Biodegradation and Metabolic Pathway of 17β-Estradiol by Rhodococcus sp. ED55
Abstract
:1. Introduction
2. Results and Discussion
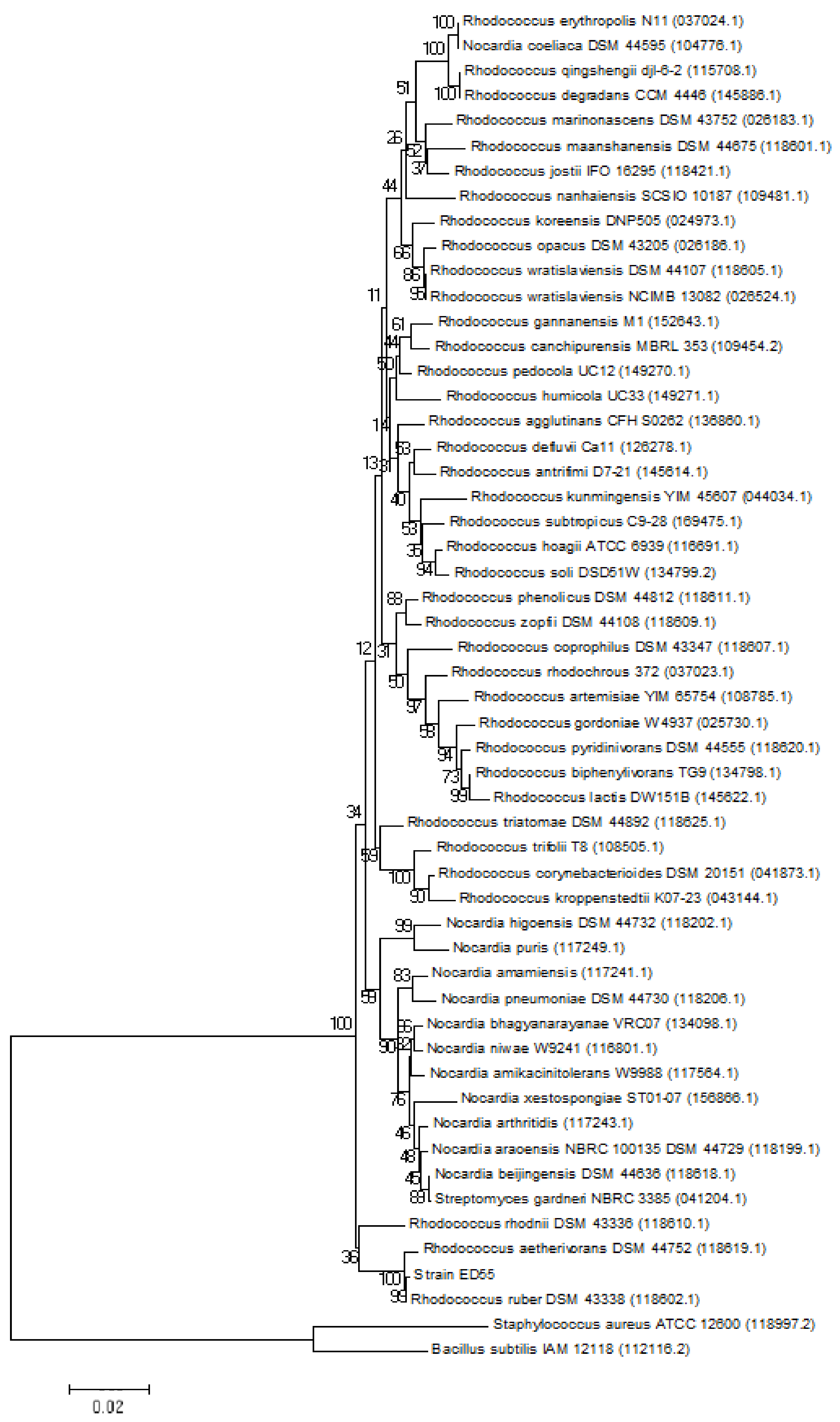
2.1. Phylogenetic Analysis of E2 Degrading Strain
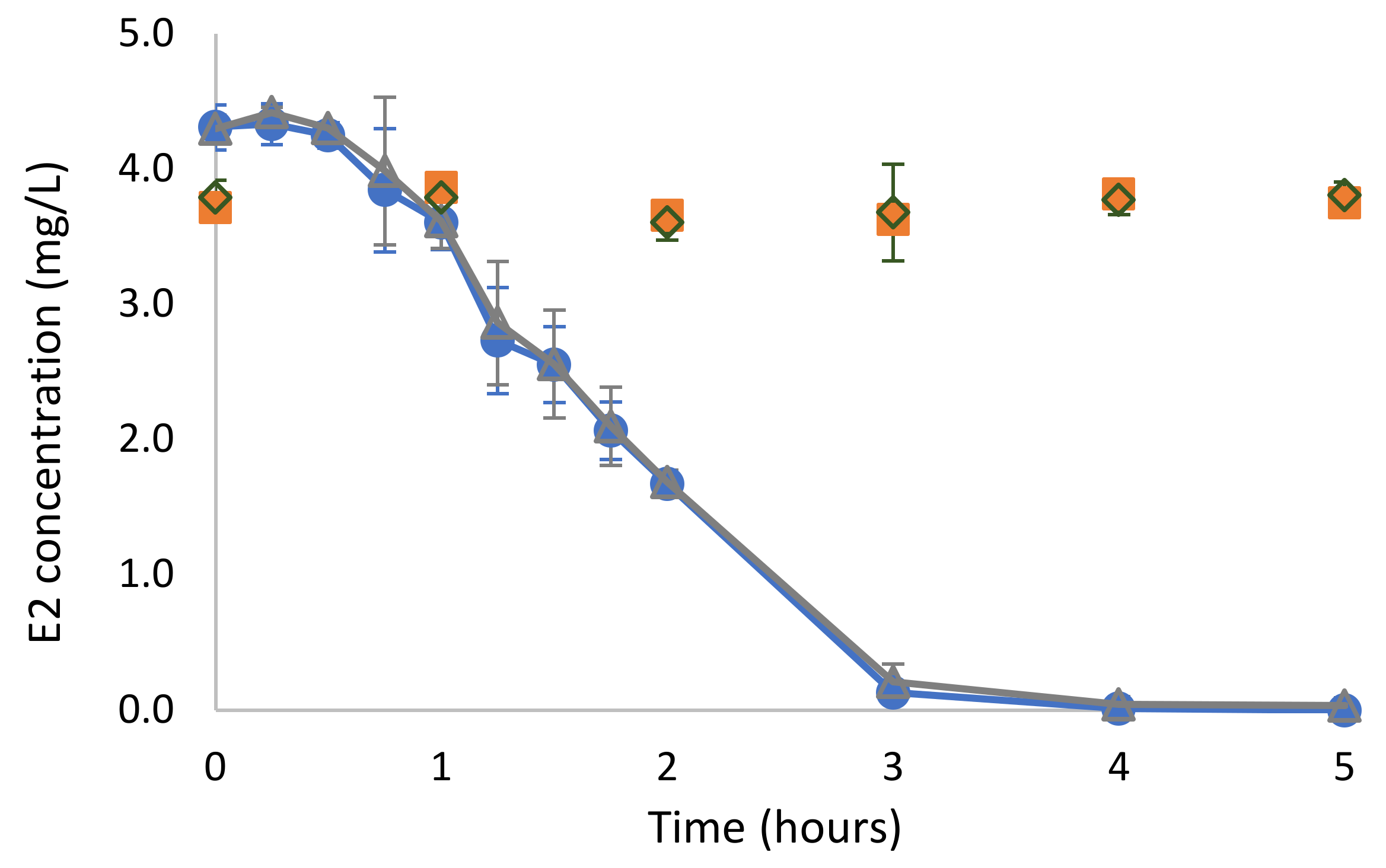
2.2. Biodegradation of E2 in Mineral Salts Medium
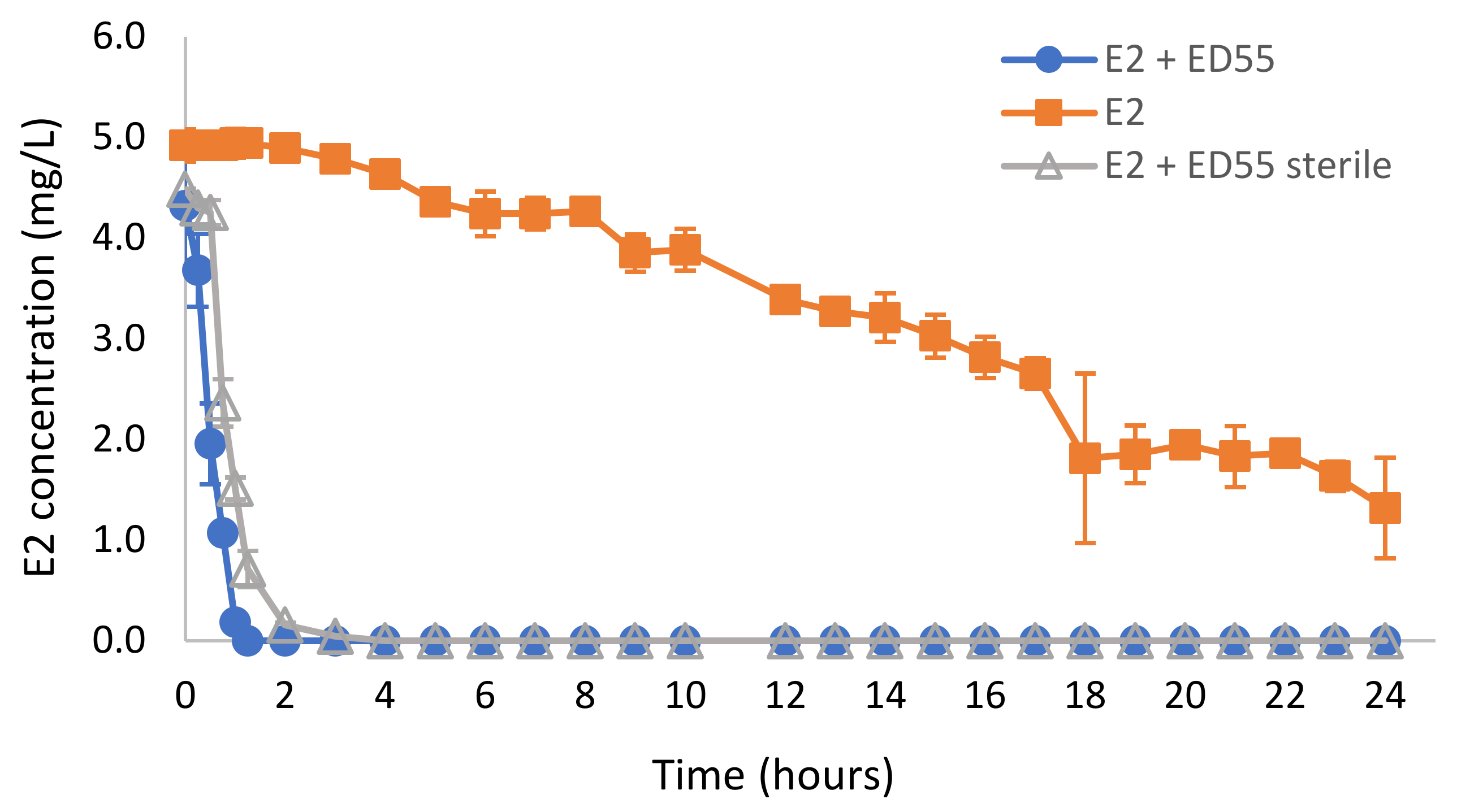
2.3. Biodegradation of E2 in Wastewater
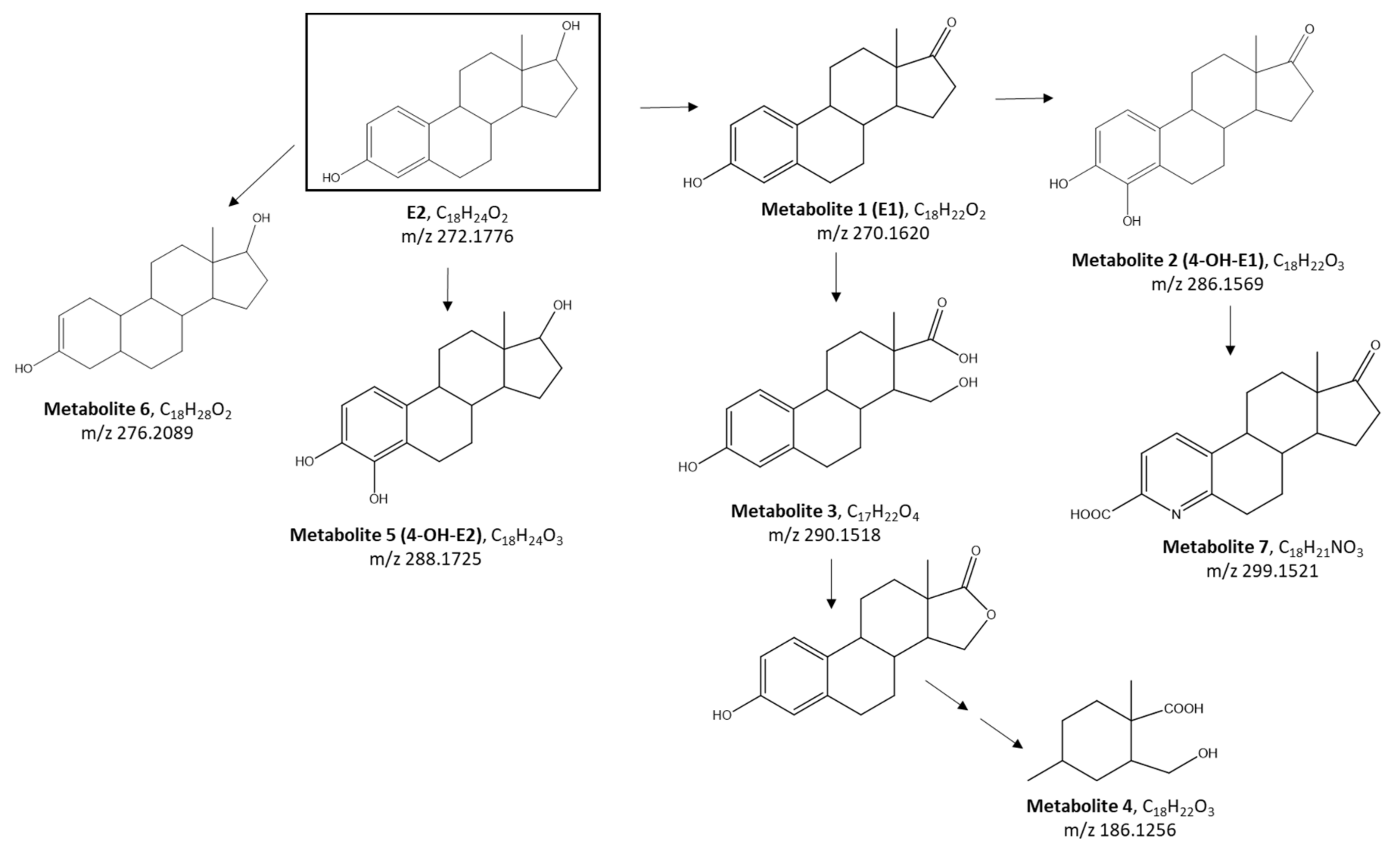
2.4. Identification of Metabolites Produced by E2 Biodegradation
2.5. Toxicity Tests
2.6. Estrogenic Activity
3. Materials and Methods
3.1. Chemicals
3.2. E2 Degrading Strain
3.3. Biodegradation of E2 in Mineral Salts Medium (MM)
3.4. Biodegradation of E2 in Wastewater
3.5. Analytical Methods and Data Processing
3.6. Toxicity Tests
3.7. Assay of Estrogenic Activity
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tiedeken, E.J.; Tahar, A.; McHugh, B.; Rowan, N.J. Monitoring, sources, receptors, and control measures for three European Union watch list substances of emerging concern in receiving waters—A 20 year systematic review. Sci. Total Environ. 2017, 574, 1140–1163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Lu, G.N.; Yin, H.; Dang, Z.; Rittmann, B. Removal of natural estrogens and their conjugates in municipal wastewater treatment plants: A critical review. Environ. Sci. Technol. 2015, 49, 5288–5300. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ying, G.G.; Zhou, L.J.; Zhang, R.Q.; Chen, Z.F.; Lai, H.J. Steroids in a typical swine farm and their release into the environment. Water Res. 2012, 46, 3754–3768. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, D.; Schrade, S.; Zähner, M.; Müller, M.; Hollender, J.; Bucheli, T.D. Occurrence and Fate of Natural Estrogens in Swiss Cattle and Pig Slurry. J. Agric. Food Chem. 2020, 68, 5545–5554. [Google Scholar] [CrossRef]
- Gao, H.; Yang, B.J.; Li, N.; Feng, L.M.; Shi, X.Y.; Zhao, W.H.; Liu, S.J. Bisphenol A and hormone-associated cancers: Current progress and perspectives. Medicine 2015, 94, e211. [Google Scholar] [CrossRef]
- Noszczyńska, M.; Piotrowska-Seget, Z. Bisphenols: Application, occurrence, safety, and biodegradation mediated by bacterial communities in wastewater treatment plants and rivers. Chemosphere 2018, 201, 214–223. [Google Scholar] [CrossRef]
- Avar, P.; Zrínyi, Z.; Maász, G.; Takátsy, A.; Lovas, S.; G-Tóth, L.; Pirger, Z. β-Estradiol and ethinyl-estradiol contamination in the rivers of the Carpathian Basin. Environ. Sci. Pollut. Res. 2016, 23, 11630–11638. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Shi, J.; Bo, T.; Meng, Y.; Zhan, X.; Zhang, M.; Zhang, Y. Distributions and ecological risk assessment of estrogens and bisphenol A in an arid and semiarid area in northwest China. Environ. Sci. Pollut. Res. 2017, 24, 7216–7225. [Google Scholar] [CrossRef]
- Pignotti, E.; Farré, M.; Barceló, D.; Dinelli, E. Occurrence and distribution of six selected endocrine disrupting compounds in surface- and groundwaters of the Romagna area (North Italy). Environ. Sci. Pollut. Res. 2017, 24, 21153–21167. [Google Scholar] [CrossRef]
- Zhou, L.J.; Zhang, B.B.; Zhao, Y.G.; Wu, Q.L. Occurrence, spatiotemporal distribution, and ecological risks of steroids in a large shallow Chinese lake, Lake Taihu. Sci. Total Environ. 2016, 557–558, 68–79. [Google Scholar] [CrossRef]
- Tiwari, M.; Sahu, S.K.; Pandit, G.G. Distribution and estrogenic potential of endocrine disrupting chemicals (EDCs) in estuarine sediments from Mumbai, India. Environ. Sci. Pollut. Res. 2016, 23, 18789–18799. [Google Scholar] [CrossRef] [PubMed]
- Nie, M.; Yang, Y.; Liu, M.; Yan, C.; Shi, H.; Dong, W.; Zhou, J.L. Environmental estrogens in a drinking water reservoir area in Shanghai: Occurrence, colloidal contribution and risk assessment. Sci. Total Environ. 2014, 487, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Badamasi, I.; Odong, R.; Masembe, C. Threats posed by xenoestrogenic chemicals to the aquatic ecosystem, fish reproduction and humans: A review. Afr. J. Aquat. Sci. 2020, 45, 243–258. [Google Scholar] [CrossRef]
- Wallis, D.J.; Truong, L.; La Du, J.; Tanguay, R.L.; Reif, D.M. Uncovering evidence for endocrine-disrupting chemicals that elicit differential susceptibility through gene-environment interactions. Toxics 2021, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, F.; Ogura, M.; Saitoh, S.; Yamazoe, A.; Yagi, O. Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J. Biosci. Bioeng. 2010, 109, 576–582. [Google Scholar] [CrossRef]
- Fernández, L.; Louvado, A.; Esteves, V.I.; Gomes, N.C.M.; Almeida, A.; Cunha, Â. Biodegradation of 17β-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media. J. Hazard. Mater. 2017, 323, 359–366. [Google Scholar] [CrossRef]
- Xiong, W.; Yin, C.; Peng, W.; Deng, Z.; Lin, S.; Liang, R. Characterization of an 17β-estradiol-degrading bacterium Stenotrophomonas maltophilia SJTL3 tolerant to adverse environmental factors. Appl. Microbiol. Biotechnol. 2020, 104, 1291–1305. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, J.; Chen, J. Isolation and characteristics of 17β-estradiol-degrading Bacillus spp. strains from activated sludge. Biodegradation 2010, 21, 729–736. [Google Scholar] [CrossRef]
- Cai, W.; Li, Y.; Niu, L.; Wang, Q.; Wu, Y. Kinetic study on the cometabolic degradation of 17β-estradiol and 17α-ethinylestradiol by an Acinetobacter sp. strain isolated from activated sludge. Desalination Water Treat. 2016, 57, 22671–22681. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Sun, M.; Ling, W.; Zhu, X. Isolation, characterization, and degradation performance of the 17β-estradiol-degrading bacterium Novosphingobium sp. E2S. Int. J. Environ. Res. Public Health 2017, 14, 115. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, J.; Xu, D.; Ling, W.; Li, S.; Chen, M. Isolation, Immobilization, and Degradation Performance of the 17β-Estradiol-Degrading Bacterium Rhodococcus sp. JX-2. Water Air Soil Pollut. 2016, 227, 422. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, P.; Liu, D.; Gao, R.; Shao, H.; Zhao, H.; Ma, Z.; Wang, D.; Huo, H. Degradation characteristics and metabolic pathway of 17β-estradiol (E2) by Rhodococcus sp. DS201. Biotechnol. Bioprocess Eng. 2016, 21, 804–813. [Google Scholar] [CrossRef]
- Yu, C.P.; Roh, H.; Chu, K.H. 17β-estradiol-degrading bacteria isolated from activated sludge. Environ. Sci. Technol. 2007, 41, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Moreira, I.S.; Lebel, A.; Peng, X.; Castro, P.M.L.; Gonçalves, D. Sediments in the mangrove areas contribute to the removal of endocrine disrupting chemicals in coastal sediments of Macau SAR, China, and harbour microbial communities capable of degrading E2, EE2, BPA and BPS. Biodegradation 2021, 32, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Meng, F.; Meng, Q.; Gao, Y.; Zhang, L.; Wang, L.; Wang, Y.; Li, X.; Huo, H. The Analysis of Estrogen-Degrading and Functional Metabolism Genes in Rhodococcus equi DSSKP-R-001. Int. J. Genom. 2020, 2020, 9369182. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Sun, K.; Yang, Z.; Ling, W. Degradation of 17β-estradiol by Novosphingobium sp. ES2-1 in aqueous solution contaminated with tetracyclines. Environ. Pollut. 2020, 260, 114063. [Google Scholar] [CrossRef]
- Roh, H.; Chu, K.H. A 17β-estradiol-utilizing bacterium, sphingomonas strain KC8: Part I—Characterization and abundance in wastewater treatment plants. Environ. Sci. Technol. 2010, 44, 4943–4950. [Google Scholar] [CrossRef]
- Iasur-Kruh, L.; Hadar, Y.; Minz, D. Isolation and bioaugmentation of an estradiol-degrading bacterium and its integration into a mature biofilm. Appl. Environ. Microbiol. 2011, 77, 3734–3740. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, Y.; Wang, C.; Niu, L.; Cai, W. Occurrence of endocrine disrupting compounds in aqueous environment and their bacterial degradation: A review. Crit. Rev. Environ. Sci. Technol. 2016, 46, 1–59. [Google Scholar] [CrossRef]
- Ma, L.; Yates, S.R. A review on structural elucidation of metabolites of environmental steroid hormones via liquid chromatography–mass spectrometry. TrAC—Trends Anal. Chem. 2018, 109, 142–153. [Google Scholar] [CrossRef]
- Chen, Y.L.; Fu, H.Y.; Lee, T.H.; Shih, C.J.; Huang, L.; Wang, Y.S.; Ismail, W.; Chiang, Y.R. Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl. Environ. Microbiol. 2018, 84, e00001-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.B.; Liu, D. Degradation of 17β-estradiol and its metabolites by sewage bacteria. Water Air Soil Pollut. 2002, 134, 351–366. [Google Scholar] [CrossRef]
- Pratush, A.; Yang, Q.; Peng, T.; Huang, T.; Hu, Z. Identification of non-accumulating intermediate compounds during estrone (E1) metabolism by a newly isolated microbial strain BH2-1 from mangrove sediments of the South China Sea. Environ. Sci. Pollut. Res. 2020, 27, 5097–5107. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Du, B.; Fan, G.; Yang, S.; Yang, L.; Zhang, M. Spatio-temporal distribution and transformation of 17α- and 17β-estradiol in sterilized soil: A column experiment. J. Hazard. Mater. 2020, 389, 122092. [Google Scholar] [CrossRef] [PubMed]
- Nakai, S.; Yamamura, A.; Tanaka, S.; Shi, J.; Nishikawa, M.; Nakashimada, Y.; Hosomi, M. Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environ. Chem. Lett. 2011, 9, 1–6. [Google Scholar] [CrossRef]
- Moreira, I.S.; Amorim, C.L.; Carvalho, M.F.; Ferreira, A.C.; Afonso, C.M.; Castro, P.M.L. Effect of the metals iron, copper and silver on fluorobenzene biodegradation by Labrys portucalensis. Biodegradation 2013, 24, 245–255. [Google Scholar] [CrossRef]
- Amorim, C.L.; Ferreira, A.C.S.; Carvalho, M.F.; Afonso, C.M.M.; Castro, P.M.L. Mineralization of 4-fluorocinnamic acid by a Rhodococcus strain. Appl. Microbiol. Biotechnol. 2014, 98, 1893–1905. [Google Scholar] [CrossRef]
- EzBioCloud, 16S-based ID. Available online: https://www.ezbiocloud.net/identify (accessed on 31 January 2022).
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Bessa, V.S.; Moreira, I.S.; Murgolo, S.; Mascolo, G.; Castro, P.M.L. Carbamazepine is degraded by the bacterial strain Labrys portucalensis F11. Sci. Total Environ. 2019, 690, 739–747. [Google Scholar] [CrossRef] [PubMed]
- GenBank Database, National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 29 April 2022).
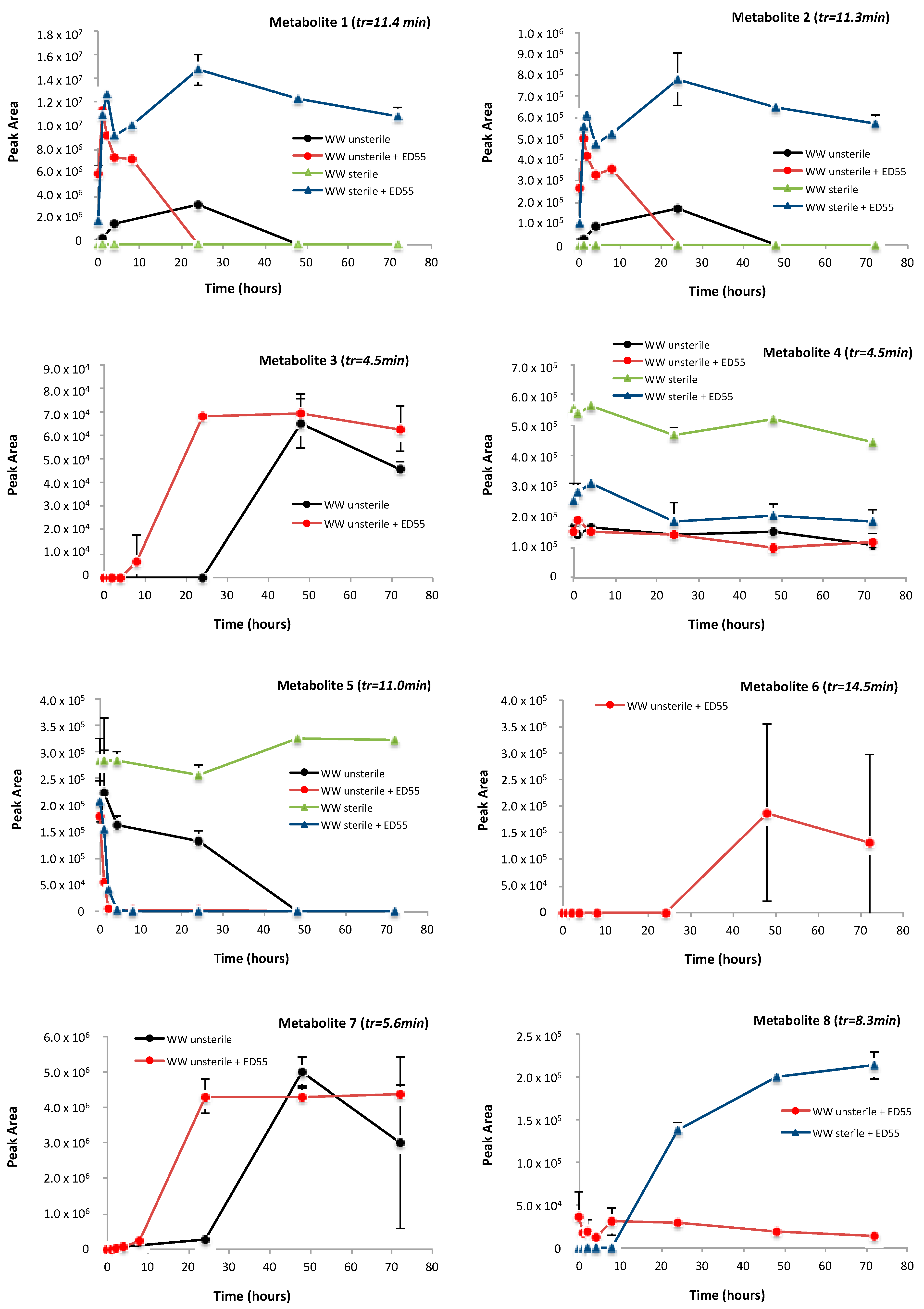

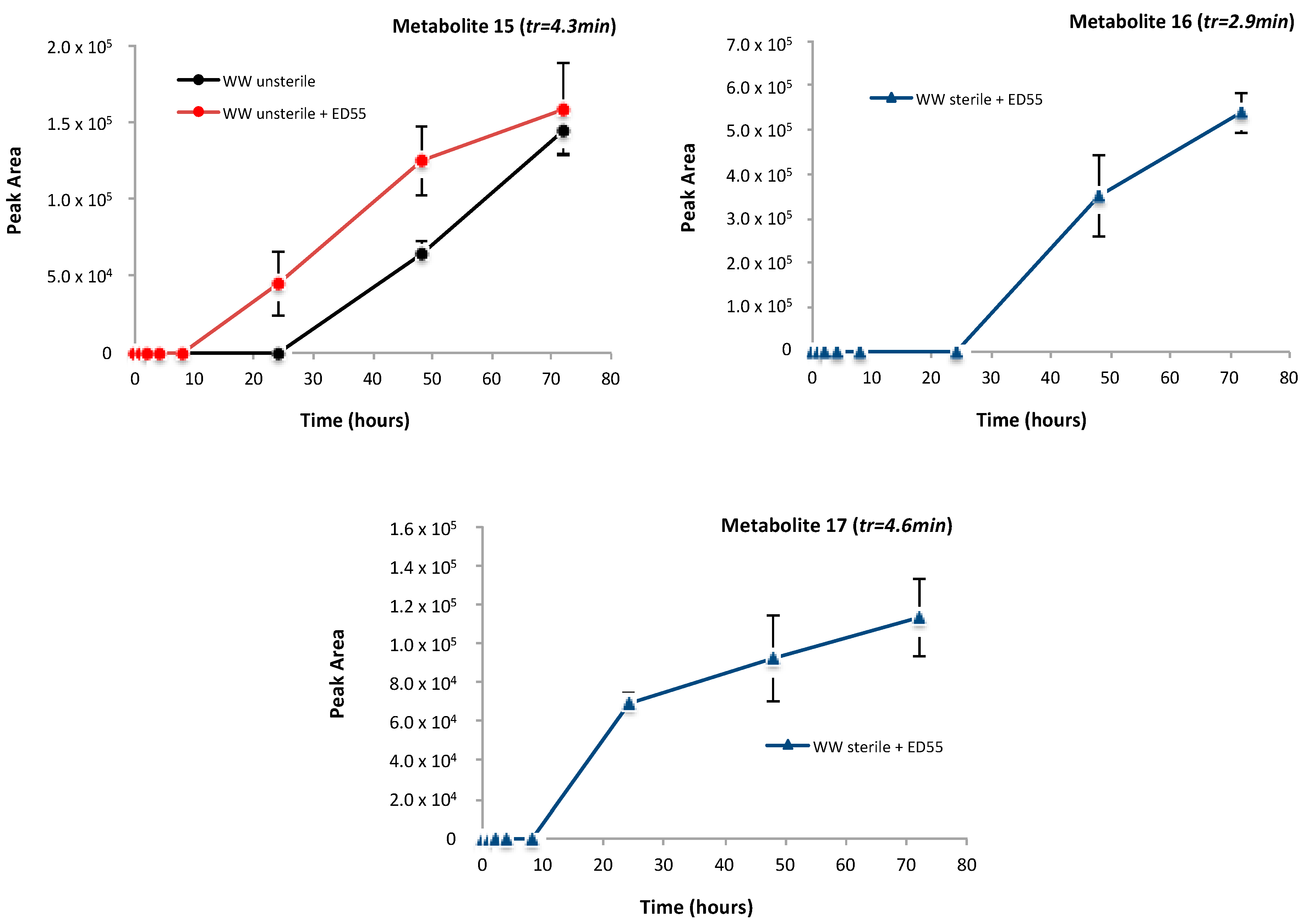
| Metabolite | Ionization Mode | Calculated m/z | Measured m/z | Error ppm | MS/MS Products | Predicted Formula |
|---|---|---|---|---|---|---|
| 1 | ESI (-) | 269.1547 | 269.1546 | −0.5 | 145.0659, 183.0891, 159.0819, 253.1238 | C18H22O2 |
| 2 | ESI (-) | 285.1496 | 285.1495 | −0.5 | 108.0214, 161.0597, 225.1273, 267.1384 | C18H22O3 |
| 3 | ESI (-) | 289.1445 | 289.1448 | 0.8 | 121.0665, 149.0975, 219.1391, 245.1538 | C17H22O4 |
| 4 | ESI (-) | 185.1183 | 185.1186 | 1.6 | 79.9556, 121.0679, 139.1107, 166.9829 | C10H18O3 |
| 5 | ESI (-) | 287.1652 | 287.1643 | −3.3 | 108.0215, 183.0808, 259.1541, 269.1542 | C18H24O3 |
| 6 | ESI (-) | 275.2016 | 275.2008 | −3.3 | 159.0774, 177.1660, 203.1895, 231.2112 | C18H28O2 |
| 7 | ESI (+) | 300.1592 | 300.1594 | −0.6 | 145.1010, 194.0971, 236.1442, 254.1538 | C18H21NO3 |
| 8 | ESI (+) | 559.2524 | 559.2538 | −2.5 | 136.0751, 283.1435, 412.1846, 531.2573 | C30H38O10 |
| 9 | ESI (-) | 329.2483 | 329.2486 | −0.9 | 163.0727, 231.2079, 251.1948, 285.2594 | C22H34O2 |
| 10 | ESI (-) | 374.2446 | 374.2449 | −0.9 | 232.1093, 290.1877 | C21H33N3O3 |
| 11 | ESI (-) | 357.2051 | 357.2044 | 1.7 | 121.0650, 186.9527, 235.1360, 274.9421 | C18H26N6O2 |
| 12 | ESI (-) | 162.0784 | 162.0785 | −0.4 | 90.0098, 100.9240, 117.0264, 146.0466 | C7H9N5 |
| 13 | ESI (-) | 150.0421 | 150.0421 | 0 | 80.0294, 90.0085, 108.0194, 133.0147 | C5H5N5O |
| 14 | ESI (+) | 329.1571 | 329.1568 | 0.9 | 75.0256, 117.0733, 243.2328, 287.2228 | C12H20N6O5 |
| 15 | ESI (+) | 658.2180 | 658.2184 | 0.6 | 171.0652, 310.0744, 456.1352, 512.1588 | C38H31N3O8 |
| 16 | ESI (+) | 113.0341 | 113.0345 | −4.2 | 70.0283, 96.0072 | C4H4N2O2 |
| 17 | ESI (+) | 364.1134 | 364.1139 | −1.4 | 152.0559, 187.0503, 230.0555, 346.1021 | C16H17N3O7 |
| WW Sterile | WW Unsterile | |
|---|---|---|
| Before | 55.5 ± 1.0 a | 50.2 ± 2.0 b |
| After (without ED55) | 49.2 ± 2.3 b | 12.6 ± 0.7 c |
| After (with ED55) | 2.3 ± 3.6 d | 0.0 ± 0.0 d |
| Germination (% Inhibition) | Root Growth (% Inhibition) | Shoot Growth (% Inhibition) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| WW Sterile | WW Unsterile | MM | WW Sterile | WW Unsterile | MM | WW Sterile | WW Unsterile | MM | |
| Before | 57.1 ± 7.1 d | 52.9 ± 8.7 cd | 28.6 ± 4.3 ab | 35.6 ± 4.6 c | 19.0 ± 2.0 b | 0.0 ± 0.0 a | 5.32 ± 0.9 ab | 12.4 ± 6.5 c | 4.37 ± 0.6 a |
| After (without ED55) | 56.6 ± 8.3 cd | 33.3 ± 2.0 b | 30.2 ± 4.6 ab | 39.7 ± 3.8 c | 0.0 ± 0.0 a | 0.0 ± 0.0 a | 5.22 ± 1.3 ab | 0.0 ± 0.0 a | 0.0 ± 0.0 a |
| After (with ED55) | 39.7 ± 2.8 bc | 34.9 ± 6.6 b | 15.9 ± 6.0 a | 5.63 ± 4.8 a | 0.0 ± 0.0 a | 5.0 ± 1.6 a | 0.0 ± 0.0 a | 0.0 ± 0.0 a | 2.34 ± 0.3 a |
| WW Sterile | WW Unsterile | MM | |
|---|---|---|---|
| Before | >400 a | >400 a | 352.2 ± 14.1 d |
| After (without ED55) | >400 a | <LOD c | >400 a |
| After (with ED55) | 126.3 ± 13.0 b | <LOD c | 124.9 ± 19.3 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, I.S.; Murgolo, S.; Mascolo, G.; Castro, P.M.L. Biodegradation and Metabolic Pathway of 17β-Estradiol by Rhodococcus sp. ED55. Int. J. Mol. Sci. 2022, 23, 6181. https://doi.org/10.3390/ijms23116181
Moreira IS, Murgolo S, Mascolo G, Castro PML. Biodegradation and Metabolic Pathway of 17β-Estradiol by Rhodococcus sp. ED55. International Journal of Molecular Sciences. 2022; 23(11):6181. https://doi.org/10.3390/ijms23116181
Chicago/Turabian StyleMoreira, Irina S., Sapia Murgolo, Giuseppe Mascolo, and Paula M. L. Castro. 2022. "Biodegradation and Metabolic Pathway of 17β-Estradiol by Rhodococcus sp. ED55" International Journal of Molecular Sciences 23, no. 11: 6181. https://doi.org/10.3390/ijms23116181
APA StyleMoreira, I. S., Murgolo, S., Mascolo, G., & Castro, P. M. L. (2022). Biodegradation and Metabolic Pathway of 17β-Estradiol by Rhodococcus sp. ED55. International Journal of Molecular Sciences, 23(11), 6181. https://doi.org/10.3390/ijms23116181







