Identification of Differentially Expressed Genes in Resistant Tetraploid Wheat (Triticum turgidum) under Sitobion avenae (F.) Infestation
Abstract
1. Introduction
2. Results
2.1. SSH Library Construction
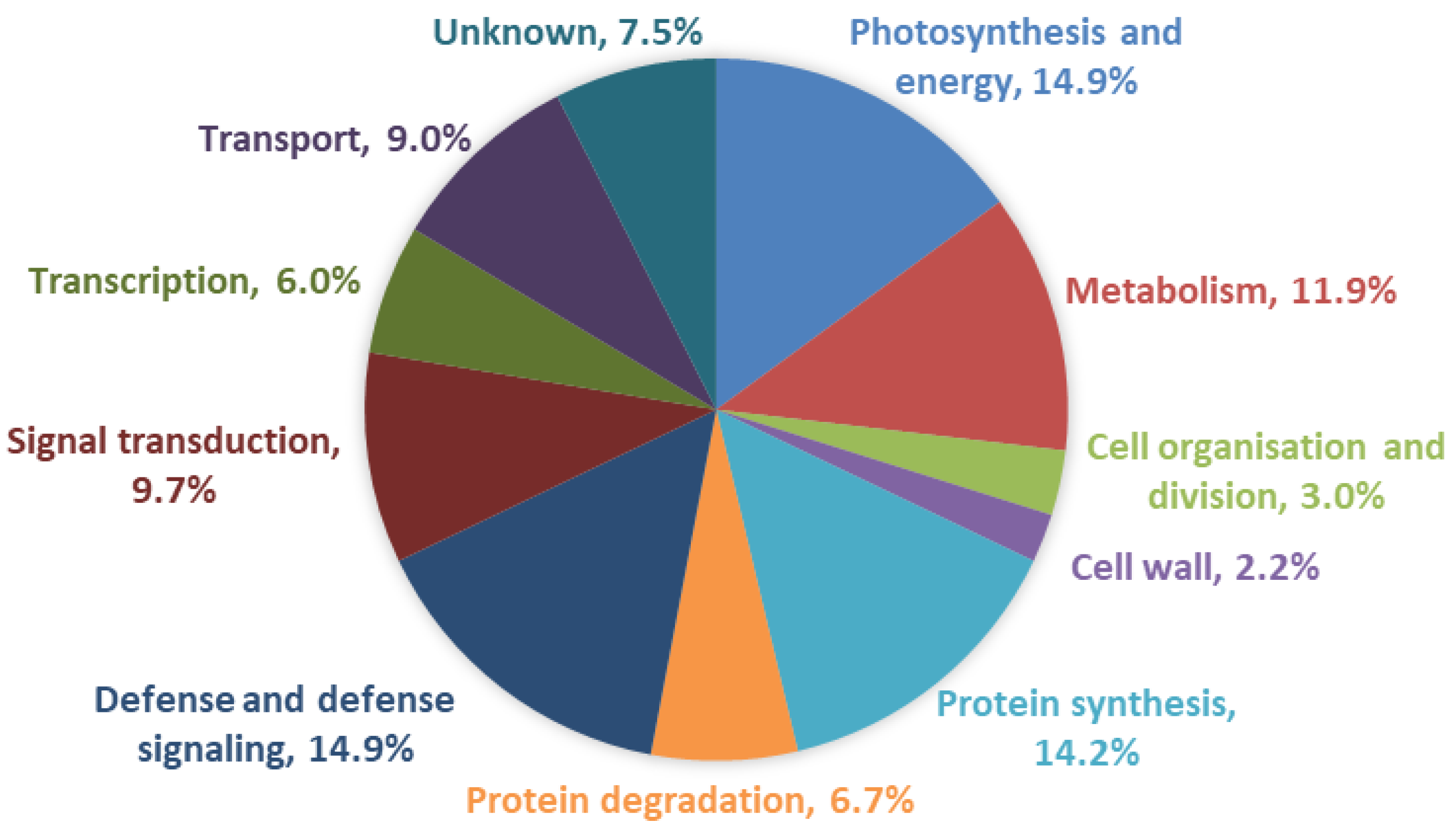
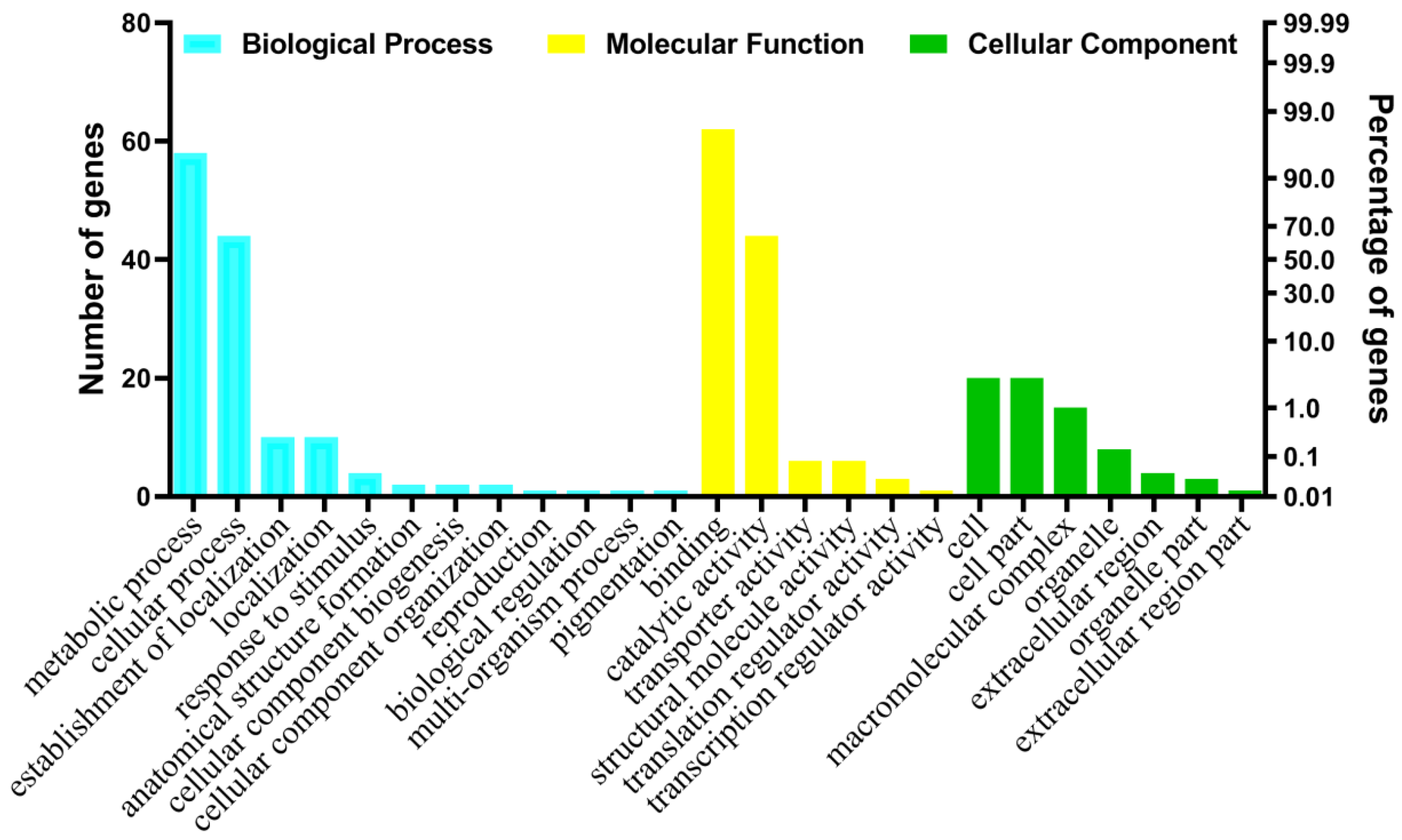
2.2. Annotation and Gene Ontology Analysis
2.3. Gene Expression Analysis
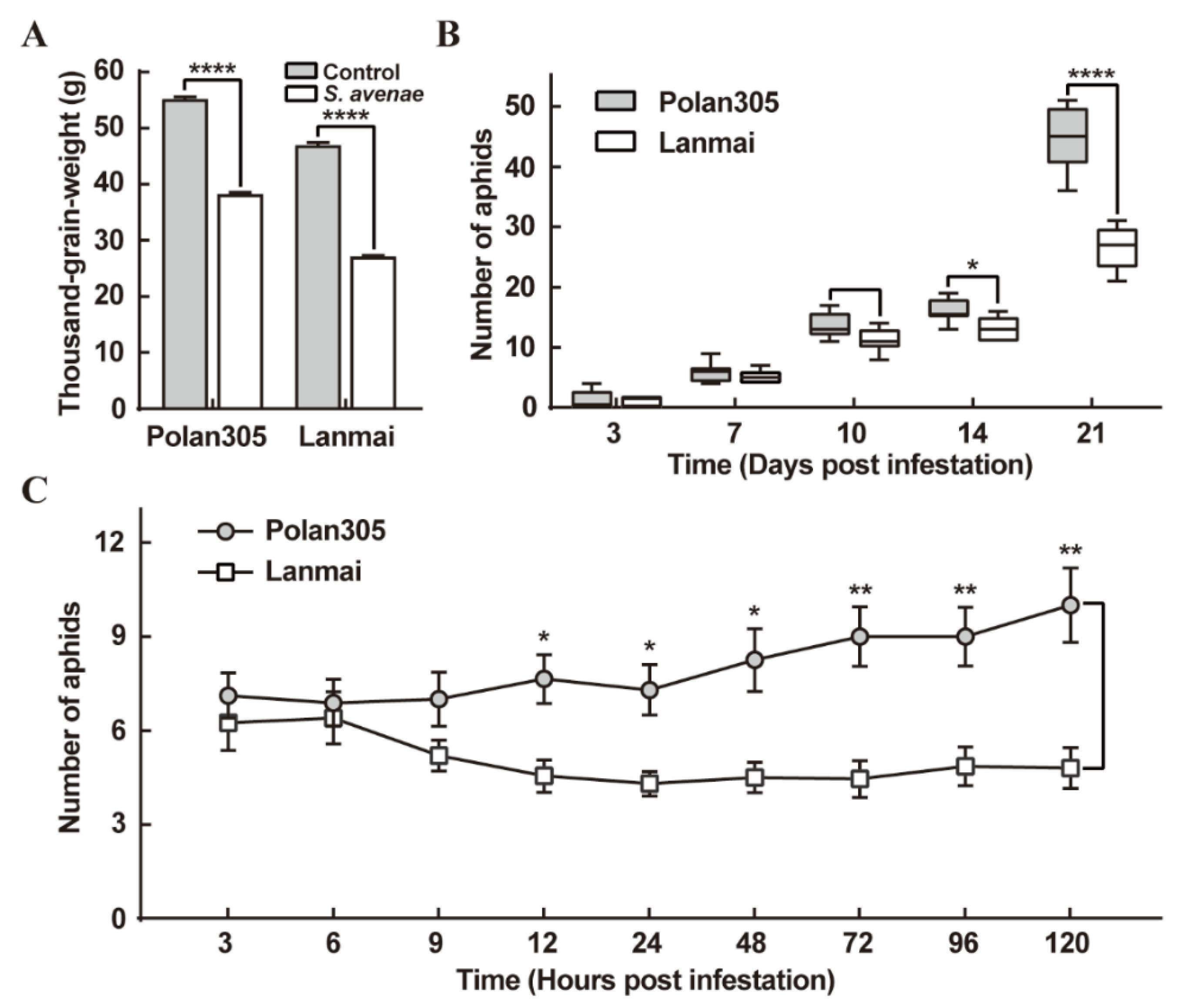
2.4. Evaluation of S. avenae Aphid Damage, Adult Fecundity and Preference
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Aphids
4.3. Aphid Infestation Biotests
4.4. RNA Isolation, Suppression Subtractive Hybridization and Library Construction
4.5. Sequencing and Sequence Analysis
4.6. Expression Profiling of Selected ESTs Using Quantitative Real-Time PCR (qRT-PCR)
4.7. Evaluation of S. avenae Damage, Performance and Preference
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almeida, M. Wheat: Genetics, Crops and Food Production; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011; pp. 1–430. [Google Scholar]
- Smith, C.M.; Boyko, E.V. The molecular bases of plant resistance and defense responses to aphid feeding. Entomol. Exp. Appl. 2007, 122, 1–16. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Z.; Chong, H.; Lu, M.; Jie, L.; Yang, Q. Statistics and analysis of crop yield losses caused by main diseases and insect pests in recent 10 years. Plant Prot. 2016, 42, 1–9. (In Chinese) [Google Scholar]
- Shahzad, M.W.; Ghani, H.; Ayyub, M.; Ali, Q.; Ahmad, H.M.; Faisal, M.; Ali, A.; Qasim, M.U. Performance of some wheat cultivars against aphid and its damage on yield and photosynthesis. J. Glob. Innov. Agric. Soc. Sci. 2019, 7, 105–109. [Google Scholar] [CrossRef]
- Ghanim, A.; Awadalla, S.; Allah, F.A.; Abdel-Aziz, A. The economic thresholds and injury levels for the English grain aphid, Sitobion avenae (F.) (Hemiptera-Aphididae) on wheat crop. J. Plant Prot. Pathol. 2018, 9, 93–95. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Garratt, M.P.D.; Bailey, A.; Potts, S.G.; Breeze, T. Economic valuation of natural pest control of the summer grain aphid in wheat in South East England. Ecosyst. Serv. 2018, 30, 149–157. [Google Scholar] [CrossRef]
- Dedryver, C.A.; Le Gallic, J.F.; Haack, L.; Halkett, F.; Outreman, Y.; Simon, J.C. Seasonal and annual genotypic variation and the effect of climate on population genetic structure of the cereal aphid Sitobion avenae in northern France. Bull Entomol. Res. 2008, 98, 159–168. [Google Scholar] [CrossRef]
- Shahzad, M.W.; Razaq, M.; Hussain, A.; Yaseen, M.; Afzal, M.; Mehmood, M.K. Yield and yield components of wheat (Triticum aestivum L.) affected by aphid feeding and sowing time at Multan, Pakistan. Pak. J. Bot. 2013, 45, 2005–2011. [Google Scholar]
- Zeb, Q.; Badshah, H.; Ali, H.; Shah, R.A.; Rehman, M. Population of aphids on different varieties/lines of wheat and their effect on yield and thousands grain weight. Sarhad J. Agric. 2011, 27, 443–450. [Google Scholar]
- National Agricultural Technology Extension Service Center. Major crops diseases and insect pests occurrence trend in the 2020. China Plant Prot. 2020, 40, 37–39; 53. [Google Scholar]
- John, F.; Saeed, N.A.; Nadeem, S.; Hamed, M. Integration of planting time and insecticide to manage aphid infestations in wheat for better crop productivity. Pak. J. Zool. 2017, 49, 1343–1351. [Google Scholar] [CrossRef]
- Sparks, T.C. Insecticide discovery: An evaluation and analysis. Pestic. Biochem. Physiol. 2013, 107, 8–17. [Google Scholar] [CrossRef]
- Painter, H.R. Insect Resistance in Crop Plants. Soil Sci. 1951, 72, 481. [Google Scholar] [CrossRef]
- Lamb, R.J.; Mackay, P.A. Tolerance of antibiotic and susceptible cereal seedlings to the aphids Metopolophium dirhodum and Rhopalosiphum padi. Ann. Appl. Biol. 2008, 127, 573–583. [Google Scholar] [CrossRef]
- Smith, C.M.; Clement, S.L. Molecular Bases of Plant Resistance to Arthropods; Springer Science & Business Media: Berlin, Germany, 2005. [Google Scholar] [CrossRef]
- Aradottir, I.G.; Martin, L.J.; Clark, J.S.; Pickett, A.J.; Smart, E.L. Searching for wheat resistance to aphids and wheat bulb fly in the historical Watkins and Gediflux wheat collections. Ann. Appl. Biol. 2017, 170, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Yang, X.F.; Wang, C.Y.; Wang, Y.J.; Zhang, H.; Ji, W.Q. Molecular mapping of resistance gene to English grain aphid (Sitobion avenae F.) in Triticum durum wheat line C273. Theor. Appl. Genet. 2012, 124, 287–293. [Google Scholar] [CrossRef]
- Wang, C.; Luo, K.; Wang, L.; Zhao, H.; Zhang, G. Molecular mapping of resistance gene to the English grain aphid, Sitobion avenae, in a Chinese wheat line XN98-10-35. Mol. Breed. 2015, 35, 203. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Wang, Y.; Zhang, H.; Ji, W. Screening and evaluation of different wheat varieties for resistance to English grain aphid Sitobion avenae at seeding and adult-plant stages. Acta Phytophylacica Sin. 2014, 41, 216–224. (In Chinese) [Google Scholar]
- El Bouhssini, M.; Street, K.; Amri, A.; Mackay, M.; Ogbonnaya, F.C.; Omran, A.; Abdalla, O.; Baum, M.; Dabbous, A.; Rihawi, F. New resistance sources to Russian wheat aphid (Diuraphis noxia) in Swedish wheat substitution and translocation lines with Rye (Secale cereale) and Leymus mollis. Plant Breed. 2011, 130, 96–97. [Google Scholar] [CrossRef]
- Razmjou, J.; Ramazani, S.; Naseri, B.; Ganbalani, G.N.; Dastjerdi, H.R. Resistance and susceptibility of various wheat varieties to Sitobion avenae (Hemiptera: Aphididae) in Iran. Appl. Entomol. Zool. 2011, 46, 455–461. [Google Scholar] [CrossRef]
- Beyer, B.M.; Haley, S.D.; Lapitan, N.L.V.; Peng, J.H.; Peairs, F.B. Inheritance of Russian wheat aphid resistance from tetraploid wheat accessions during transfer to hexaploid wheat. Euphytica 2011, 179, 247–255. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Shavit, R.; Distelfeldb, A.; Christensenc, S.A.; Tzin, V. Exploring the metabolic variation between domesticated and wild tetraploid wheat genotypes in response to corn leaf aphid infestation. Plant Signal. Behav. 2018, 13, ee1486148. [Google Scholar] [CrossRef]
- Navabi, Z.; Shiran, B.; Assad, M.T. Microsatellite mapping of a Russian wheat aphid resistance gene on chromosome 7B of an Iranian tetraploid wheat line: Preliminary results. Cereal Res. Commun. 2004, 32, 451–457. [Google Scholar] [CrossRef]
- Liu, X.L.; Lu, B.Y.; Wang, C.Y.; Wang, Y.J.; Zhang, H.; Tian, Z.R.; Ji, W.Q. Identification of Sitobion avenae F. Resistance and Genetic Diversity of Wheat Landraces from Qinling Mountains, China. Cereal Res. Commun. 2018, 46, 104–113. [Google Scholar] [CrossRef]
- Ferry, N.; Stavroulakis, S.; Guan, W.; Davison, G.M.; Bell, H.A.; Weaver, R.J.; Down, R.E.; Gatehouse, J.A.; Gatehouse, A.M.R. Molecular interactions between wheat and cereal aphid (Sitobion avenae) analysis of changes to the wheat proteome. Proteomics 2011, 11, 1985–2002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fu, Y.; Fan, J.; Li, Q.; Francis, F.; Chen, J. Comparative transcriptome and histological analyses of wheat in response to phytotoxic aphid Schizaphis graminum and non-phytotoxic aphid Sitobion avenae feeding. BMC Plant Biol. 2019, 19, 547. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Chen, J.L.; Cheng, D.F.; Sun, J.R.; Liu, Y.; Tian, Z. Biochemical and molecular characterizations of Sitobion avenae-induced wheat defense responses. Crop Prot. 2009, 28, 435–442. [Google Scholar] [CrossRef]
- Smith, C.M.; Liu, X.; Wang, L.J.; Liu, X.; Chen, M.S.; Starkey, S.; Bai, J. Aphid Feeding Activates Expression of a Transcriptome of oxylipin-based defense signals in wheat involved in resistance to herbivory. J. Chem. Ecol. 2010, 36, 260–276. [Google Scholar] [CrossRef][Green Version]
- Guan, W.; Ferry, N.; Edwards, M.G.; Bell, H.A.; Othman, H.; Gatehouse, J.A.; Gatehouse, A.M.R. Proteomic analysis shows that stress response proteins are significantly up-regulated in resistant diploid wheat (Triticum monococcum) in response to attack by the grain aphid (Sitobion avenae). Mol. Breed. 2015, 35, 57. [Google Scholar] [CrossRef]
- Botha, A.M.; Eck, L.V.; Burger, N.F.V.; Swanevelder, Z.H. Near-isogenic lines of Triticum aestivum with distinct modes of resistance exhibit dissimilar transcriptional regulation during Diuraphis noxia feeding. Biol. Open 2014, 3, 1116–1126. [Google Scholar] [CrossRef]
- Reddy, S.K.; Weng, Y.; Rudd, J.C.; Akhunova, A.; Liu, S. Transcriptomics of induced defense responses to greenbug aphid feeding in near isogenic wheat lines. Plant Sci. 2013, 212, 26–36. [Google Scholar] [CrossRef]
- Li, F.Q.; Peng, J.H. Genetic and Association Mapping Study of English Grain Aphid resistance and tolerance in bread wheat germplasm. J. Integr. Agric. 2014, 13, 40–53. [Google Scholar] [CrossRef]
- Xu, L.; Hou, Q.; Zhao, Y.; Ni, Z.; Liang, H.; Liang, R. Biochemical responses of resistant and susceptible wheat cultivars to English grain aphid (Sitobion avenae F.) at grain-filling stage. Acad. J. Biotechnol. 2016, 4, 276–284. [Google Scholar]
- Lan, H.; Zhang, Z.; Wu, J.; Cao, H.; Liu, T.X. Performance and transcriptomic response of the English grain aphid, Sitobion avenae, feeding on resistant and susceptible wheat cultivars. J. Integr. Agric. 2021, 20, 178–190. [Google Scholar] [CrossRef]
- Luo, K.; Yao, X.J.; Luo, C.; Hu, X.S.; Wang, C.P.; Wang, Y.; Hu, Z.Q.; Zhang, G.S.; Zhao, H.Y. Biological and Morphological features associated with English grain aphid and bird cherry-oat aphid tolerance in winter wheat line XN98-10-35. J. Plant Growth Regul. 2019, 38, 46–54. [Google Scholar] [CrossRef]
- Havlíčková, H. Differences in level of tolerance to cereal aphids in five winter wheat cultivars. Rostl. Vyrob. UZPI 1998, 43, 593–596. [Google Scholar]
- Divol, F.; Vilaine, F.; Thibivilliers, S.; Amselem, J.; Palauqui, J.C.; Kusiak, C.; Dinant, S. Systemic response to aphid infestation by Myzus persicae in the phloem of Apium graveolens. Plant Mol. Biol. 2005, 57, 517–540. [Google Scholar] [CrossRef]
- Voelckel, C.; Weisser, W.W.; Baldwin, I.T. An analysis of plant–aphid interactions by different microarray hybridization strategies. Mol. Ecol. 2004, 13, 3187–3195. [Google Scholar] [CrossRef]
- Tetreault, H.M.; Grover, S.; Scully, E.D.; Gries, T.; Palmer, N.A.; Sarath, G.; Louis, J.; Sattler, S.E. Global responses of resistant and susceptible Sorghum (Sorghum bicolor) to sugarcane aphid (Melanaphis sacchari). Front. Plant Sci. 2019, 10, 145. [Google Scholar] [CrossRef]
- Botha, A.M.; Swanevelder, Z.H.; Lapitan, N.L.V. Transcript profiling of wheat genes expressed during feeding by two different biotypes of Diuraphis noxia. Environ. Entomol. 2010, 39, 1206–1231. [Google Scholar] [CrossRef]
- Park, S.J.; Huang, Y.; Ayoubi, P. Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 2006, 223, 932–947. [Google Scholar] [CrossRef]
- Boyko, E.V.; Smith, C.M.; Thara, V.K.; Bruno, Y.D.; Starkey, S.R.; Klaahsen, D.L. Molecular basis of plant gene expression during aphid invasion wheat Pto- and Pti-like sequences are involved in interactions between wheat and Russian wheat aphid (Homoptera: Aphididae). J. Econ. Entomol. 2006, 99, 1430–1445. [Google Scholar] [CrossRef]
- Liu, F.; Kang, Z.; Tan, X.; Fan, Y.; Tian, H.; Liu, T. Physiology and defense responses of wheat to the infestation of different cereal aphids. J. Integr. Agric. 2020, 19, 1464–1474. [Google Scholar] [CrossRef]
- Zhai, Y.; Li, P.; Mei, Y.; Chen, M.; Chen, X.; Xu, H.; Zhou, X.; Dong, H.; Zhang, C.; Jiang, W. Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J. Exp. Bot. 2017, 68, 4153–4169. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Shao, Y.; Jiang, J.; Ren, L.; Chen, F.; Fang, W.; Guan, Z.; Chen, S. Gene expression profiles responses to aphid feeding in chrysanthemum (Chrysanthemum morifolium). MC Genom. 2014, 15, 1050. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Does, D.V.D.; Zamioudis, C.; Leon-Reyes, A.; Wees, S.C.M.V. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Delp, G.; Gradin, T.; Åhman, I.; Jonsson, L.M.V. Microarray analysis of the interaction between the aphid Rhopalosiphum padi between susceptible and partially resistant barley lines. Mol. Genet. Genom. 2009, 281, 233–248. [Google Scholar] [CrossRef]
- Iizasa, S.; Iizasa, E.; Matsuzaki, S.; Tanaka, H.; Kodama, Y.; Watanabe, K.; Nagano, Y. Arabidopsis LBP/BPI related-1 and -2 bind to LPS directly and regulate PR1 expression. Sci. Rep. 2016, 6, 27527. [Google Scholar] [CrossRef]
- Zytynska, S.E.; Jourdie, V.; Naseeb, S.; Delneri, D.; Preziosi, R.F. Induced expression of defence-related genes in barley is specific to aphid genotype. Biol. J. Linn. Soc. 2016, 117, 672–685. [Google Scholar] [CrossRef]
- Losvik, A.; Beste, L.; Glinwood, R.; Ivarson, E.; Stephens, J.; Zhu, L.H.; Jonsson, L. Overexpression and down-regulation of barley lipoxygenase LOX2.2 affects jasmonate-regulated genes and aphid fecundity. Int. J. Mol. Sci. 2017, 18, 2765. [Google Scholar] [CrossRef]
- Pingault, L.; Varsani, S.; Palmer, N.; Ray, S.; Williams, W.P.; Luthe, D.S.; Ali, J.G.; Sarath, G.; Louis, J. Transcriptomic and volatile signatures associated with maize defense against corn leaf aphid. BMC Plant Biol. 2021, 21, 138. [Google Scholar] [CrossRef]
- Casaretto, J.A.; Corcuera, L.J. Proteinase inhibitor accumulation in aphid-infested barley leaves. Phytochemistry 1998, 49, 2279–2286. [Google Scholar] [CrossRef]
- Losvik, A.; Beste, L.; Jennifer, S.; Jonsson, L.; Leandro, P. Overexpression of the aphid-induced serine protease inhibitor CI2c gene in barley affects the generalist green peach aphid, not the specialist bird cherry-oat aphid. PLoS ONE 2018, 3, e0193816. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, M.; Feng, Y.; Li, H.; Wang, Y.; Ma, Y.; Li, M.; An, F.; Guo, H. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Foyer, G.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; del Rio, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Sytykiewicz, H.; Bar, H.; Semenenko, V.; Ukasik, I.; Sprawka, I.; Sempruch, C.; Leszczyński, B. Expression patterns of genes involved in Ascorbate-Glutathione Cycle in aphid-infested maize (Zea mays L.) seedlings. Int. J. Mol. Sci. 2016, 17, 268. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Klie, S.; Nikoloski, Z. The choice between MapMan and gene ontology for automated gene function prediction in plant science. Front. Genet. 2012, 3, 115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Kou, X.; Bai, S.; Luo, Y.; Wang, Z.; Xie, L.; Deng, P.; Zhang, H.; Wang, C.; Wang, Y.; et al. Identification of Differentially Expressed Genes in Resistant Tetraploid Wheat (Triticum turgidum) under Sitobion avenae (F.) Infestation. Int. J. Mol. Sci. 2022, 23, 6012. https://doi.org/10.3390/ijms23116012
Liu X, Kou X, Bai S, Luo Y, Wang Z, Xie L, Deng P, Zhang H, Wang C, Wang Y, et al. Identification of Differentially Expressed Genes in Resistant Tetraploid Wheat (Triticum turgidum) under Sitobion avenae (F.) Infestation. International Journal of Molecular Sciences. 2022; 23(11):6012. https://doi.org/10.3390/ijms23116012
Chicago/Turabian StyleLiu, Xinlun, Xudan Kou, Shichao Bai, Yufeng Luo, Zhenyu Wang, Lincai Xie, Pingchuan Deng, Hong Zhang, Changyou Wang, Yajuan Wang, and et al. 2022. "Identification of Differentially Expressed Genes in Resistant Tetraploid Wheat (Triticum turgidum) under Sitobion avenae (F.) Infestation" International Journal of Molecular Sciences 23, no. 11: 6012. https://doi.org/10.3390/ijms23116012
APA StyleLiu, X., Kou, X., Bai, S., Luo, Y., Wang, Z., Xie, L., Deng, P., Zhang, H., Wang, C., Wang, Y., Zhao, J., & Ji, W. (2022). Identification of Differentially Expressed Genes in Resistant Tetraploid Wheat (Triticum turgidum) under Sitobion avenae (F.) Infestation. International Journal of Molecular Sciences, 23(11), 6012. https://doi.org/10.3390/ijms23116012





