The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA
Abstract
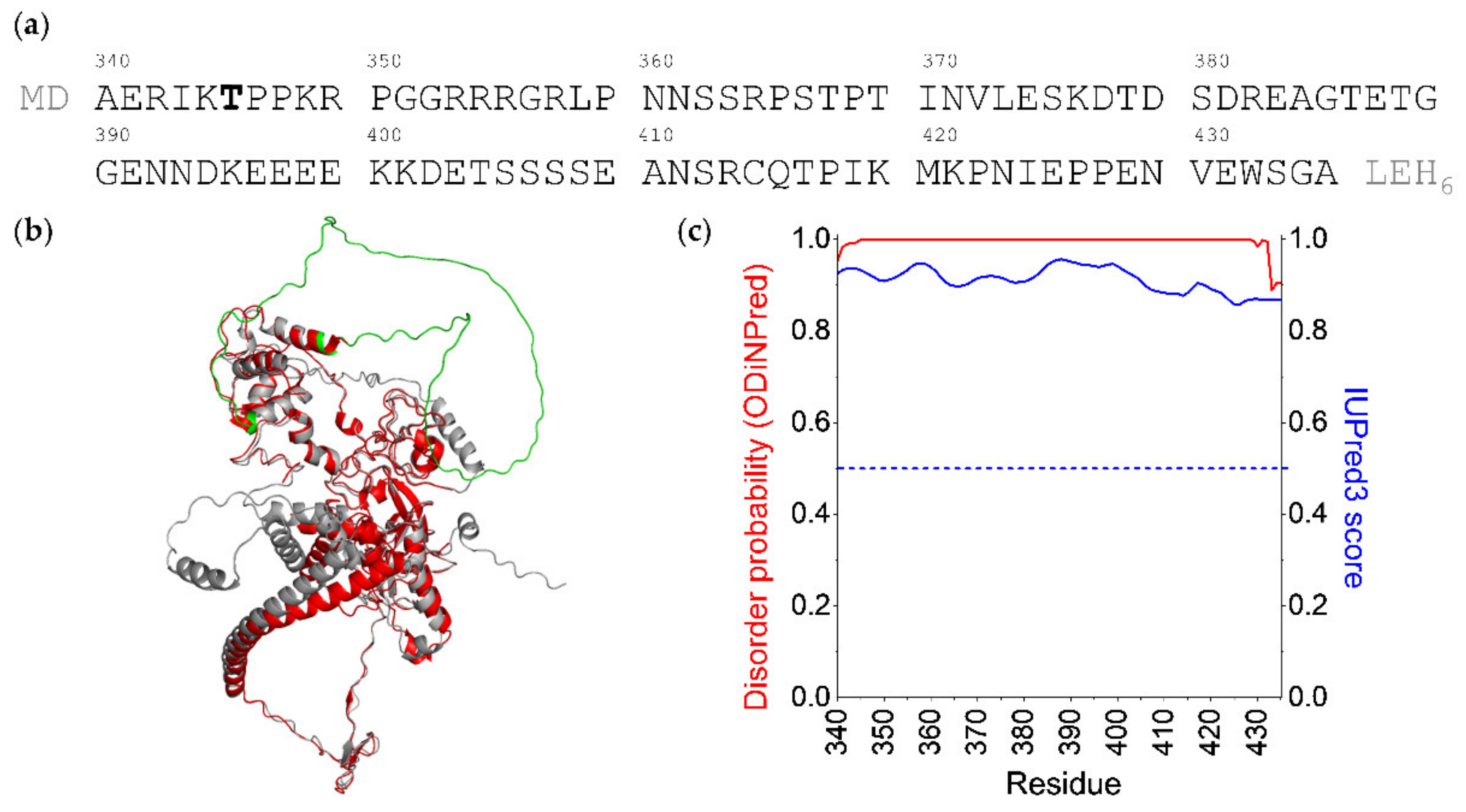
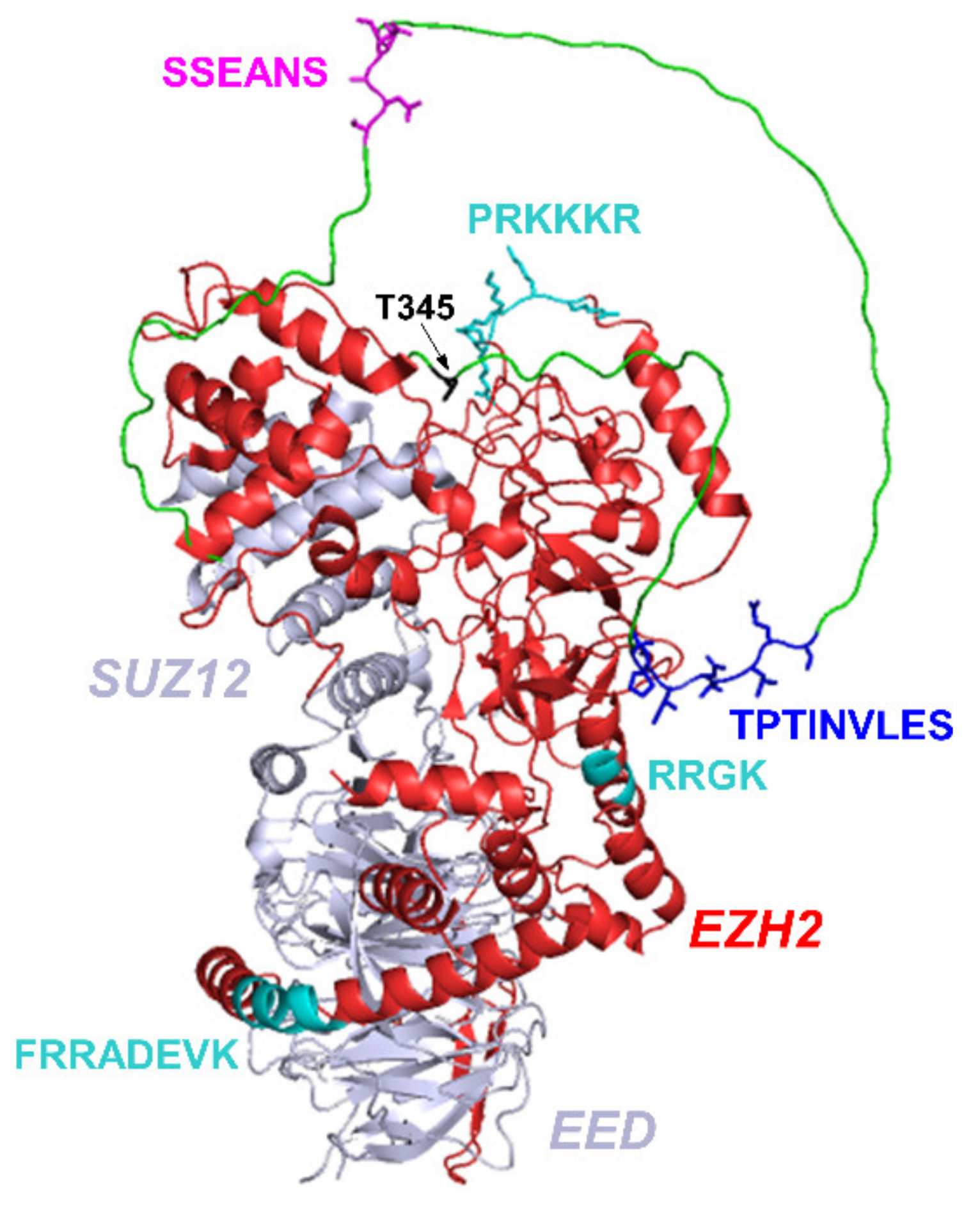
:1. Introduction
2. Results
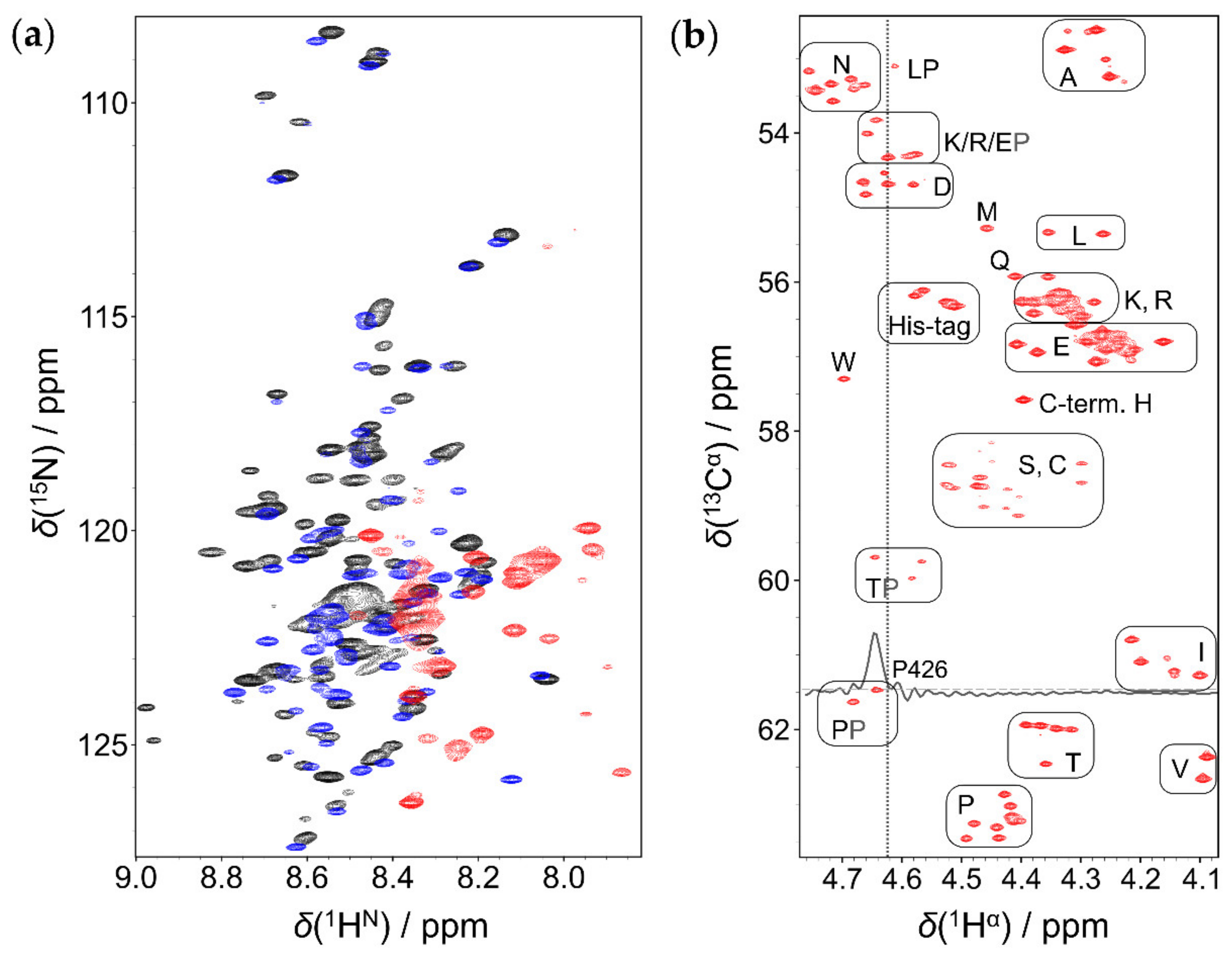
2.1. Comparison of 1HN and 1Hα Detected Approaches
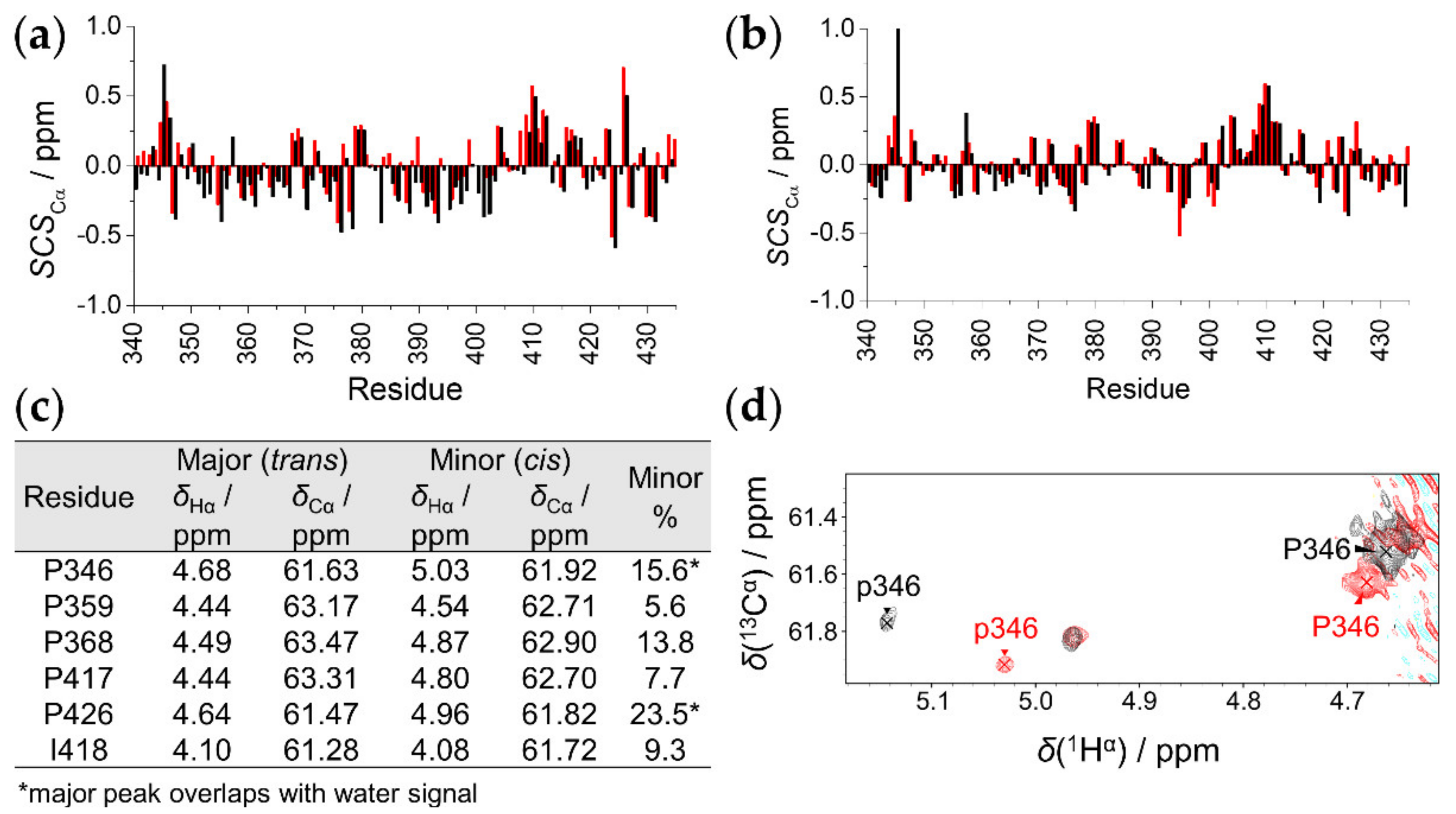
2.2. Secondary Structural Propensities
2.3. The Effect of T8D Mutation and Proline Isomers
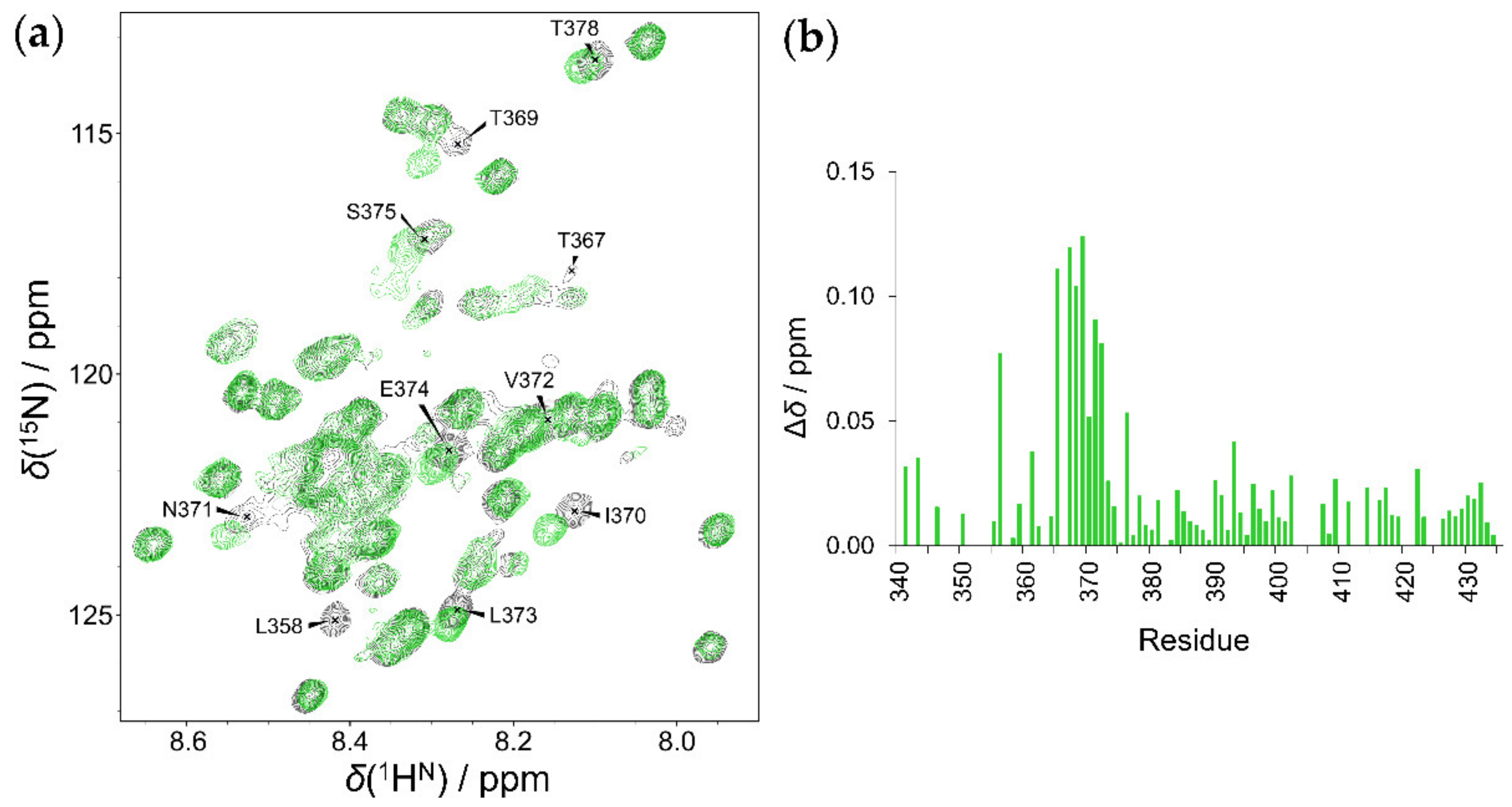
2.4. Interaction of HOTAIR140 with the EZH2 Loop
3. Discussion
4. Materials and Methods
4.1. EZH2 Loop Expression and Purification
4.2. RNA Transcription and Purification
4.3. NMR Measurements
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Su, M.; Xiao, Y.; Tang, J.; Wu, J.; Ma, J.; Tian, B.; Zhou, Y.; Wang, H.; Yang, D.; Liao, Q.-J.; et al. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J. Cancer 2018, 9, 4156–4165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathiyalagan, P.; Keating, S.T.; Du, X.-J.; El-Osta, A. Interplay of chromatin modifications and non-coding RNAs in the heart. Epigenetics 2014, 9, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Cifuentes-Rojas, C.; Hernandez, A.J.; Sarma, K.; Lee, J.T. Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Mol. Cell 2014, 55, 171–185. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Murat, P.; Matak-Vinkovic, D.; Murrell, A.; Balasubramanian, S. Binding Interactions between Long Noncoding RNA HOTAIR and PRC2 Proteins. Biochemistry 2013, 52, 9519–9527. [Google Scholar] [CrossRef] [PubMed]
- Justin, N.; Zhang, Y.; Tarricone, C.; Martin, S.R.; Chen, S.; Underwood, E.; De Marco, V.; Haire, L.F.; Walker, P.A.; Reinberg, D.; et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016, 7, 11316. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide Identification of Polycomb-Associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, S.; Son, J.; Shen, S.S.; Reinberg, D.; Bonasio, R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1258–1264. [Google Scholar] [CrossRef] [Green Version]
- Davidovich, C.; Zheng, L.; Goodrich, K.J.; Cech, T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [Google Scholar] [CrossRef] [Green Version]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef] [Green Version]
- Kanhere, A.; Viiri, K.; Araújo, C.C.; Rasaiyaah, J.; Bouwman, R.D.; Whyte, W.A.; Pereira, C.F.; Brookes, E.; Walker, K.; Bell, G.W.; et al. Short RNAs Are Transcribed from Repressed Polycomb Target Genes and Interact with Polycomb Repressive Complex-2. Mol. Cell 2010, 38, 675–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xie, Y.; Li, L.; He, Y.; Zheng, D.; Yu, P.; Yu, L.; Tang, L.; Wang, Y.; Wang, Z. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr. Gene Ther. 2018, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Li, G.; Son, J.; Xu, C.-F.; Margueron, R.; Neubert, T.A.; Reinberg, D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang Jordon, K.; Lan, F.; Shi, Y.; Segal, E.; Chang Howard, Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltran, M.; Tavares, M.; Justin, N.; Khandelwal, G.; Ambrose, J.; Foster, B.M.; Worlock, K.B.; Tvardovskiy, A.; Kunzelmann, S.; Herrero, J.; et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat. Struct. Mol. Biol. 2019, 26, 899–909. [Google Scholar] [CrossRef]
- Wang, X.; Goodrich, K.J.; Gooding, A.R.; Naeem, H.; Archer, S.; Paucek, R.D.; Youmans, D.T.; Cech, T.R.; Davidovich, C. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol. Cell 2017, 65, 1056–1067.e1055. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Bolanos, B.; Gong, L.; Liu, W.; Goodrich, K.J.; Yang, X.; Chen, S.; Gooding, A.R.; Maegley, K.A.; Gajiwala, K.S.; et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. eLife 2017, 6, e31558. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, B.; Wapinski, O.L.; Tsai, M.C.; Qu, K.; Zhang, J.; Carlson, J.C.; Lin, M.; Fang, F.; Gupta, R.A.; et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013, 5, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Bohrer, L.R.; Rai, A.N.; Pan, Y.; Gan, L.; Zhou, X.; Bagchi, A.; Simon, J.A.; Huang, H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat. Cell Biol. 2010, 12, 1108–1114. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef] [PubMed]
- Dass, R.; Mulder, F.A.A.; Nielsen, J.T. ODiNPred: Comprehensive prediction of protein order and disorder. Sci. Rep. 2020, 10, 14780. [Google Scholar] [CrossRef]
- Balcerak, A.; Trebinska-Stryjewska, A.; Konopinski, R.; Wakula, M.; Grzybowska, E.A. RNA-protein interactions: Disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biol. 2019, 9, 190096. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [Green Version]
- Beckmann, B.M.; Horos, R.; Fischer, B.; Castello, A.; Eichelbaum, K.; Alleaume, A.M.; Schwarzl, T.; Curk, T.; Foehr, S.; Huber, W.; et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015, 6, 10127. [Google Scholar] [CrossRef]
- Bodor, A. Intrinsic structural disorder of proteins: From prediction to experimental identification. In Amino Acids, Peptides and Proteins, 1st ed.; Ryadnov, M., Hudecz, F., Eds.; The Royal Society of Chemistry: London, UK, 2021; Volume 44, pp. 64–114. [Google Scholar] [CrossRef]
- Theillet, F.-X.; Kalmar, L.; Tompa, P.; Han, K.-H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins 2013, 1, e24360. [Google Scholar] [CrossRef] [Green Version]
- Sebák, F.; Ecsédi, P.; Bermel, W.; Luy, B.; Nyitray, L.; Bodor, A. Selective 1Hα NMR Methods Reveal Functionally Relevant Proline cis/trans Isomers in Intrinsically Disordered Proteins: Characterization of Minor Forms, Effects of Phosphorylation, and Occurrence in Proteome. Angew. Chem. Int. Ed. Engl. 2022, 61, e202108361. [Google Scholar] [CrossRef]
- Connelly, G.P.; Bai, Y.; Jeng, M.F.; Englander, S.W. Isotope effects in peptide group hydrogen exchange. Proteins 1993, 17, 87–92. [Google Scholar] [CrossRef]
- Bodor, A.; Haller, J.D.; Bouguechtouli, C.; Theillet, F.X.; Nyitray, L.; Luy, B. Power of Pure Shift HαCα Correlations: A Way to Characterize Biomolecules under Physiological Conditions. Anal. Chem. 2020, 92, 12423–12428. [Google Scholar] [CrossRef] [PubMed]
- Haller, J.D.; Bodor, A.; Luy, B. Real-time pure shift measurements for uniformly isotope-labeled molecules using X-selective BIRD homonuclear decoupling. J. Magn. Reson. 2019, 302, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Kanelis, V.; Donaldson, L.; Muhandiram, D.R.; Rotin, D.; Forman-Kay, J.D.; Kay, L.E. Sequential assignment of proline-rich regions in proteins: Application to modular binding domain complexes. J. Biomol. NMR 2000, 16, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Mäntylahti, S.; Aitio, O.; Hellman, M.; Permi, P. HA-detected experiments for the backbone assignment of intrinsically disordered proteins. J. Biomol. NMR 2010, 47, 171–181. [Google Scholar] [CrossRef]
- Mäntylahti, S.; Tossavainen, H.; Hellman, M.; Permi, P. An intraresidual i(HCA)CO(CA)NH experiment for the assignment of main-chain resonances in 15N, 13C labeled proteins. J. Biomol. NMR 2009, 45, 301. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E.; Xu, G.Y.; Singer, A.U.; Muhandiram, D.R.; Forman-Kay, J.D. A Gradient-Enhanced HCCH-TOCSY Experiment for Recording Side-Chain 1H and 13C Correlations in H2O Samples of Proteins. J. Magn. Reson. 1993, 101, 333–337. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Poulsen, F.M. Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 2011, 50, 157–165. [Google Scholar] [CrossRef]
- Nielsen, J.T.; Mulder, F.A.A. POTENCI: Prediction of temperature, neighbor and pH-corrected chemical shifts for intrinsically disordered proteins. J. Biomol. NMR 2018, 70, 141–165. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Nørholm, A.B.; Hendus-Altenburger, R.; Pedersen, S.F.; Poulsen, F.M.; Kragelund, B.B. Temperature-dependent structural changes in intrinsically disordered proteins: Formation of α-helices or loss of polyproline II? Protein Sci. 2010, 19, 1555–1564. [Google Scholar] [CrossRef]
- Marsh, J.A.; Singh, V.K.; Jia, Z.; Forman-Kay, J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 2006, 15, 2795–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, N.J.; Williamson, M.P. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 1997, 9, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Reimer, U.; Scherer, G.; Drewello, M.; Kruber, S.; Schutkowski, M.; Fischer, G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 1998, 279, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta Proteins Proteom. 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [Green Version]
- Dudás, E.F.; Pálfy, G.; Menyhárd, D.K.; Sebák, F.; Ecsédi, P.; Nyitray, L.; Bodor, A. Tumor-Suppressor p53TAD1-60 Forms a Fuzzy Complex with Metastasis-Associated S100A4: Structural Insights and Dynamics by an NMR/MD Approach. ChemBioChem 2020, 21, 3087–3095. [Google Scholar] [CrossRef]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef]
- Zeke, A.; Schád, É.; Horváth, T.; Abukhairan, R.; Szabó, B.; Tantos, A. Deep structural insights into RNA-binding disordered protein regions. Wiley Interdiscip. Rev. RNA 2022, e1714. [Google Scholar] [CrossRef]
- Yan, J.; Dutta, B.; Hee, Y.T.; Chng, W.-J. Towards understanding of PRC2 binding to RNA. RNA Biol. 2019, 16, 176–184. [Google Scholar] [CrossRef] [Green Version]
- Mori, S.; Abeygunawardana, C.; Johnson, M.O.; Vanzijl, P.C.M. Improved Sensitivity of HSQC Spectra of Exchanging Protons at Short Interscan Delays Using a New Fast HSQC (FHSQC) Detection Scheme That Avoids Water Saturation. J. Magn. Reson. B 1995, 108, 94–98. [Google Scholar] [CrossRef]
- Schanda, P.; Brutscher, B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005, 127, 8014–8015. [Google Scholar] [CrossRef]
- Schanda, P.; Van Melckebeke, H.; Brutscher, B. Speeding Up Three-Dimensional Protein NMR Experiments to a Few Minutes. J. Am. Chem. Soc. 2006, 128, 9042–9043. [Google Scholar] [CrossRef] [PubMed]
- Lescop, E.; Schanda, P.; Brutscher, B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007, 187, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, T.; Maurer, M.; Sattler, M.; Schwendinger, M.G.; Glaser, S.J.; Griesinger, C. PLUSH TACSY: Homonuclear planar TACSY with two-band selective shaped pulses applied to Cα,C’ transfer and Cβ, Caromatic correlations. J. Biomol. NMR 1996, 8, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Ottiger, M.; Bax, A. An Empirical Correlation between Amide Deuterium Isotope Effects on 13Cα Chemical Shifts and Protein Backbone Conformation. J. Am. Chem. Soc. 1997, 119, 8070–8075. [Google Scholar] [CrossRef]
- Wang, A.C.; Grzesiek, S.; Tschudin, R.; Lodi, P.J.; Bax, A. Sequential backbone assignment of isotopically enriched proteins in D2O by deuterium-decoupled HA(CA)N and HA(CACO)N. J. Biomol. NMR 1995, 5, 376–382. [Google Scholar] [CrossRef]
- Ogura, K.; Kumeta, H.; Inagaki, F. Structure determination of proteins in 2H2O solution aided by a deuterium-decoupled 3D HCA(N)CO experiment. J. Biomol. NMR 2010, 47, 243–248. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabó, C.L.; Szabó, B.; Sebák, F.; Bermel, W.; Tantos, A.; Bodor, A. The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA. Int. J. Mol. Sci. 2022, 23, 6150. https://doi.org/10.3390/ijms23116150
Szabó CL, Szabó B, Sebák F, Bermel W, Tantos A, Bodor A. The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA. International Journal of Molecular Sciences. 2022; 23(11):6150. https://doi.org/10.3390/ijms23116150
Chicago/Turabian StyleSzabó, Csenge Lilla, Beáta Szabó, Fanni Sebák, Wolfgang Bermel, Agnes Tantos, and Andrea Bodor. 2022. "The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA" International Journal of Molecular Sciences 23, no. 11: 6150. https://doi.org/10.3390/ijms23116150
APA StyleSzabó, C. L., Szabó, B., Sebák, F., Bermel, W., Tantos, A., & Bodor, A. (2022). The Disordered EZH2 Loop: Atomic Level Characterization by 1HN- and 1Hα-Detected NMR Approaches, Interaction with the Long Noncoding HOTAIR RNA. International Journal of Molecular Sciences, 23(11), 6150. https://doi.org/10.3390/ijms23116150






