 replication depends on altered lysosomal pH and verified roles of host cell hydrolases, it is still unclear whether lysosomal deacidification of cells is actually caused by SARS-CoV-2 infection [67]. Alternatively, it was postulated that infected cells may actively deacidify lysosomes to promote the release of newly generated SARS-CoV-2 from cells as a mechanism for relieving viral pressure within infected cells [67]. Consequently, researchers speculate that lysosomal deacidification may be a consequence of the action of specific SARS-CoV-2 proteins, while other researchers suggest that lysosomal deacidification is indirectly caused by too much cargo loading and/or disruption of proton pump or ion channel trafficking functions [67,68,69]. In short, it is necessary to explore the mechanism underlying SARS-CoV-2 infection-associated lysosome deacidification in order to devise strategies to re-acidify lysosomes so they can destroy viruses, inhibit virus egress, and restore antigen-presenting cell (APC) function [67].
5. Conclusions
This review summarizes the correlation between lysosomes and virus replication by describing multiple roles played by lysosomes during SARS-CoV-2 replication and by providing potential mechanisms to explain how SARS-CoV-2 infection leads to lysosomal dysfunction. However, the detailed mechanisms underlying virus-induced lysosome dysfunction must be explored further through investigations of SARS-CoV-2-induced active and passive lysosome deacidification mechanisms within infected cells, as must the effects of lysosomal reacidification on SARS-CoV-2 egress and antigen presentation in APCs. Moreover, here the significance of CTSL activity and lysosomal pH modulation in COVID-19 disease are also discussed, with a focus on exploiting lysosomotropic molecules for use as anti-SARS-CoV-2 agents that act to specifically modulate lysosomal pH. Nevertheless, any strategy based on the alteration of lysosomal pH must take into account the different roles played by lysosomal acidification and deacidification during different stages of viral replication before the therapeutic potential of lysosome pH-modulating agents can be tapped to guide the development of clinical treatments to block virus infection.
In addition, the novel oral anti-SARS-CoV-2 drugs are potentially immense and may bring new hope for COVID-19 treatment and recovery [18]. Recently, the oral antivirals of COVID-19 were shown to generally function against viral enzyme structures such as RNA-dependent RNA polymerase (RdRp), main protease (3CLpro), and papain-like protease (PLpro), which are important for viral replication [8,9,18]. Paxlovid targets the SARS-CoV-2 main protease referred to as 3CLpro or nsp5 protease [11]. Molnupiravir is used as a competitive alternative substrate and targets SARS-CoV-2 RdRp [10]. Thus, both oral antivirals can prevent the SARS-CoV-2 virus from copying itself directly. Although they are less affected by SARS-CoV-2 mutation, due to the continual occurrence of mutations of SARS-CoV-2 genomes, a worthwhile strategy for designing SARS-CoV-2 antiviral drugs might be to block a general cellular mechanism involved in the virus replication process instead of inhibiting specific viral proteins crucially needed for SARS-CoV-2 infection. For example, based on lysosome roles in SARS-CoV-2 replication, targeting lysosome-based replication of SARS-CoV-2 may be a prevention strategy for COVID-19, as it affects virus entry, virus RNA release, and progeny virus release. Interestingly, whether the oral drugs for treatment of COVID-19, such as Paxlovid and molnupiravir, are lysosomotropic molecularly, and can indirectly affect the anti-SARS-CoV-2 effects by modulating lysosomal function, warrants further study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23116188/s1.
Author Contributions
Y.L. and G.W. conceived the work and figures; Y.L. wrote the manuscript; Z.W. and Y.C. contributed materials and analysis tools; D.S., F.G. and W.H. revised the manuscript for valuable intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
The National Natural Science Foundation of China (Grant 32172804 to Y.L., Grant 32172805 to F.G. and Grant 32172828 to W.H.), the China Postdoctoral Science Foundation (Grant 2020M670858 to Y.L.), Scientific and Technological Project of Jilin Province (Grant 20210202041NC to W.H.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Fenghua Hu, an Associate Professor at Weill Institute (Cornell University), for the initial discussions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CoVs | Coronaviruses |
| βCoV | Betacoronavirus |
| COVID-19 | Coronavirus disease 2019 |
| SARS-CoV | Severe acute respiratory syndrome coronavirus |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| CTSL | Cathepsin L |
| ACE2 | Angiotensin-converting enzyme 2 |
| PM | Plasma membrane |
| V-ATPase | Vacuolar H+-ATPase |
| CQ | Chloroquine |
| HCQ | Hydroxychloroquine |
| APC | Antigen-presenting cell |
| PLGA | Poly (lactic-co-glycolic acid) |
| FDA | The United States Food and Drug Administration |
| ORFs | Open reading frames |
References
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus Infections-More Than Just the Common Cold. JAMA 2020, 323, 707–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- V’Kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Benincasa, G.; Criscuolo, C.; Faenza, M.; Liberato, C.; Rusciano, M. Immune reactivity during COVID-19: Implications for treatment. Immunol. Lett. 2021, 231, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Blaess, M.; Kaiser, L.; Sauer, M.; Csuk, R.; Deigner, H.P. COVID-19/SARS-CoV-2 Infection: Lysosomes and Lysosomotropism Implicate New Treatment Strategies and Personal Risks. Int. J. Mol. Sci. 2020, 21, 4953. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, F.; Ganesan, S.; James, A.; Zaher, W.A. Understanding perception and acceptance of Sinopharm vaccine and vaccination against COVID-19 in the UAE. BMC Public Health 2021, 21, 1602. [Google Scholar] [CrossRef]
- Singh, L.; Bansal, S.; Bode, L.; Budak, C.; Chi, G.; Kawintiranon, K.; Padden, C.; Vanarsdall, R.; Vraga, E.; Wang, Y. A first look at COVID-19 information and misinformation sharing on Twitter. arXiv 2020, arXiv:2003.13907. [Google Scholar]
- Kumar, S.; Chandele, A.; Sharma, A. Current status of therapeutic monoclonal antibodies against SARS-CoV-2. PLoS Pathog. 2021, 17, e1009885. [Google Scholar] [CrossRef]
- Kim, S. COVID-19 Drug Development. J. Microbiol. Biotechnol. 2022, 32, 1–5. [Google Scholar] [CrossRef]
- Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study—Merck.com. 2021. Available online: https://www.merck.com/news/ (accessed on 10 October 2021).
- COVID-19 Antiviral Efforts | Pfizer. Available online: https://www.pfizer.com/science/coronavirus/antiviral-efforts (accessed on 10 October 2021).
- Food and Druga Administration (FDA). Emergency Use Authorization 105. 22 December 2021. Paxlovid (Nirmatrelvir Co-Packaged with Ritonavir) for the Treatment of Mild-to-Moderate Coronavirus Disease 2019 (COVID-19) in Certain Adults and Pediatric Patients. Available online: https://www.fda.gov/media/155049/download (accessed on 28 May 2022).
- Menendez-Arias, L. Decoding molnupiravir-induced mutagenesis in SARS-CoV-2. J. Biol. Chem. 2021, 297, 100867. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports. BMJ 2021, 375, n2422. [Google Scholar] [CrossRef] [PubMed]
- Pfizer’s Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 EPIC-HR Study. Available online: https://www.pfizer.com/news/pressrelease/press-release-detail (accessed on 28 May 2022).
- Fishbane, S.; Hirsch, J.S.; Nair, V. Special Considerations for Paxlovid Treatment Among Transplant Recipients with SARS-CoV-2 Infection. Am. J. Kidney Dis. 2022, 79, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef]
- Wen, W.; Chen, C.; Tang, J.; Wang, C.; Zhou, M.; Cheng, Y.; Zhou, X.; Wu, Q.; Zhang, X.; Feng, Z.; et al. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: A meta-analysis. Ann. Med. 2022, 54, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.W.; Xie, Y.; Tang, L.S.; Pu, D.; Zhu, Y.J.; Liu, J.Y.; Ma, X.L. Therapeutic targets and interventional strategies in COVID-19: Mechanisms and clinical studies. Signal Transduct. Target. Ther. 2021, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. βCoronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14. [Google Scholar] [CrossRef]
- Eichel, H.J.; Bukovsky, J. Intracellular distribution pattern of rat liver glutamic-oxalacetic transaminase. Nature 1961, 191, 243–245. [Google Scholar] [CrossRef]
- Saftig, P.; Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- De Duve, C. The lysosome turns fifty. Nat. Cell Biol. 2005, 7, 847–849. [Google Scholar] [CrossRef]
- Braulke, T.; Bonifacino, J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta 2009, 1793, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Luzio, J.P.; Hackmann, Y.; Dieckmann, N.M.; Griffiths, G.M. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb. Perspect. Biol. 2014, 6, a016840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Wang, X. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 2021, 220, e202102001. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Shirihai, O.S.; Grinstaff, M.W. Modulating lysosomal pH: A molecular and nanoscale materials design perspective. J. Life Sci. 2020, 2, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Bright, N.A.; Davis, L.J.; Luzio, J.P. Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity. Curr. Biol. 2016, 26, 2233–2245. [Google Scholar] [CrossRef] [Green Version]
- Tancini, B.; Buratta, S.; Delo, F.; Sagini, K.; Chiaradia, E.; Pellegrino, R.M.; Emiliani, C.; Urbanelli, L. Lysosomal Exocytosis: The Extracellular Role of an Intracellular Organelle. Membranes 2020, 10, 406. [Google Scholar] [CrossRef]
- Medina, D.L.; Fraldi, A.; Bouche, V.; Annunziata, F.; Mansueto, G.; Spampanato, C.; Puri, C.; Pignata, A.; Martina, J.A.; Sardiello, M.; et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 2011, 21, 421–430. [Google Scholar] [CrossRef]
- Samie, M.; Wang, X.; Zhang, X.; Goschka, A.; Li, X.; Cheng, X.; Gregg, E.; Azar, M.; Zhuo, Y.; Garrity, A.G.; et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell 2013, 26, 511–524. [Google Scholar] [CrossRef] [Green Version]
- Maeda, F.Y.; van Haaren, J.J.; Langley, D.B.; Christ, D.; Andrews, N.W.; Song, W. Surface-associated antigen induces permeabilization of primary mouse B-cells and lysosome exocytosis facilitating antigen uptake and presentation to T-cells. Elife 2021, 10, e66984. [Google Scholar] [CrossRef]
- Westman, J.; Plumb, J.; Licht, A.; Yang, M.; Allert, S.; Naglik, J.R.; Hube, B.; Grinstein, S.; Maxson, M.E. Calcium-dependent ESCRT recruitment and lysosome exocytosis maintain epithelial integrity during Candida albicans invasion. Cell Rep. 2022, 38, 110187. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Castrejon-Jimenez, N.S.; Leyva-Paredes, K.; Hernandez-Gonzalez, J.C.; Luna-Herrera, J.; Garcia-Perez, B.E. The role of autophagy in bacterial infections. Biosci. Trends 2015, 9, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.; Gao, Y.; Zhan, S.; Ge, W. Quantitative proteomic profiling for clarification of the crucial roles of lysosomes in microbial infections. Mol. Immunol. 2017, 87, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Fernandez de Castro, I.; Tenorio, R.; Ortega-Gonzalez, P.; Knowlton, J.J.; Zamora, P.F.; Lee, C.H.; Fernandez, J.J.; Dermody, T.S.; Risco, C. A modified lysosomal organelle mediates nonlytic egress of reovirus. J. Cell Biol. 2020, 219, e201910131. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, A.Z.; Kiyani, D.A.; Hamid, S.M.; Saalim, M.; Fahim, A.; Jalal, N. Spiking dependence of SARS-CoV-2 pathogenicity on TMPRSS2. J. Med. Virol. 2021, 93, 4205–4218. [Google Scholar] [CrossRef]
- Lei, C.; Qian, K.; Li, T.; Zhang, S.; Fu, W.; Ding, M.; Hu, S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020, 11, 2070. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Shang, J.; Yang, Y.; Liu, C.; Wan, Y.; Geng, Q.; Wang, M.; Baric, R.; Li, F. Lysosomal Proteases Are a Determinant of Coronavirus Tropism. J. Virol. 2018, 92, e01504-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxfield, F.R. Role of endosomes and lysosomes in human disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016931. [Google Scholar] [CrossRef] [Green Version]
- Savini, M.; Zhao, Q.; Wang, M.C. Lysosomes: Signaling Hubs for Metabolic Sensing and Longevity. Trends Cell Biol. 2019, 29, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Gang, X.; He, G.; Liu, Y.; Wang, Y.; Zhao, X.; Wang, G. The Molecular Mechanisms Underlying Mitochondria-Associated Endoplasmic Reticulum Membrane-Induced Insulin Resistance. Front. Endocrinol. 2020, 11, 592129. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, J.; Janikiewicz, J.; Michalska, B.; Patalas-Krawczyk, P.; Perrone, M.; Ziolkowski, W.; Duszynski, J.; Pinton, P.; Dobrzyn, A.; Wieckowski, M.R. Interaction of Mitochondria with the Endoplasmic Reticulum and Plasma Membrane in Calcium Homeostasis, Lipid Trafficking and Mitochondrial Structure. Int. J. Mol. Sci. 2017, 18, 1576. [Google Scholar] [CrossRef] [PubMed]
- Weissmann, G.; Dukor, P. The role of lysosomes in immune responses. Adv. Immunol. 1970, 12, 283–331. [Google Scholar]
- Davidson, S.M.; Vander Heiden, M.G. Critical Functions of the Lysosome in Cancer Biology. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 481–507. [Google Scholar] [CrossRef]
- Ducatelle, R.; Hoorens, J. Significance of lysosomes in the morphogenesis of coronaviruses. Arch. Virol. 1984, 79, 1–12. [Google Scholar] [CrossRef]
- Andrews, N.W. Regulated secretion of conventional lysosomes. Trends Cell Biol. 2000, 10, 316–321. [Google Scholar] [CrossRef]
- Andrews, N.W. Membrane repair and immunological danger. EMBO Rep. 2005, 6, 826–830. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zheng, Q.; Sun, L.; Ji, M.; Li, Y.; Deng, H.; Zhang, H. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell 2021, 56, 3250–3263.e5. [Google Scholar] [CrossRef] [PubMed]
- Guntuku, L.; Gangasani, J.K.; Thummuri, D.; Borkar, R.M.; Manavathi, B.; Ragampeta, S.; Vaidya, J.R.; Sistla, R.; Vegi, N.G.M. IITZ-01, a novel potent lysosomotropic autophagy inhibitor, has single-agent antitumor efficacy in triple-negative breast cancer in vitro and in vivo. Oncogene 2019, 38, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Mindell, J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motyka, B.; Korbutt, G.; Pinkoski, M.J.; Heibein, J.A.; Caputo, A.; Hobman, M.; Barry, M.; Shostak, I.; Sawchuk, T.; Holmes, C.F.; et al. Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell 2000, 103, 491–500. [Google Scholar] [CrossRef] [Green Version]
- Reczek, D.; Schwake, M.; Schroder, J.; Hughes, H.; Blanz, J.; Jin, X.; Brondyk, W.; Van Patten, S.; Edmunds, T.; Saftig, P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell 2007, 131, 770–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, C.C.; Vacca, F.; Gruenberg, J. Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 2014, 31, 2–10. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, L. Autophagic lysosome reformation. Exp. Cell Res. 2013, 319, 142–146. [Google Scholar] [CrossRef]
- Gegg, M.E.; Schapira, A.H. Mitochondrial dysfunction associated with glucocerebrosidase deficiency. Neurobiol. Dis. 2016, 90, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Goronzy, J.J.; Weyand, C.M. Autophagy in autoimmune disease. J. Mol. Med. 2015, 93, 707–717. [Google Scholar] [CrossRef] [Green Version]
- Ge, W.; Li, D.; Gao, Y.; Cao, X. The Roles of Lysosomes in Inflammation and Autoimmune Diseases. Int. Rev. Immunol. 2015, 34, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Melino, G.; Shi, Y. Actively or passively deacidified lysosomes push beta-coronavirus egress. Cell Death Dis. 2021, 12, 235. [Google Scholar] [CrossRef] [PubMed]
- Westerbeck, J.W.; Machamer, C.E. The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus. J. Virol. 2019, 93, e00015-19. [Google Scholar] [CrossRef] [Green Version]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Chiaradia, E.; Urbanelli, L.; Emiliani, C. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int. J. Mol. Sci. 2020, 21, 2576. [Google Scholar] [CrossRef] [Green Version]
- Alesi, N.; Akl, E.W.; Khabibullin, D.; Liu, H.J.; Nidhiry, A.S.; Garner, E.R.; Filippakis, H.; Lam, H.C.; Shi, W.; Viswanathan, S.R.; et al. TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat. Commun. 2021, 12, 4245. [Google Scholar] [CrossRef]
- Fujishima, A.; Imai, Y.; Nomura, T.; Fujisawa, Y.; Yamamoto, Y.; Sugawara, T. The crystal structure of human cathepsin L complexed with E-64. FEBS Lett. 1997, 407, 47–50. [Google Scholar] [CrossRef]
- Liu, T.; Luo, S.; Libby, P.; Shi, G.P. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020, 213, 107587. [Google Scholar] [CrossRef]
- Bode, L.; Dietrich, D.E.; Spannhuth, C.W.; Ludwig, H. Prominent Efficacy of Amantadine against Human Borna Disease Virus Infection In Vitro and In Vivo. Comment on Fink et al. Amantadine Inhibits SARS-CoV-2 In Vitro. Viruses 2021, 13, 539. Viruses 2022, 14, 494. [Google Scholar]
- Cortes-Borra, A.; Aranda-Abreu, G.E. Amantadine in the prevention of clinical symptoms caused by SARS-CoV-2. Pharmacol. Rep. 2021, 73, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Fink, K.; Nitsche, A.; Neumann, M.; Grossegesse, M.; Eisele, K.H.; Danysz, W. Amantadine Inhibits SARS-CoV-2 In Vitro. Viruses 2021, 13, 539. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Pan, T.; Huang, F.; Ying, R.; Liu, J.; Fan, H.; Zhang, J.; Liu, W.; Lin, Y.; Yuan, Y.; et al. Glycopeptide Antibiotic Teicoplanin Inhibits Cell Entry of SARS-CoV-2 by Suppressing the Proteolytic Activity of Cathepsin L. Front. Microbiol. 2022, 13, 884034. [Google Scholar] [CrossRef] [PubMed]
- Jacquin, E.; Leclerc-Mercier, S.; Judon, C.; Blanchard, E.; Fraitag, S.; Florey, O. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy 2017, 13, 854–867. [Google Scholar] [CrossRef]
- Ahmadi, A.R.; Ayazi-Nasrabadi, R. Astaxanthin protective barrier and its ability to improve the health in patients with COVID-19. Iran. J. Microbiol. 2021, 13, 434–441. [Google Scholar] [CrossRef]
- Gomes, C.P.; Fernandes, D.E.; Casimiro, F.; da Mata, G.F.; Passos, M.T.; Varela, P.; Mastroianni-Kirsztajn, G.; Pesquero, J.B. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front. Cell Infect. Microbiol. 2020, 10, 589505. [Google Scholar] [CrossRef]
- Wang, S.Q.; Du, Q.S.; Zhao, K.; Li, A.X.; Wei, D.Q.; Chou, K.C. Virtual screening for finding natural inhibitor against cathepsin-L for SARS therapy. Amino Acids 2007, 33, 129–135. [Google Scholar] [CrossRef]
- Kang, S.N.; Lee, J.S.; Park, J.H.; Cho, J.H.; Cho, K.K.; Lee, O.H.; Kim, I.S. In vitro anti-osteoporosis properties of diverse Korean Drynariae rhizoma phenolic extracts. Nutrients 2014, 6, 1737–1751. [Google Scholar] [CrossRef] [Green Version]
- De Duve, C.; de Barsy, T.; Poole, B.; Trouet, A.; Tulkens, P.; Van Hoof, F. Commentary. Lysosomotropic agents. Biochem. Pharmacol. 1974, 23, 2495–2531. [Google Scholar] [CrossRef]
- Batiha, G.E.; Shaheen, H.M.; Al-Kuraishy, H.M.; Teibo, J.O.; Akinfe, O.A.; Al-Garbee, A.I.; Teibo, T.K.A.; Kabrah, S.M. Possible mechanistic insights into iron homeostasis role of the action of 4-aminoquinolines (chloroquine/hydroxychloroquine) on COVID-19 (SARS-CoV-2) infection. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 7565–7584. [Google Scholar]
- Schrezenmeier, E.; Dorner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, M.X.; Lu, G.D.; Shen, H.M.; Zhou, J. Hydroxychloroquine/Chloroquine as Therapeutics for COVID-19: Truth under the Mystery. Int. J. Biol. Sci. 2021, 17, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Saghir, S.A.M.; AlGabri, N.A.; Alagawany, M.M.; Attia, Y.A.; Alyileili, S.R.; Elnesr, S.S.; Shafi, M.E.; Al-Shargi, O.Y.A.; Al-Balagi, N.; Alwajeeh, A.S.; et al. Chloroquine and Hydroxychloroquine for the Prevention and Treatment of COVID-19: A Fiction, Hope or Hype? An Updated Review. Ther. Clin. Risk Manag. 2021, 17, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Blaess, M.; Kaiser, L.; Sommerfeld, O.; Rentschler, S.; Csuk, R.; Deigner, H.P. Rational Drug Repurposing: Focus on Lysosomotropism, Targets in Disease Process, Drug Profile, and Pulmonary Tissue Accumulation in SARS-CoV-2 Infection/COVID-19. Front. Pharmacol. 2020, 11, 584881. [Google Scholar] [CrossRef]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Tran, B.N.; Cheng, Y.S.; Xu, M.; Pradhan, M.; Henderson, M.; Zhu, W.; et al. The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infect. Dis. 2021, 7, 1389–1408. [Google Scholar] [CrossRef]
- Meneses Calderon, J.; Figueroa Flores, M.D.R.; Paniagua Coria, L.; Briones Garduno, J.C.; Meneses Figueroa, J.; Vargas Contretas, M.J.; De la Cruz Avila, L.; Diaz Meza, S.; Ramirez Chacon, R.; Padmanabhan, S.; et al. Nitazoxanide against COVID-19 in three explorative scenarios. J. Infect. Dev. Ctries. 2020, 14, 982–986. [Google Scholar] [CrossRef]
- Chiappori, A.; Williams, C.; Northfelt, D.W.; Adams, J.W.; Malik, S.; Edelman, M.J.; Rosen, P.; Van Echo, D.A.; Berger, M.S.; Haura, E.B. Obatoclax mesylate, a pan-bcl-2 inhibitor, in combination with docetaxel in a phase 1/2 trial in relapsed non-small-cell lung cancer. J. Thorac. Oncol. 2014, 9, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wang, J.; Hassan, A.; Lee, C.H.; Xie, X.S.; Li, X. Molecular basis of V-ATPase inhibition by bafilomycin A1. Nat. Commun. 2021, 12, 1782. [Google Scholar] [CrossRef]
- Hart, P.D.; Young, M.R. Ammonium chloride, an inhibitor of phagosome-lysosome fusion in macrophages, concurrently induces phagosome-endosome fusion, and opens a novel pathway: Studies of a pathogenic mycobacterium and a nonpathogenic yeast. J. Exp. Med. 1991, 174, 881–889. [Google Scholar] [CrossRef]
- Dabydeen, S.A.; Meneses, P.I. The role of NH4Cl and cysteine proteases in Human Papillomavirus type 16 infection. Virol. J. 2009, 6, 109. [Google Scholar] [CrossRef] [Green Version]
- Shang, C.; Zhuang, X.; Zhang, H.; Li, Y.; Zhu, Y.; Lu, J.; Ge, C.; Cong, J.; Li, T.; Tian, M.; et al. Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virol. J. 2021, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Shirihai, O.S.; Grinstaff, M.W. Degradable Nanoparticles Restore Lysosomal pH and Autophagic Flux in Lipotoxic Pancreatic Beta Cells. Adv. Healthc. Mater. 2019, 8, e1801511. [Google Scholar] [CrossRef] [PubMed]
- Cunha, A.; Gaubert, A.; Latxague, L.; Dehay, B. PLGA-Based Nanoparticles for Neuroprotective Drug Delivery in Neurodegenerative Diseases. Pharmaceutics 2021, 13, 1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jin, L.; Wu, Z.; Xie, Y.; Zhang, P.; Wang, Q.; Yan, S.; Chen, B.; Liang, H.; Naman, C.B.; et al. PLGA-PEG Nanoparticles Facilitate In Vivo Anti-Alzheimer’s Effects of Fucoxanthin, a Marine Carotenoid Derived from Edible Brown Algae. J. Agric. Food Chem. 2021, 69, 9764–9777. [Google Scholar] [CrossRef] [PubMed]
- Pourhajibagher, M.; Azimi, M.; Haddadi-Asl, V.; Ahmadi, H.; Gholamzad, M.; Ghorbanpour, S.; Bahador, A. Robust antimicrobial pho todynamic therapy with curcumin-poly (lactic-co-glycolic acid) nanoparticles against COVID-19: A preliminary in vitro study in Vero cell line as a model. Photodiagnosis Photodyn. Ther. 2021, 34, 102286. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).