Carbon Metabolism of a Soilborne Mn(II)-Oxidizing Escherichia coli Isolate Implicated as a Pronounced Modulator of Bacterial Mn Oxidation
Abstract
1. Introduction
2. Results
2.1. Effects of Different Carbon Source Materials on the MnOA of E. coli MB266
2.2. Different Metabolic Pathways Have Dissimilar Promoting Effects on the MnOA
2.3. Effect of Simultaneous Knockout of Several Carbon Metabolic Genes on Mn Oxide Formation
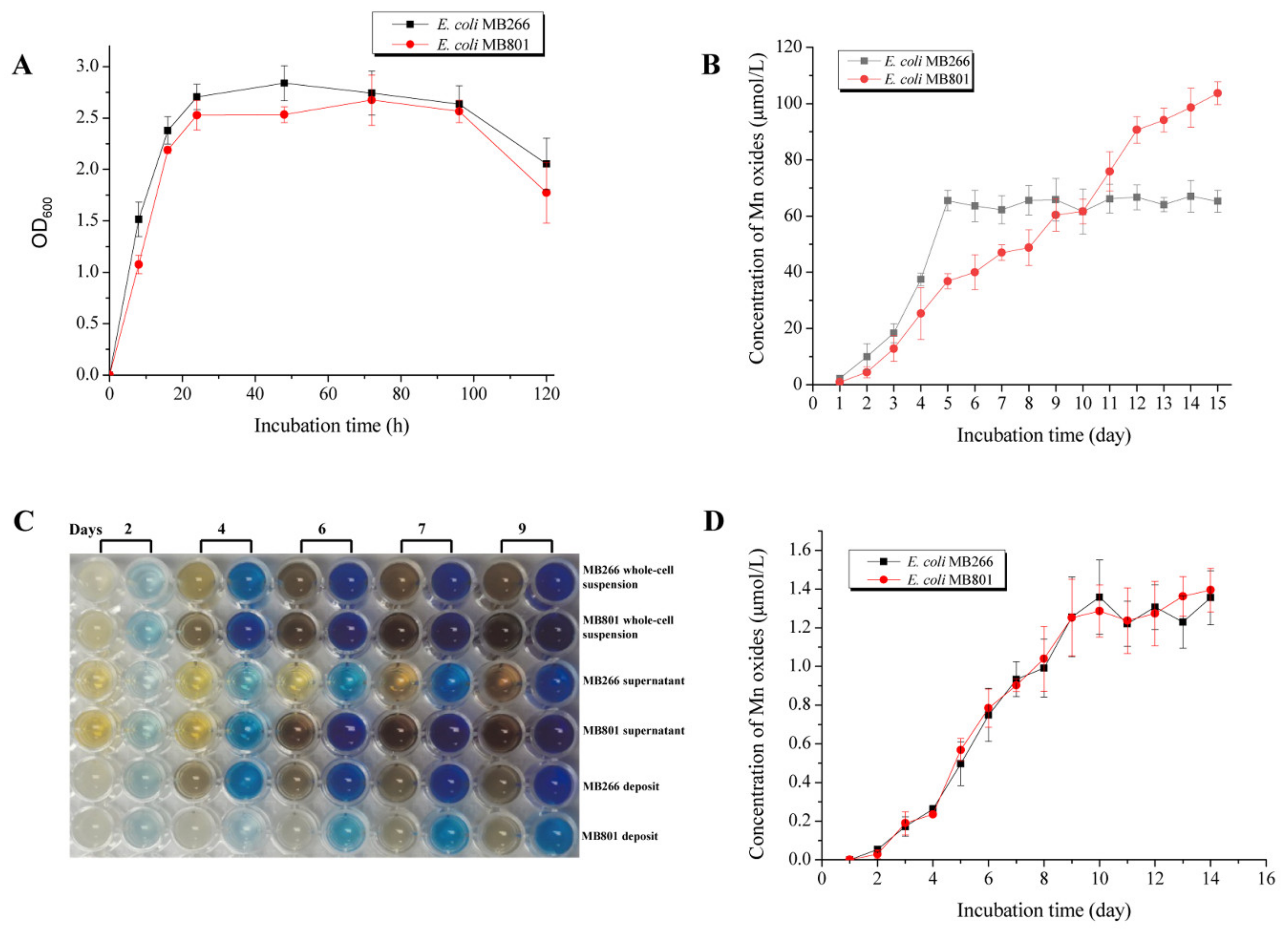
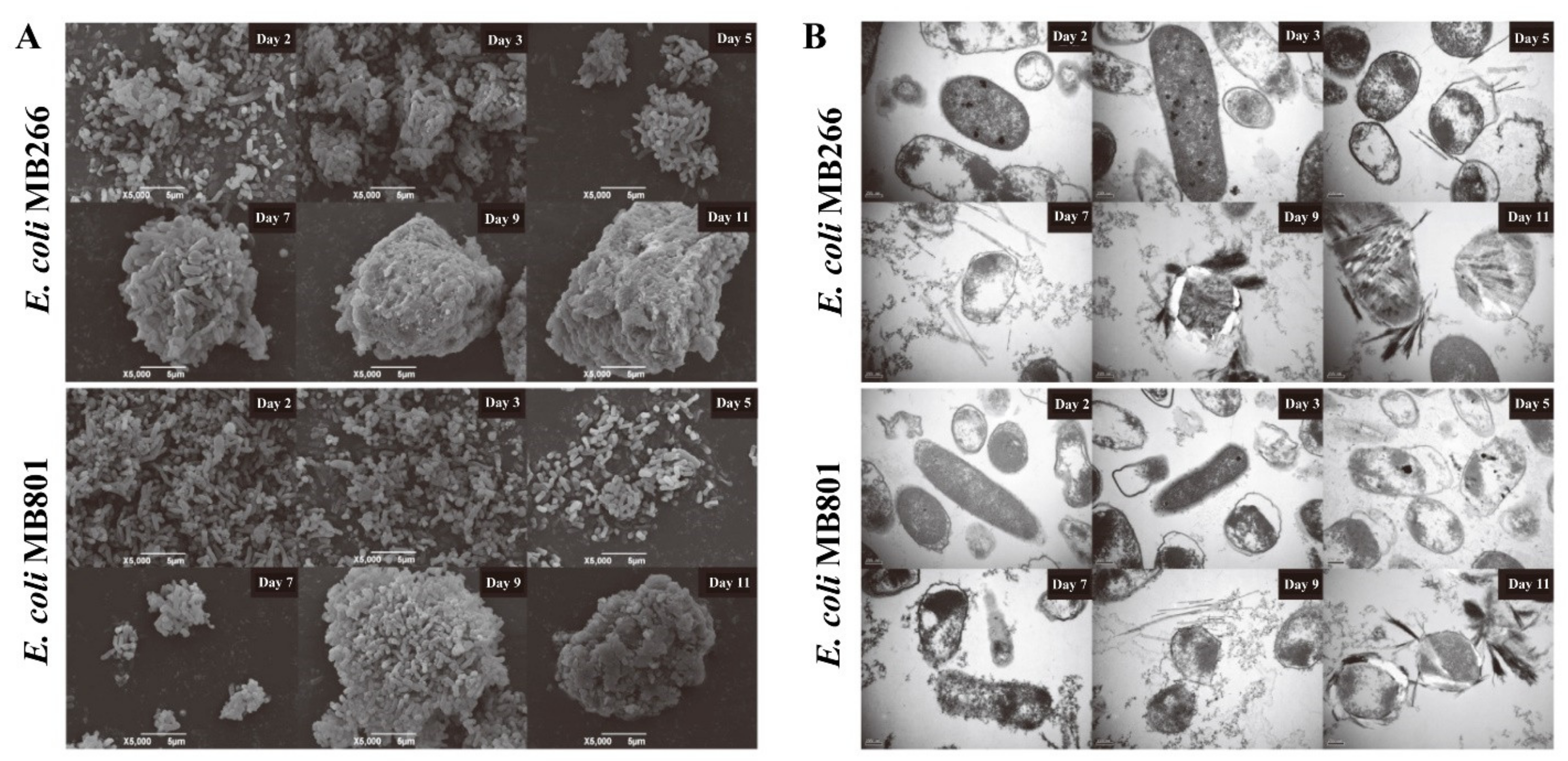
2.4. Comparative Characterization of Mn Oxide Aggregate Structures Formed in MB801 and MB266
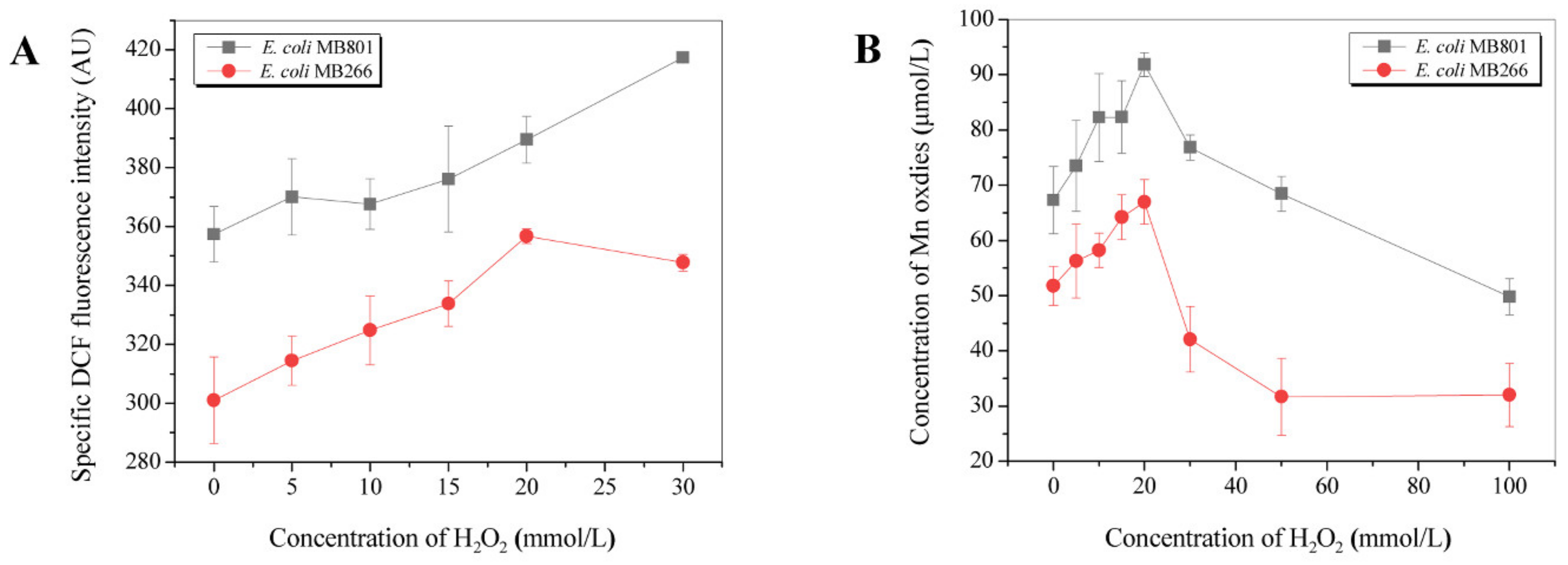
2.5. Influence of Carbon Metabolism on MnODA Implicated in Intracellular ROS Levels
3. Discussion
3.1. Variances in MnOA Were Highly Related to the Overall Carbon Metabolic Level of the Cells
3.2. Carbon Metabolism Can Modulate MnODA by Affecting the Resorption of Mn Oxides and the Formation of Aggregates
3.3. Effect of Carbon Metabolism on Mn Oxide Aggregate Formation Was Highly Related to the Intracellular ROS Level
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Plasmid Construction and Transformation
4.3. Gene Disruption
4.4. Analytical Assays
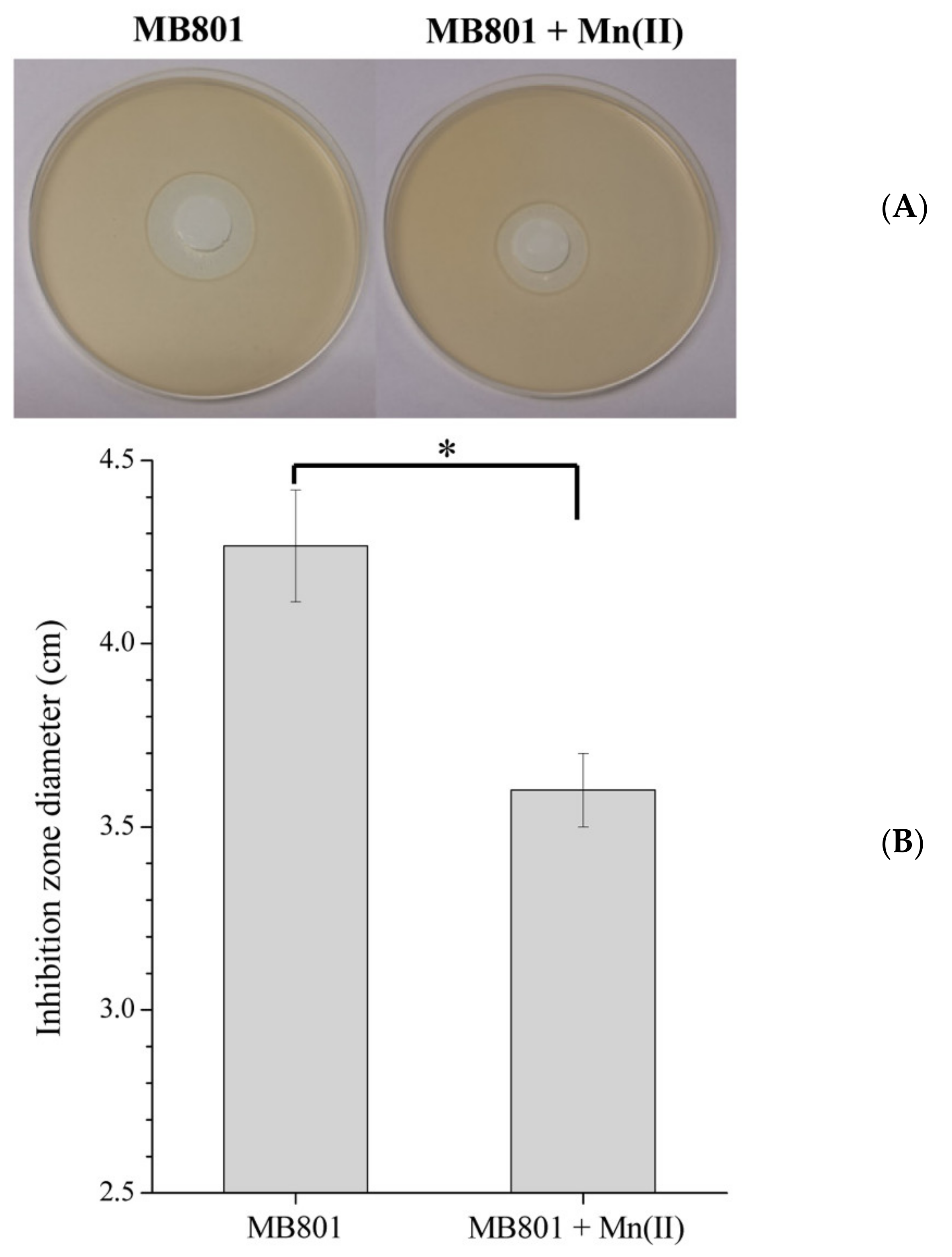
4.5. H2O2 Inhibition Zone Measurement
4.6. Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM) Assays
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
| Mn | Manganese |
| MnODA | Mn oxide deposit amount |
| MnOA | Mn(II)-oxidizing activity |
| BioMnOx | Biogenic manganese oxides |
| ROS | Reactive oxygen species |
| TCA | Tricarboxylic acid |
| LBB | Leucoberbelin blue |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Ace | Acetate |
| Oxa | Sodium oxalate |
| Glu | Glucose |
| Frc | Fructose |
| Man | Mannose |
| Kga | Ketoglutarate |
| Ben | Benzoate |
| CYT-C | Cytochrome c |
| SEM | Scanning electron microscopy |
| TEM | Transmission electron microscopy |
References
- Tebo, B.M.; Johnson, H.A.; McCarthy, J.K.; Templeton, A.S. Geomicrobiology of Manganese(II) Oxidation. Trends Microbiol. 2005, 13, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Tebo, B.M.; Bargar, J.R.; Clement, B.G.; Dick, G.J.; Murray, K.J.; Parker, D.; Verity, R.; Webb, S.M. Biogenic Manganese Oxides: Properties and Mechanisms of Formation. Annu. Rev. Earth Planet. Sci. 2004, 32, 287–328. [Google Scholar] [CrossRef]
- Hansel, C.; Learman, D.R. Geomicrobiology of Manganese. In Ehrlich’s Geomicrobiology; Ehrlich, H.L., Newman, D.K., Kappler, A., Eds.; CRC Press: Boca Roton, FL, USA, 2015; pp. 401–452. [Google Scholar] [CrossRef]
- Parker, D.L.; Sposito, G.; Tebo, B.M. Manganese(III) Binding to a Pyoverdine Siderophore Produced by a Manganese(II)-Oxidizing Bacterium. Geochim. Cosmochim. Acta 2004, 68, 4809–4820. [Google Scholar] [CrossRef]
- Francis, C.A.; Tebo, B.M. Enzymatic Manganese(II) Oxidation by Metabolically Dormant Spores of Diverse Bacillus Species. Appl. Environ. Microbiol. 2002, 68, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Y.; Qin, Z.; Luo, P.; Ma, Z.; Tan, M.; Kang, H.; Huang, Z. A Novel Manganese Oxidizing Bacterium-Aeromonas hydrophila Strain DS02: Mn(II) Oxidization and Biogenic Mn Oxides Generation. J. Hazard Mater. 2019, 367, 539–545. [Google Scholar] [CrossRef]
- Yu, H.; Leadbetter, J.R. Bacterial Chemolithoautotrophy via Manganese Oxidation. Nature 2020, 583, 453–458. [Google Scholar] [CrossRef]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.; Kemner, K.M.; et al. Protein Oxidation Implicated as the Primary Determinant of Bacterial Radioresistance. PLoS Biol. 2007, 5, e92. [Google Scholar] [CrossRef]
- Tabita, F.R. Microbial Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase: A Different Perspective. Photosynth. Res. 1999, 60, 1–28. [Google Scholar] [CrossRef]
- Dick, G.J.; Podell, S.; Johnson, H.A.; Rivera-Espinoza, Y.; Bernier-Latmani, R.; McCarthy, J.K.; Torpey, J.W.; Clement, B.G.; Gaasterland, T.; Tebo, B.M. Genomic Insights into Mn(II) Oxidation by the Marine Alphaproteobacterium Aurantimonas sp. Strain SI85-9A1. Appl. Environ. Microbiol. 2008, 74, 2646–2658. [Google Scholar] [CrossRef]
- Elzinga, E.J. Reductive Transformation of Birnessite by Aqueous Mn(II). Environ. Sci. Technol. 2011, 45, 6366–6372. [Google Scholar] [CrossRef]
- Tao, L.; Stich, T.A.; Butterfield, C.N.; Romano, C.A.; Spiro, T.G.; Tebo, B.M.; Casey, W.H.; Britt, R.D. Mn(II) Binding and Subsequent Oxidation by the Multicopper Oxidase MnxG Investigated by Electron Paramagnetic Resonance Spectroscopy. J. Am. Chem. Soc. 2015, 137, 10563–10575. [Google Scholar] [CrossRef] [PubMed]
- Soldatova, A.V.; Romano, C.A.; Tao, L.; Stich, T.A.; Casey, W.H.; Britt, R.D.; Tebo, B.M.; Spiro, T.G. Mn(II) Oxidation by the Multicopper Oxidase Complex Mnx: A Coordinated Two-Stage Mn(II)/(III) and Mn(III)/(IV) Mechanism. J. Am. Chem. Soc. 2017, 139, 11381–11391. [Google Scholar] [CrossRef]
- Wright, M.H.; Farooqui, S.M.; White, A.R.; Greene, A.C. Production of Manganese Oxide Nanoparticles by Shewanella Species. Appl. Environ. Microbiol. 2016, 82, 5402–5409. [Google Scholar] [CrossRef]
- Chaput, D.L.; Fowler, A.J.; Seo, O.; Duhn, K.; Hansel, C.M.; Santelli, C.M. Mn Oxide Formation by Phototrophs: Spatial and Temporal Patterns, with Evidence of an Enzymatic Superoxide-Mediated Pathway. Sci. Rep. 2019, 9, 18244. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Hoeller, S.; Meier, B.; Peiter, E. Manganese in Plants: From Acquisition to Subcellular Allocation. Front. Plant Sci. 2020, 11, 300. [Google Scholar] [CrossRef]
- Pospisil, P. Production of Reactive Oxygen Species by Photo system II as a Response to Light and Temperature Stress. Front. Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, X.; Kang, J.; Duan, X.; Wang, S. Persulfate Activation on Crystallographic Manganese Oxides: Mechanism of Singlet Oxygen Evolution for Nonradical Selective Degradation of Aqueous Contaminants. Environ. Sci. Technol. 2019, 53, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese(II) Oxidation by Manganese Peroxidase from the Basidiomycete Phanerochaete Chrysosporium. Kinetic Mechanism and Role of Chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef]
- Hansel, C.M.; Zeiner, C.A.; Santelli, C.M.; Webb, S.M. Mn(II) Oxidation by an Ascomycete Fungus Is Linked to Superoxide Production during Asexual Reproduction. Proc. Natl. Acad. Sci. USA 2012, 109, 12621–12625. [Google Scholar] [CrossRef]
- Li, H.P.; Daniel, B.; Creeley, D.; Grandbois, R.; Zhang, S.; Xu, C.; Ho, Y.F.; Schwehr, K.A.; Kaplan, D.I.; Santschi, P.H.; et al. Superoxide Production by a Manganese-Oxidizing Bacterium Facilitates Iodide Oxidation. Appl. Environ. Microbiol. 2014, 80, 2693–2699. [Google Scholar] [CrossRef]
- Myers, C.R.; Nealson, K.H. Bacterial Manganese Reduction and Growth with Manganese Oxide as the Sole Electron Acceptor. Science 1988, 240, 1319–1321. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Phillips, E.J.P. Novel Mode of Microbial Energy-Metabolism—Organic-Carbon Oxidation Coupled to Dissimilatory Reduction of Iron or Manganese. Appl. Environ. Microbiol. 1988, 54, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Bosma, E.F.; Rau, M.H.; van Gijtenbeek, L.A.; Siedler, S. Regulation and Distinct Physiological Roles of Manganese in Bacteria. FEMS Microbiol. Rev. 2021, 45, fuab08. [Google Scholar] [CrossRef] [PubMed]
- Waters, L.S. Bacterial Manganese Sensing and Homeostasis. Curr. Opin. Chem. Biol. 2020, 55, 96–102. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Z.; Chen, H.; Liu, J.; Liu, C.; Ni, H.; Zhao, C.; Ali, M.; Liu, F.; Li, L. Surface Mn(II) Oxidation Actuated by a Multicopper Oxidase in a Soil Bacterium Leads to the Formation of Manganese Oxide Minerals. Sci. Rep. 2015, 5, 10895. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, Z.; Zhang, Z.; Chen, H.; Liu, J.; Ali, M.; Liu, F.; Li, L. Population Structure of Manganese-Oxidizing Bacteria in Stratified Soils and Properties of Manganese Oxide Aggregates under Manganese-Complex Medium Enrichment. PLoS ONE 2013, 8, e73778. [Google Scholar] [CrossRef]
- Liu, J.; Gu, T.; Sun, X.; Li, L.; Xiao, F.; Wang, Z.; Li, L. Synthesis of MnO/C/Co3O4 Nanocomposites by a Mn2+-Oxidizing Bacterium as a Biotemplate for Lithium-Ion Batteries. Sci. Technol. Adv. Mater. 2021, 22, 429–440. [Google Scholar] [CrossRef]
- Liu, J.; Gu, T.; Li, L.; Li, L. Synthesis of MnO/C/NiO-Doped Porous Multiphasic Composites for Lithium-Ion Batteries by Biomineralized Mn Oxides from Engineered Pseudomonas putida Cells. Nanomaterials 2021, 11, 361. [Google Scholar] [CrossRef]
- Miller, A.Z.; Dionísio, A.; Sequeira Braga, M.A.; Hernández-Mariné, M.; Afonso, M.J.; Muralha, V.S.F.; Herrera, L.K.; Raabe, J.; Fernandez-Cortes, A.; Cuezva, S.; et al. Biogenic Mn Oxide Minerals Coating in a Subsurface Granite Environment. Chem. Geol. 2012, 322–323, 181–191. [Google Scholar] [CrossRef]
- Henkel, J.V.; Dellwig, O.; Pollehne, F.; Herlemann, D.P.R.; Leipe, T.; Schulz-Vogt, H.N. A Bacterial Isolate from the Black Sea Oxidizes Sulfide with Manganese(IV) Oxide. Proc. Natl. Acad. Sci. USA 2019, 116, 12153–12155. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Wang, J.P.; Liu, J.; Chen, H.; Li, M.S.; Li, L. Mechanistic Insights into Manganese Oxidation of a Soil-Borne Mn(II)-Oxidizing Escherichia coli Strain by Global Proteomic and Genetic Analyses. Sci. Rep. 2017, 7, 1352. [Google Scholar] [CrossRef] [PubMed]
- Krumbein, W.E.; Altmann, H.J. A New Method for the Detection and Enumeration of Manganese Oxidizing and Reducing Microorganisms. Helgoländer Wiss. Meeresunters. 1973, 25, 347–356. [Google Scholar] [CrossRef]
- Ramakrishnan, T.; Adelberg, E.A. Regulatory Mechanisms in the Biosynthesis of Isoleucine and Valine. I. Genetic Derepression of Enzyme Formation. J. Bacteriol. 1964, 87, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Ornston, L.N.; Ornston, M.K. Regulation of Glyoxylate Metabolism in Escherichia coli K-12. J. Bacteriol. 1969, 98, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Raman, B.; Nandakumar, M.P.; Muthuvijayan, V.; Marten, M.R. Proteome Analysis to Assess Physiological Changes in Escherichia coli Grown under Glucose-Limited Fed-Batch Conditions. Biotechnol. Bioeng. 2005, 92, 384–392. [Google Scholar] [CrossRef]
- Langley, D.; Guest, J.R. Biochemical Genetics of the Alpha-Keto Acid Dehydrogenase Complexes of Escherichia coli K12: Isolation and Biochemical Properties of Deletion Mutants. J. Gen. Microbiol. 1977, 99, 263–276. [Google Scholar] [CrossRef]
- Tarmy, E.M.; Kaplan, N.O. Chemical Characterization of D-Lactate Dehydrogenase from Escherichia coli B. J. Biol. Chem. 1968, 243, 2579–2586. [Google Scholar] [CrossRef]
- Lopez-Campistrous, A.; Semchuk, P.; Burke, L.; Palmer-Stone, T.; Brokx, S.J.; Broderick, G.; Bottorff, D.; Bolch, S.; Weiner, J.H.; Ellison, M.J. Localization, Annotation, and Comparison of the Escherichia coli K-12 Proteome under Two States of Growth. Mol. Cell. Proteom. MCP 2005, 4, 1205–1209. [Google Scholar] [CrossRef]
- Diaz, J.M.; Hansel, C.M.; Voelker, B.M.; Mendes, C.M.; Andeer, P.F.; Zhang, T. WIdespread Production of Extracellular Superoxide by Heterotrophic Bacteria. Science 2013, 340, 1223–1226. [Google Scholar] [CrossRef]
- Learman, D.R.; Voelker, B.M.; Vazquez-Rodriguez, A.I.; Hansel, C.M. Formation of Manganese Oxides by Bacterially Generated Superoxide. Nat. Geosci. 2011, 4, 95–98. [Google Scholar] [CrossRef]
- Culotta, V.C. Superoxide Dismutase, Oxidative Stress, and Cell Metabolism. Curr. Top Cell. Regul. 2000, 36, 117–132. [Google Scholar] [PubMed]
- Niederhoffer, E.C.; Naranjo, C.M.; Bradley, K.L.; Fee, J.A. Control of Escherichia coli Superoxide Dismutase (sodA and sodB) Genes by the Ferric Uptake Regulation (fur) Locus. J. Bacteriol. 1990, 172, 1930–1938. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Johnson, H.A.; Tebo, B.M. In Vitro Studies Indicate a Quinone Is Involved in Bacterial Mn(II) Oxidation. Arch. Microbiol. 2008, 189, 59–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-Guided Editing of Bacterial Genomes using CRISPR-Cas Systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef]
- Su, T.; Liu, F.; Gu, P.; Jin, H.; Chang, Y.; Wang, Q.; Liang, Q.; Qi, Q. A CRISPR-Cas9 Assisted Non-Homologous End-Joining Strategy for One-step Engineering of Bacterial Genome. Sci. Rep. 2016, 6, 37895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Adikaram, P.; Pandey, M.; Genis, A.; Simonds, W.F. Optimization of Genome Editing through CRISPR-Cas9 Engineering. Bioengineered 2016, 7, 166–174. [Google Scholar] [CrossRef]
- Bannikov, A.V.; Lavrov, A.V. CRISPR/CAS9, the King of Genome Editing Tools. Mol. Biol. 2017, 51, 582–594. [Google Scholar] [CrossRef]
- Della, M.; Palmbos, P.L.; Tseng, H.M.; Tonkin, L.M.; Daley, J.M.; Topper, L.M.; Pitcher, R.S.; Tomkinson, A.E.; Wilson, T.E.; Doherty, A.J. Mycobacterial Ku and Ligase Proteins Constitute a Two-Component NHEJ Repair Machine. Science 2004, 306, 683–685. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Singh, S.P.; Häder, D.P.; Sinha, R.P. Detection of Reactive Oxygen Species (ROS) by the Oxidant-Sensing Probe 2’,7’-Dichlorodihydrofluorescein Diacetate in the Cyanobacterium Anabaena variabilis PCC 7937. Biochem. Biophys. Res. Commun. 2010, 397, 603–607. [Google Scholar] [CrossRef]
- Francis, C.A.; Casciotti, K.L.; Tebo, B.M. Localization of Mn(II)-Oxidizing Activity and the Putative Multicopper Oxidase, MnxG, to the Exosporium of the Marine Bacillus sp. Strain SG-1. Arch. Microbiol. 2002, 178, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Michalak, I.; Chojnacka, K.; Marycz, K. Using ICP-OES and SEM-EDX in Biosorption Studies. Mikrochim. Acta 2011, 172, 65–74. [Google Scholar] [CrossRef] [PubMed]

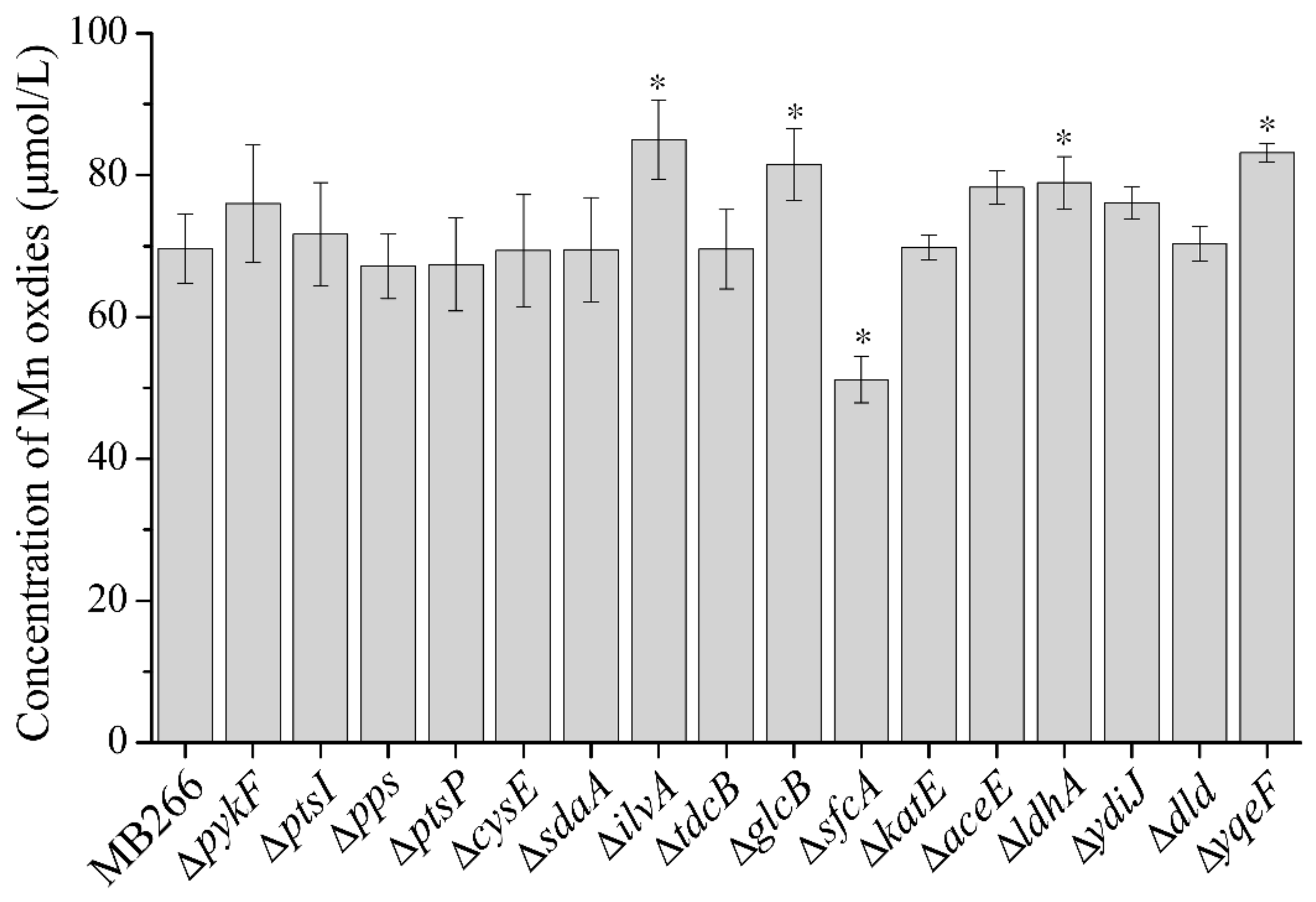
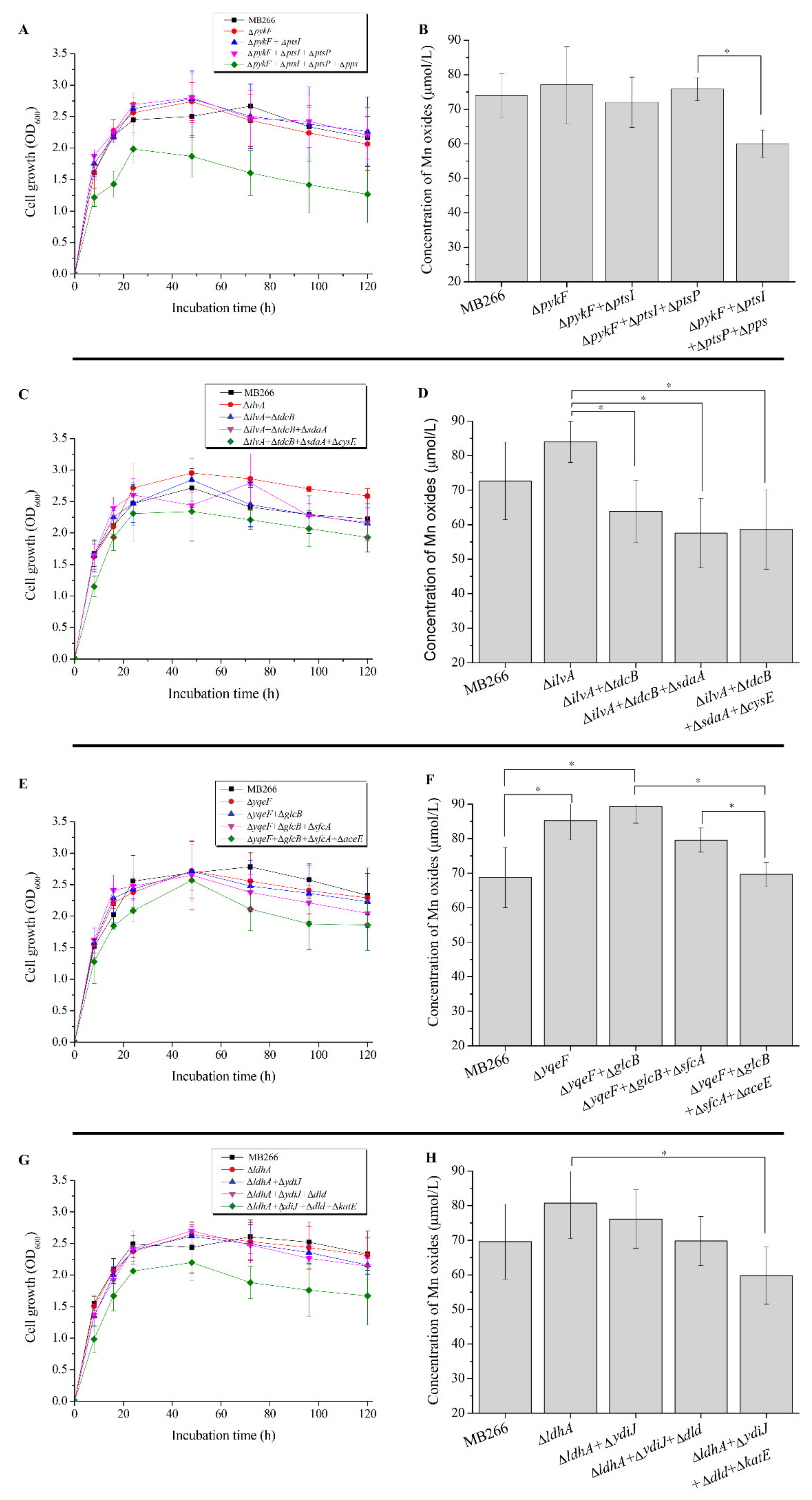
| Protein (Gene) | Predicted Function/Activity | Expression Fold Change |
|---|---|---|
| PykF (pykF) | Pyruvate kinase I | 1.171 |
| PtsI (ptsI) | PEP-protein phosphotransferase of PTS system | 1.135 |
| Pps (pps) | Phosphoenolpyruvate synthase | 1.11 |
| PtsP (ptsP) | Fused PEP-protein phosphotransferase (enzyme I) of PTS system | 0.879 |
| CysE (cysE) | Serine acetyltransferase | 0.824 |
| SdaA (sdaA) | L-Serine deaminase I | 0.907 |
| IlvA (ilvA) | Threonine deaminase | 0.754 |
| TdcB (tdcB) | Catabolic threonine dehydratase, PLP-dependent | 1.189 |
| GlcB (glcB) | Malate synthase G | 0.81 |
| SfcA (sfcA) | Malate dehydrogenase, NAD-requiring | 0.816 |
| KatE (katE) | Hydroperoxidase HPII (III) | 1.155 |
| AceE (aceE) | Pyruvate dehydrogenase | 0.872 |
| LdhA (ldhA) | Fermentative D-Lactate dehydrogenase | 1.173 |
| YdiJ (ydiJ) | FAD-linked oxidoreductase | 0.848 |
| YqeF (yqeF) | Acyltransferase | 1.252 |
| Dld (dld) | NADH independent D-lactate dehydrogenase | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, T.; Tong, Z.; Zhang, X.; Wang, Z.; Zhang, Z.; Hwang, T.-S.; Li, L. Carbon Metabolism of a Soilborne Mn(II)-Oxidizing Escherichia coli Isolate Implicated as a Pronounced Modulator of Bacterial Mn Oxidation. Int. J. Mol. Sci. 2022, 23, 5951. https://doi.org/10.3390/ijms23115951
Gu T, Tong Z, Zhang X, Wang Z, Zhang Z, Hwang T-S, Li L. Carbon Metabolism of a Soilborne Mn(II)-Oxidizing Escherichia coli Isolate Implicated as a Pronounced Modulator of Bacterial Mn Oxidation. International Journal of Molecular Sciences. 2022; 23(11):5951. https://doi.org/10.3390/ijms23115951
Chicago/Turabian StyleGu, Tong, Zhenghu Tong, Xue Zhang, Zhiyong Wang, Zhen Zhang, Tzann-Shun Hwang, and Lin Li. 2022. "Carbon Metabolism of a Soilborne Mn(II)-Oxidizing Escherichia coli Isolate Implicated as a Pronounced Modulator of Bacterial Mn Oxidation" International Journal of Molecular Sciences 23, no. 11: 5951. https://doi.org/10.3390/ijms23115951
APA StyleGu, T., Tong, Z., Zhang, X., Wang, Z., Zhang, Z., Hwang, T.-S., & Li, L. (2022). Carbon Metabolism of a Soilborne Mn(II)-Oxidizing Escherichia coli Isolate Implicated as a Pronounced Modulator of Bacterial Mn Oxidation. International Journal of Molecular Sciences, 23(11), 5951. https://doi.org/10.3390/ijms23115951






