The Formation of Hollow Trait in Cucumber (Cucumis sativus L.) Fruit Is Controlled by CsALMT2
Abstract
:1. Introduction
2. Results
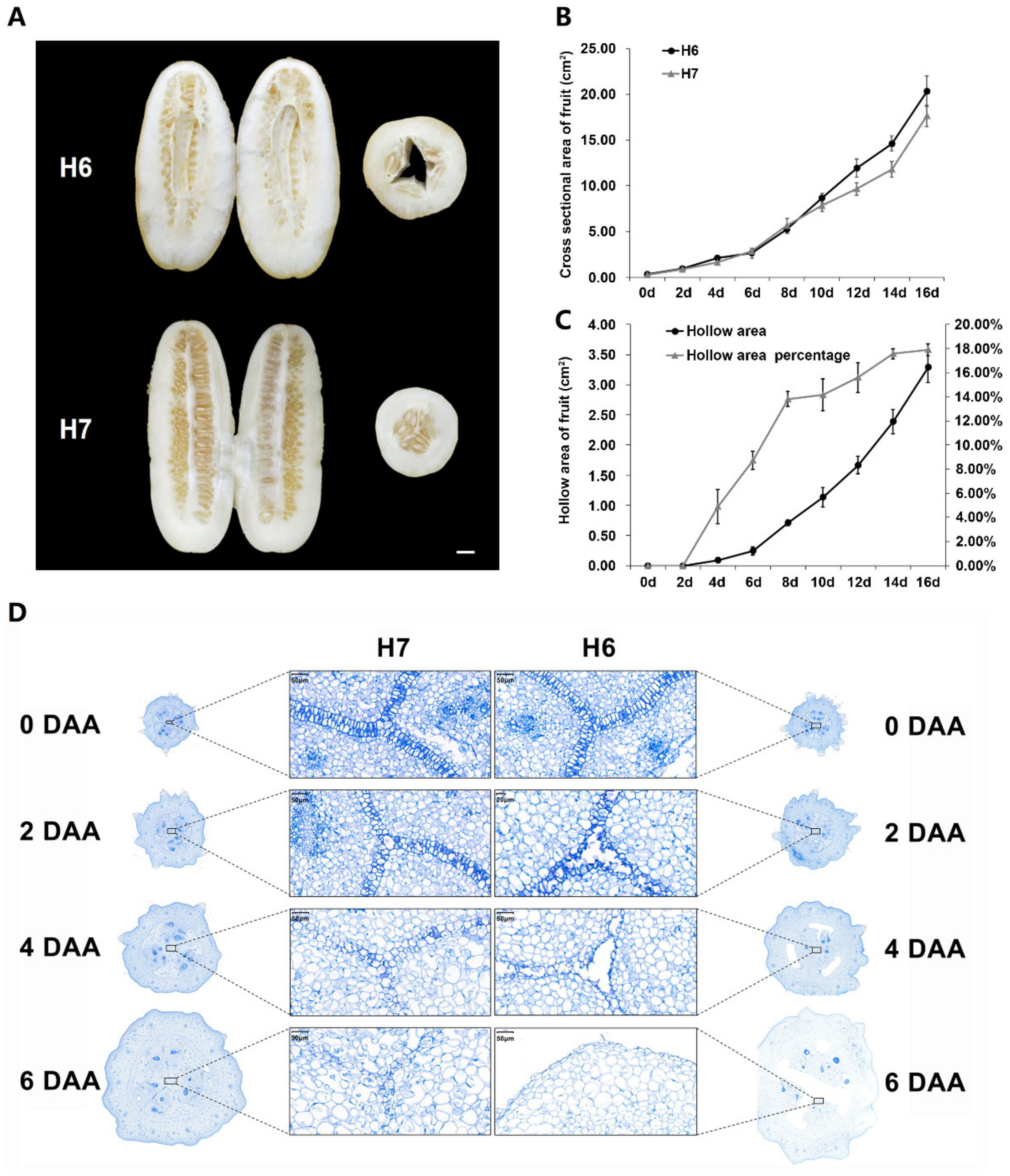
2.1. Phenotypic Characterization of H6 and H7
2.2. Cytological Observations of H6 and H7
2.3. Genetic Analysis of Hollow Traits in Cucumber Fruit
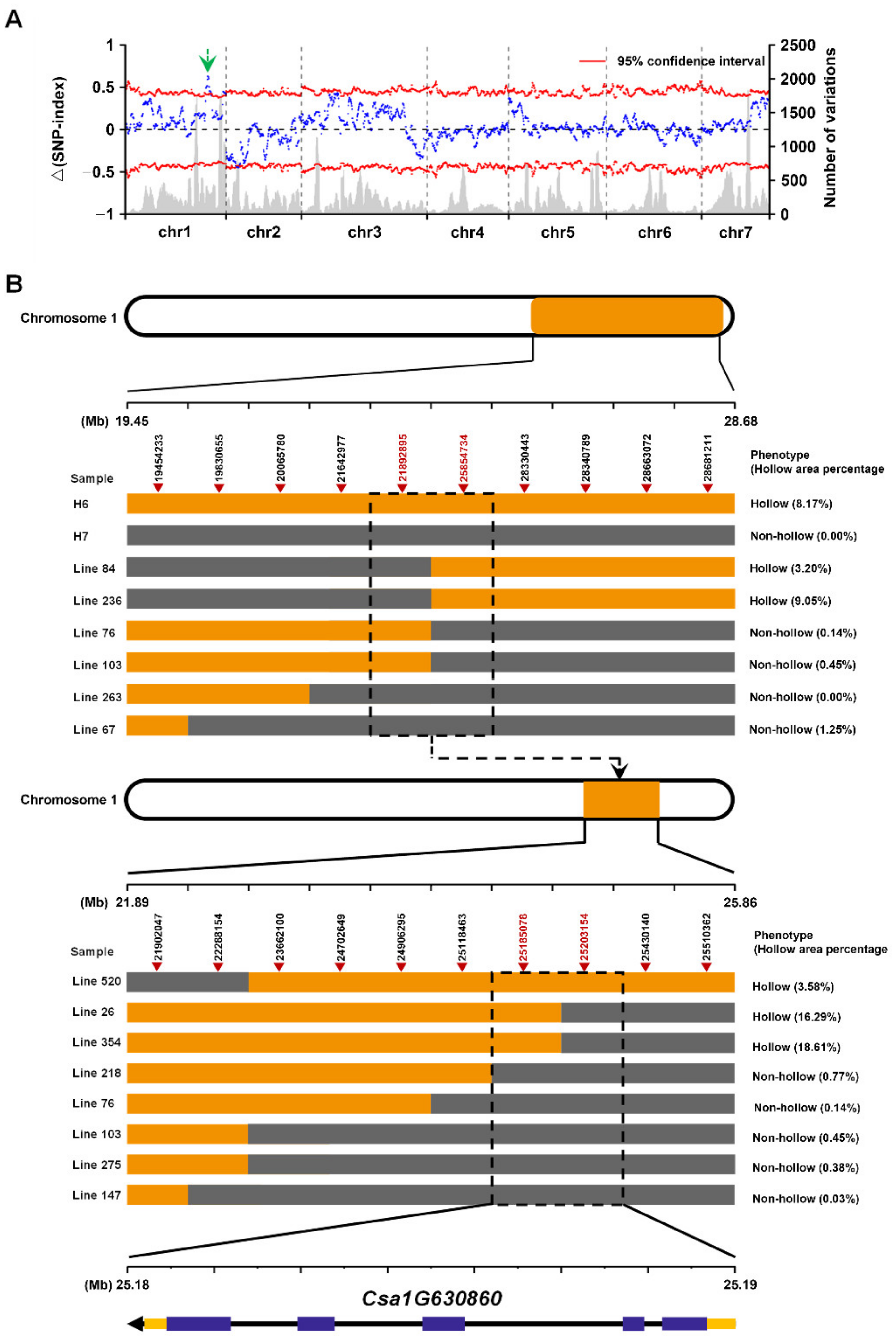
2.4. Whole Genome Resequencing Based on BSA Analysis
2.5. Fine Mapping and Identification of the Causal Gene
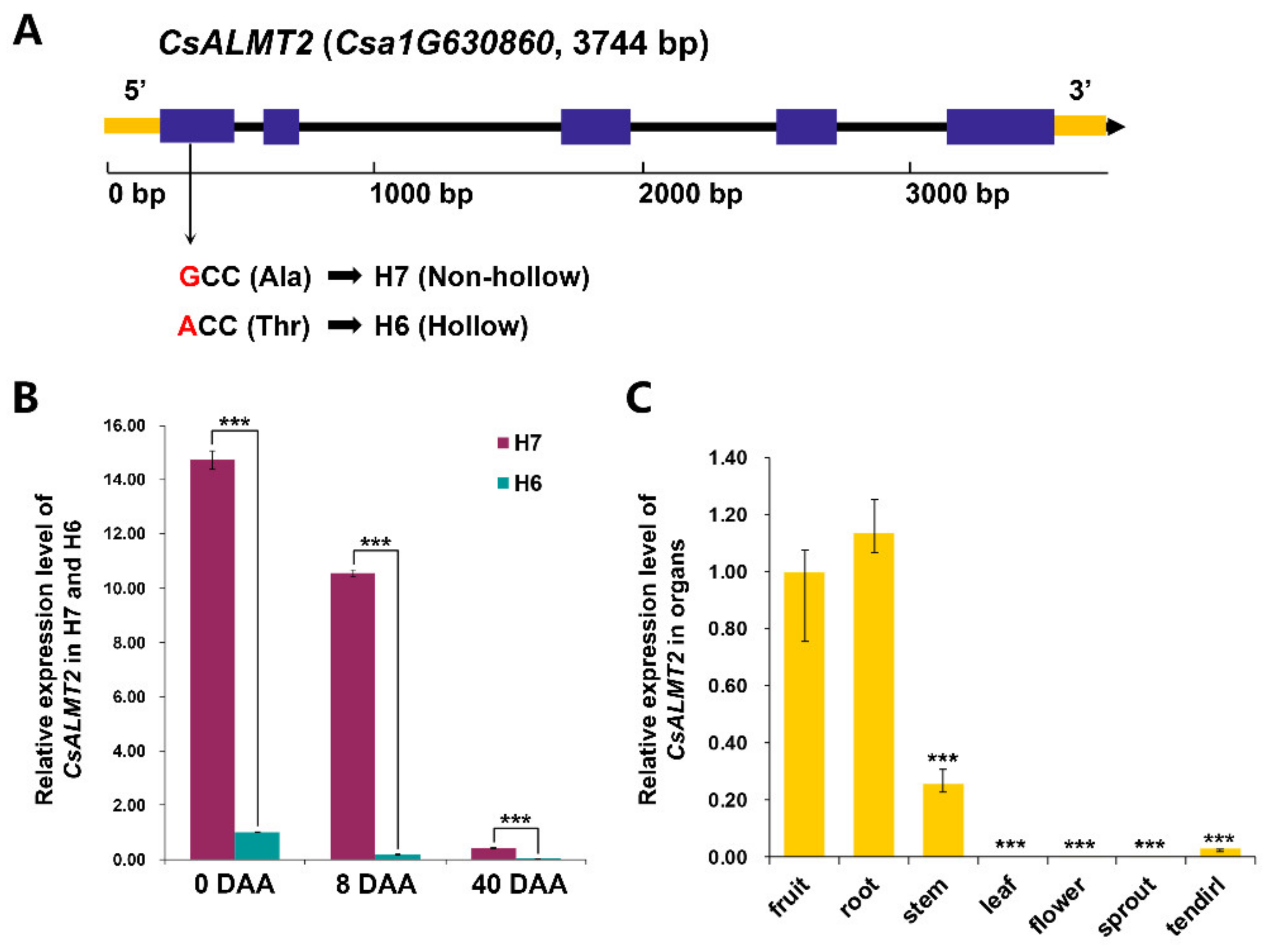
2.6. Sequence and Cis-Element Prediction of CsALMT2
2.7. Expression Characteristic Analysis of CsALMT2
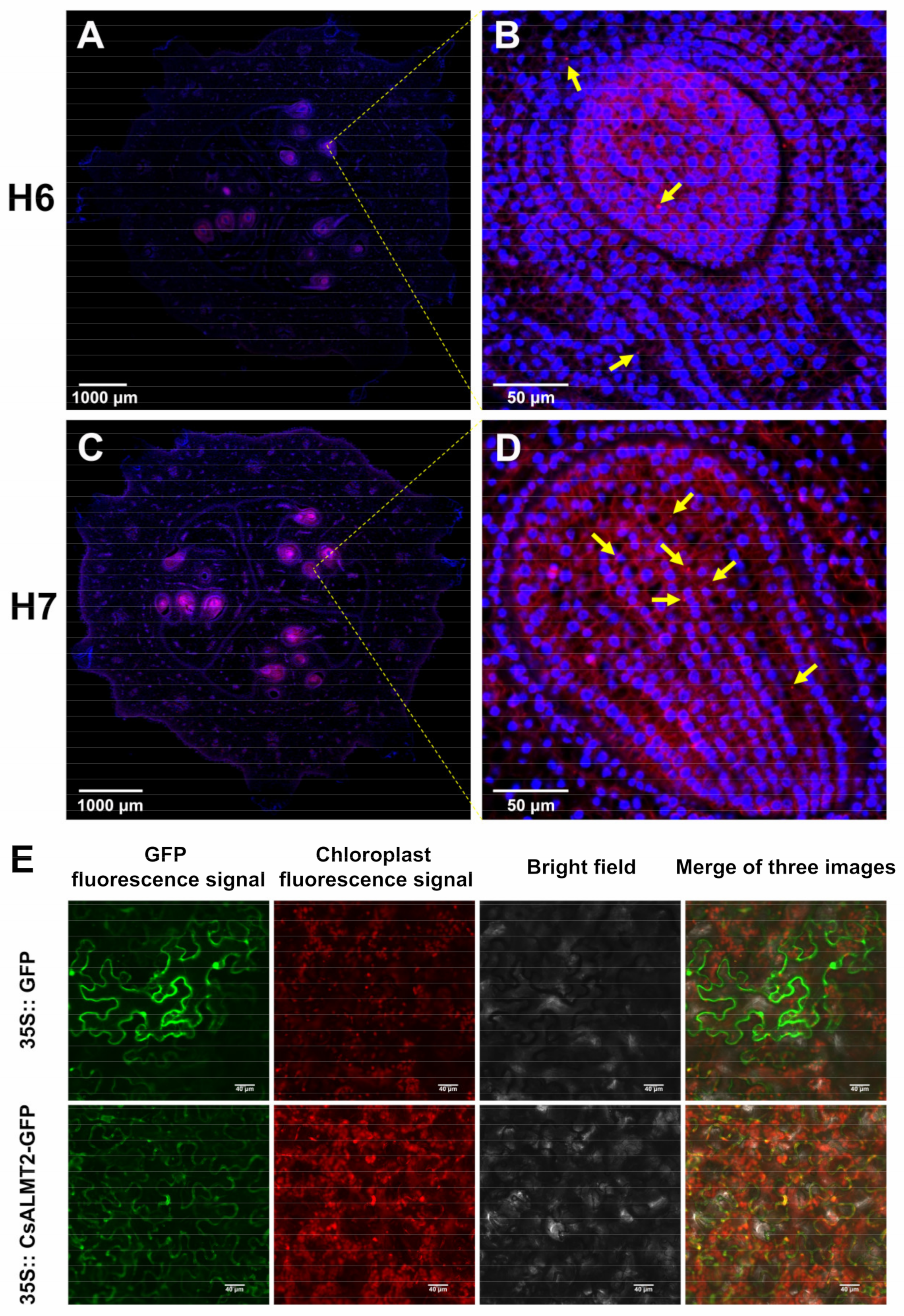
2.8. Fluorescence In Situ Hybridization
2.9. Subcellular Localization of CsALMT2-Encoding Protein
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Crossing
4.1.1. Plant Materials
4.1.2. Construction of Genetic Population
4.1.3. Identification of Hollow Trait of New Open Field Cucumber Varieties in China
4.2. Investigation on Hollow Trait of Cucumber Fruit
4.3. Determination of Population Hollow Rate
4.4. Paraffin Sectioning
4.5. Whole Genome Resequencing
4.6. SNP Calling and Filtering
4.7. SNP Genotyping by KASP
4.8. Domain and Cis-Element Prediction
4.9. RNA Isolation and qRT-PCR
4.10. Fluorescence In Situ Hybridization
4.11. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gebretsadik, K.; Qiu, X.Y.; Dong, S.Y.; Miao, H.; Bo, K.L. Molecular research progress and improvement approach of fruit quality traits in cucumber. Appl. Genet. 2021, 134, 3535–3552. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Zhang, J.P.; Mu, Z.H.; Wang, Y.J.; Wen, C.L.; Wu, T.; Yu, C.; Li, Z.; Wang, H.S. Recent progress on the molecular breeding of Cucumis sativus L. in China. Appl. Genet. 2020, 133, 1777–1790. [Google Scholar] [CrossRef] [PubMed]
- Colle, M.; Weng, Y.Q.; Kang, Y.Y.; Ophir, R.; Sherman, A.; Grumet, R. Variation in cucumber (Cucumis sativus L.) fruit size and shape results from multiple components acting pre-anthesis and post-pollination. Planta 2017, 246, 641–658. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.P.; Wang, Y.H.; McGregor, C.; Liu, S.; Luan, F.S.; Gao, M.L.; Weng, Y.Q. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef]
- Chu, Y.H.; Jang, J.C.; Huang, Z.J.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 2019, 3, e00142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Zhang, K.; Bai, J.R.; Lu, J.H.; Lu, X.X.; Hu, J.L.; Pan, C.Y.; He, S.M.; Yuan, J.L.; Zhang, Y.Y.; et al. All-flesh fruit in tomato is controlled by reduced expression dosage of AFF through a structural variant mutation in the promoter. J. Exp. Bot. 2022, 73, 123–138. [Google Scholar] [CrossRef]
- Che, G.; Zhang, X.L. Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 2019, 47, 38–46. [Google Scholar] [CrossRef]
- Kamiuchi, Y.; Yamamoto, K.; Furutani, M.; Tasaka, M.; Aida, M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 2014, 5, 165. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.H.; Jiang, B.; Dymerski, R.; Xu, X.W.; Weng, Y.Q. Quantitative trait loci for horticulturally important traits defining the Sikkim cucumber, Cucumis sativus var. sikkimensis. Appl. Genet. 2021, 134, 229–247. [Google Scholar] [CrossRef]
- Davies, J.W.; Cocking, E.C. Changes in carbohydrates, proteins and nucleic acids during cellular development in tomato fruit locule tissue. Planta 1965, 67, 242–253. [Google Scholar] [CrossRef]
- Cheng, G.W.; Huber, D.J. Alterations in structural polysaccharides during liquefaction of tomato locule tissue. Plant Physiol. 1996, 111, 447–457. [Google Scholar] [CrossRef] [Green Version]
- Frary, A.; Nesbitt, T.C.; Frary, A.; Grandillo, S.; van der Knaap, E.; Cong, B.; Liu, J.P.; Meller, J.; Elber, R.; Alpert, K.B.; et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Okuhara, K.; Kohyama, K. Visualization of planar stress distributions in cucumber cultivars using a multiple-point sheet sensor. J. Sci. Food Agric. 2004, 84, 1091–1096. [Google Scholar] [CrossRef]
- Li, S.; Pan, Y.P.; Wen, C.L.; Li, Y.H.; Liu, X.F.; Zhang, X.L.; Behera, T.K.; Xing, G.M.; Weng, Y.Q. Integrated analysis in bi-parental and natural populations reveals CsCLAVATA3 (CsCLV3) underlying carpel number variations in cucumber. Appl. Genet. 2016, 129, 1007–1022. [Google Scholar] [CrossRef]
- Che, G.; Gu, R.; Zhao, J.Y.; Liu, X.F.; Song, X.F.; Zi, H.L.; Cheng, Z.H.; Shen, J.J.; Wang, Z.Y.; Liu, R.Y.; et al. Gene regulatory network of carpel number variation in cucumber. Development 2020, 147, dev184788. [Google Scholar] [CrossRef]
- Hooker, J.D. Cucumis sativus var. sikkimensis. Curtis’s Bot. Mag. 1876, 32, 6206. [Google Scholar]
- Wilson, J.E.; Baker, L.R. Inheritance of carpel separation in mature fruits of pickling cucumbers. J. Am. Soc. Hortic. Sci. 1976, 101, 66–69. [Google Scholar]
- Elkner, K. The influence of different moisture contents and nitrogen levels on the formation of empty cavities in cucumbers (Cucumis sativus L.). Acta Agrobot. 1982, 35, 61–68. [Google Scholar] [CrossRef]
- Liu, B.; Liu, X.W.; Yang, S.; Chen, C.H.; Xue, S.D.; Cai, Y.L.; Wang, D.D.; Yin, S.; Gai, X.S.; Ren, H.Z. Silencing of the gibberellin receptor homolog, CsGID1a, affects locule formation in cucumber (Cucumis sativus) fruit. New Phytol. 2016, 210, 551–563. [Google Scholar] [CrossRef] [Green Version]
- Falconer, D.S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet. 1965, 29, 51–76. [Google Scholar] [CrossRef]
- Hao, N.; Du, Y.L.; Li, H.Y.; Wang, C.; Wang, C.; Gong, S.Y.; Zhou, S.G.; Wu, T. CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus L.). Appl. Genet. 2018, 131, 1659–1669. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Eduardo, I.; Arus, P.; Monforte, A.J.; Obando, J.; Fernandez-Trujillo, J.P.; Martinez, J.A.; Alarcon, A.L.; Alvarez, J.M.; van der Knaap, E. Estimating the genetic architecture of fruit quality traits in melon using a genomic library of near isogenic lines. J. Am. Soc. Hortic. Sci. 2007, 132, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Monforte, A.J.; Diaz, A.; Cano-Delgado, A.; van der Knaap, E. The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J. Exp. Bot. 2014, 65, 4625–4637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, K.; Carr, K.M.; Grumet, R. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genom. 2012, 13, 518. [Google Scholar] [CrossRef] [Green Version]
- Gillaspy, G.; Ben-David, H.; Gruissem, W. Fruits: A developmental perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar] [CrossRef] [Green Version]
- Fu, F.Q.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Spatio-temporal changes in cell division, endoreduplication and expression of cell cycle-related genes in pollinated and plant growth substances-treated ovaries of cucumber. Plant Biol. 2010, 12, 98–107. [Google Scholar] [CrossRef]
- Yang, X.Y.; Wang, Y.; Jiang, W.J.; Liu, X.L.; Zhang, X.M.; Yu, H.J.; Huang, S.W.; Liu, G.Q. Characterization and expression profiling of cucumber kinesin genes during early fruit development: Revealing the roles of kinesins in exponential cell production and enlargement in cucumber fruit. J. Exp. Bot. 2013, 64, 4541–4557. [Google Scholar] [CrossRef] [Green Version]
- Kovermann, P.; Meyer, S.; Hörtensteiner, S.; Picco, C.; Scholz-Starke, J.; Ravera, S.; Lee, Y.; Martinoia, E. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007, 52, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.J.; Baker, A.; Muench, S.P. The varied functions of aluminium-activated malate transporters-much more than aluminium resistance. Biochem. Soc. Trans. 2016, 44, 856–862. [Google Scholar] [CrossRef]
- Sharma, T.; Dreyer, I.; Kochian, L.; Piñeros, M.A. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.M.; Alam, S.M.; Anwar, R.; Ali, S.; Shi, M.; Liang, D.D.; Lin, Z.M.; Chen, F.X. Genome-Wide Identification, Characterization and Expression Profiling of Aluminum-Activated Malate Transporters in Eriobotrya japonica Lindl. Horticulturae 2021, 7, 441. [Google Scholar] [CrossRef]
- Sasaki, T.; Yamamoto, Y.; Ezaki, B.; Kasuara, M.; Ahn, S.J.; Ryan, P.R.; Delhaize, E.; Matsumoto, H. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004, 37, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, É.C.D.; Pinto-Maglio, C.A.F. Cytogenetic mapping of the ALMT (aluminum-activated malate transporter) gene in wheat genotypes. Sci. Agric. 2019, 77, e20190012. [Google Scholar] [CrossRef]
- Furuichi, T.; Sasaki, T.; Tsuchiya, Y.; Ryan, P.R.; Delhaize, E.; Yamamoto, Y. An extracellular hydrophilic carboxy-terminal domain regulates the activity of TaALMT1, the aluminum-activated malate transport protein of wheat. Plant J. 2010, 64, 47–55. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef] [Green Version]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [Green Version]
- De Angeli, A.; Zhang, J.; Meyer, S.; Martinoia, E. AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 2013, 4, 1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.S.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.W.; An, F.; Wang, L.F.; Guo, D.; Xie, G.H.; Liu, Z.F. Genome-wide identification of aluminum-activated malate transporter (ALMT) gene family in rubber trees (Hevea brasiliensis) highlights their involvement in aluminum detoxification. Forests 2020, 11, 142. [Google Scholar] [CrossRef] [Green Version]
- Piñeros, M.A.; Cancado, G.M.A.; Maron, L.G.; Lyi, S.M.; Menossi, M.; Kochian, L.V. Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: The case of ZmALMT1-an anion-selective transporter. Plant J. 2008, 53, 352–367. [Google Scholar] [CrossRef]
- Ligaba, A.; Maron, L.; Shaff, J.; Kochian, L.; Piñeros, M. Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 2012, 35, 1185–1200. [Google Scholar] [CrossRef]
- Xu, M.Y.; Gruber, B.D.; Delhaize, E.; White, R.G.; James, R.A.; You, J.F.; Yang, Z.M.; Ryan, P.R. The barley anion channel, HvALMT1, has multiple roles in guard cell physiology and grain metabolism. Physiol. Plant 2015, 153, 183–193. [Google Scholar] [CrossRef]
- Collins, N.C.; Shirley, N.J.; Saeed, M.; Pallotta, M.; Gustafson, J.P. An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.). Genetics 2008, 179, 669–682. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.Y.; Piñeros, M.A.; Tian, J.; Yao, Z.F.; Sun, L.L.; Liu, J.P.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.R.; Liu, L.; Su, H.; Guo, L.Q.; Zhang, J.L.; Li, Y.F.; Xu, J.Y.; Zhang, X.C.; Guo, Y.D.; Zhang, N. Jasmonate and aluminum crosstalk in tomato: Identification and expression analysis of WRKYs and ALMTs during JA/Al-regulated root growth. Plant Physiol. Biochem. 2020, 154, 409–418. [Google Scholar] [CrossRef]
- Sasaki, T.; Tsuchiya, Y.; Ariyoshi, M.; Nakano, R.; Ushijima, K.; Kubo, Y.; Mori, L.C.; Higashiizumi, E.; Galis, L.; Yamamoto, Y. Two members of the aluminum-activated malate transporter family, SlALMT4 and SlALMT5, are expressed during fruit development, and the overexpression of SlALMT5 alters organic acid contents in seeds in tomato (Solanum lycopersicum). Plant Cell Physiol. 2016, 57, 2367–2379. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Wang, X.; Hu, T.X.; Zhang, F.X.; Wang, B.; Li, C.X.; Yang, T.X.; Li, H.X.; Lu, Y.G.; Giovannoni, J.J.; et al. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 2017, 29, 2249–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jawad, U.M.; Gao, L.; Gebremeskel, H.; Safdar, L.B.; Yuan, P.L.; Zhao, S.J.; Lu, X.Q.; He, N.; Zhu, H.J.; Liu, W.J. Expression pattern of sugars and organic acids regulatory genes during watermelon fruit development. Sci. Hortic. 2020, 265, 109102. [Google Scholar] [CrossRef]
- Clark, M. Plant Molecular Biology: A Laboratory Manual; Springer: Berlin, Germany, 1997. [Google Scholar]
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.W.; Xie, B.Y.; Ni, P.X.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Thijs, G.; Rombauts, S.; Lescot, M.; Marchal, K.; De Moor, B.; Moreau, Y.; Rouzé, P. Detection of cis-acting regulatory elements in plants: A Gibbs sampling approach. In Proceedings of the Second International Conference on Bioinformatics of Genome Regulation and Structure, Novosibirsk, Russia, 7–11 August 2000; Volume 1, pp. 118–126. [Google Scholar]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, G.; Chen, C.; Liu, X.; Yang, K.; Wang, C.; Lu, X.; Tian, Y.; Chen, H. The Formation of Hollow Trait in Cucumber (Cucumis sativus L.) Fruit Is Controlled by CsALMT2. Int. J. Mol. Sci. 2022, 23, 6173. https://doi.org/10.3390/ijms23116173
Zhou G, Chen C, Liu X, Yang K, Wang C, Lu X, Tian Y, Chen H. The Formation of Hollow Trait in Cucumber (Cucumis sativus L.) Fruit Is Controlled by CsALMT2. International Journal of Molecular Sciences. 2022; 23(11):6173. https://doi.org/10.3390/ijms23116173
Chicago/Turabian StyleZhou, Geng, Chen Chen, Xiaohong Liu, Kankan Yang, Chong Wang, Xiangyang Lu, Yun Tian, and Huiming Chen. 2022. "The Formation of Hollow Trait in Cucumber (Cucumis sativus L.) Fruit Is Controlled by CsALMT2" International Journal of Molecular Sciences 23, no. 11: 6173. https://doi.org/10.3390/ijms23116173
APA StyleZhou, G., Chen, C., Liu, X., Yang, K., Wang, C., Lu, X., Tian, Y., & Chen, H. (2022). The Formation of Hollow Trait in Cucumber (Cucumis sativus L.) Fruit Is Controlled by CsALMT2. International Journal of Molecular Sciences, 23(11), 6173. https://doi.org/10.3390/ijms23116173






