Molecular Classification and Overcoming Therapy Resistance for Acute Myeloid Leukemia with Adverse Genetic Factors
Abstract
1. Introduction
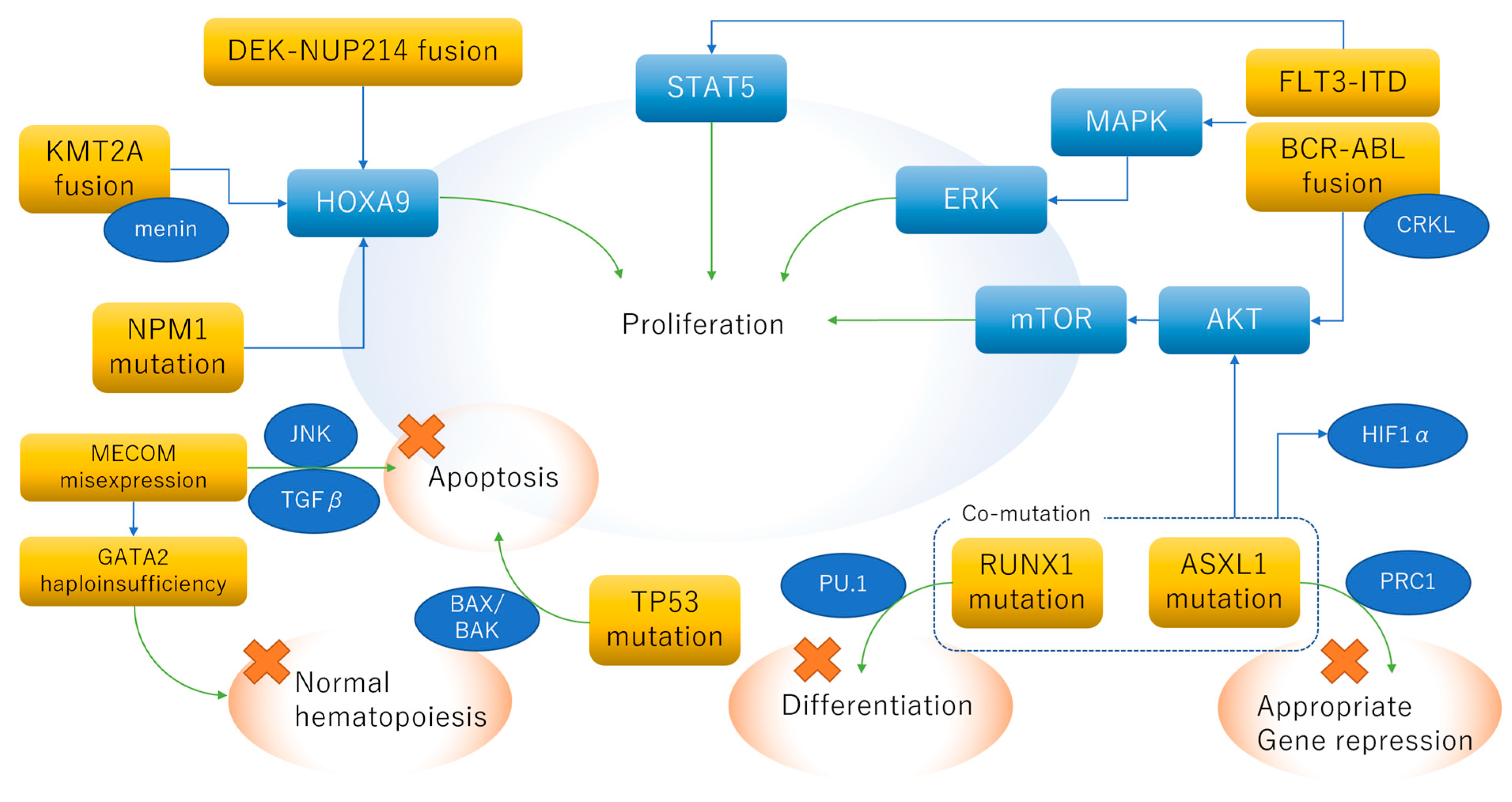
2. Biology of Adverse Genetic Abnormalities
2.1. DEK-NUP214 Fusion
2.2. KMT2A (MLL) Rearrangement
2.3. BCR-ABL1 Fusion
2.4. Haploinsufficiency of GATA2 and Mis-Expression of MECOM
2.5. Complex Karyotype
2.6. FLT3-ITD with Wild-Type NPM1
2.7. RUNX1 Loss-of-Function Mutation
2.8. ASXL1 Loss-of-Function Mutation
2.9. TP53 Loss-of-Function Mutation
3. Real-World Etiology of Adverse Genetic Abnormalities
3.1. Ad Hoc Analysis of the Japan Adult Leukemia Study Group (JALSG) AML201 Study
3.2. Hematologic Malignancy (HM)-SCREEN JAPAN 01 Study
3.2.1. Study Design
3.2.2. ELN Classification
3.2.3. Conventional Versus NGS-Based Classification
3.2.4. Clinical Outcome
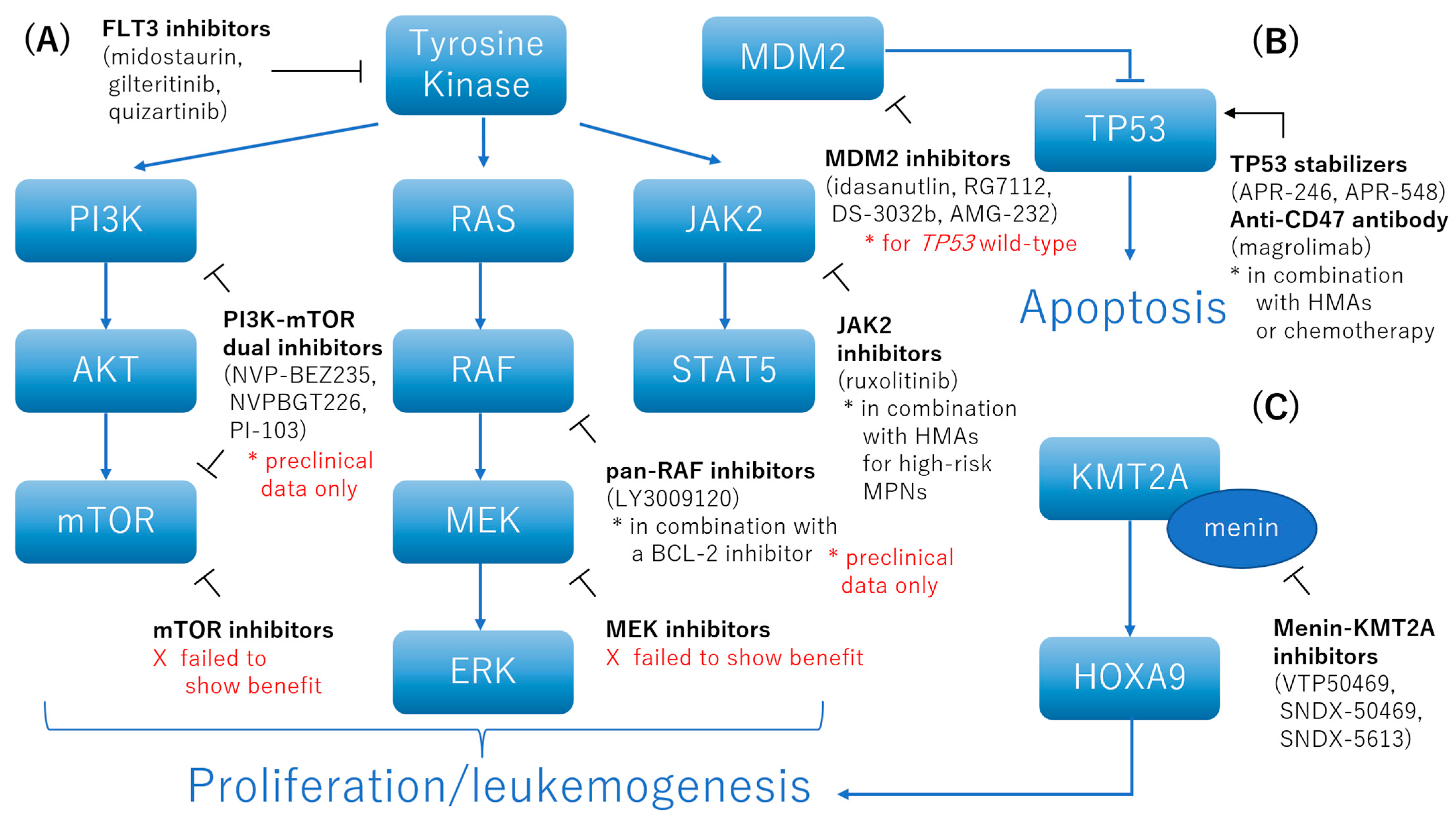
4. How to Deal with Specific Adverse Genetic Factors
4.1. FLT3 Inhibitors
4.2. Inhibition of the AKT, MAPK, and STAT Pathways
4.3. Menin-KMT2A Inhibitors
4.4. TP53 Stabilizers
4.5. Anti-CD47 Antibody
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Pratcorona, M.; Brunet, S.; Nomdedéu, J.; Ribera, J.M.; Tormo, M.; Duarte, R.; Escoda, L.; Guàrdia, R.; Queipo de Llano, M.P.; Salamero, O.; et al. Favorable Outcome of Patients with Acute Myeloid Leukemia Harboring a Low-Allelic Burden FLT3-ITD Mutation and Concomitant NPM1 Mutation: Relevance to Post-Remission Therapy. Blood 2013, 121, 2734–2738. [Google Scholar] [CrossRef] [PubMed]
- Paschka, P.; Schlenk, R.F.; Gaidzik, V.I.; Herzig, J.K.; Aulitzky, T.; Bullinger, L.; Späth, D.; Teleanu, V.; Kündgen, A.; Köhne, C.-H.; et al. ASXL1 Mutations in Younger Adult Patients with Acute Myeloid Leukemia: A Study by the German-Austrian Acute Myeloid Leukemia Study Group. Haematologica 2015, 100, 324–330. [Google Scholar] [CrossRef]
- Allen, C.; Hills, R.K.; Lamb, K.; Evans, C.; Tinsley, S.; Sellar, R.; O’Brien, M.; Yin, J.L.; Burnett, A.K.; Linch, D.C.; et al. The Importance of Relative Mutant Level for Evaluating Impact on Outcome of KIT, FLT3 and CBL Mutations in Core-Binding Factor Acute Myeloid Leukemia. Leukemia 2013, 27, 1891–1901. [Google Scholar] [CrossRef]
- Jourdan, E.; Boissel, N.; Chevret, S.; Delabesse, E.; Renneville, A.; Cornillet, P.; Blanchet, O.; Cayuela, J.-M.; Recher, C.; Raffoux, E.; et al. Prospective Evaluation of Gene Mutations and Minimal Residual Disease in Patients with Core Binding Factor Acute Myeloid Leukemia. Blood 2013, 121, 2213–2223. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K.; on behalf of the National Cancer Research Institute Adult Leukaemia Working Group. Refinement of Cytogenetic Classification in Acute Myeloid Leukemia: Determination of Prognostic Significance of Rare Recurring Chromosomal Abnormalities among 5876 Younger Adult Patients Treated in the United Kingdom Medical Research Council Trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Slovak, M.L.; Gundacker, H.; Bloomfield, C.D.; Dewald, G.; Appelbaum, F.R.; Larson, R.A.; Tallman, M.S.; Bennett, J.M.; Stirewalt, D.L.; Meshinchi, S.; et al. A Retrospective Study of 69 Patients with t(6;9)(P23;Q34) AML Emphasizes the Need for a Prospective, Multicenter Initiative for Rare “poor Prognosis” Myeloid Malignancies. Leukemia 2006, 20, 1295–1297. [Google Scholar] [CrossRef]
- Hu, H.; Scholten, I.; Gruss, C.; Knippers, R. The Distribution of the DEK Protein in Mammalian Chromatin. Biochem. Biophys. Res. Commun. 2007, 358, 1008–1014. [Google Scholar] [CrossRef]
- Riveiro-Falkenbach, E.; Soengas, M.S. Control of Tumorigenesis and Chemoresistance by the DEK Oncogene. Clin. Cancer Res. 2010, 16, 2932–2938. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; van Deursen, J.; van Baal, S.; Reynolds, A.; Davis, D.; Murti, K.G.; Fransen, J.; Grosveld, G. The Human Homologue of Yeast CRM1 Is in a Dynamic Subcomplex with CAN/Nup214 and a Novel Nuclear Pore Component Nup88. EMBO J. 1997, 16, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Fichtman, B.; Harel, T.; Biran, N.; Zagairy, F.; Applegate, C.D.; Salzberg, Y.; Gilboa, T.; Salah, S.; Shaag, A.; Simanovsky, N.; et al. Pathogenic Variants in NUP214 Cause “Plugged” Nuclear Pore Channels and Acute Febrile Encephalopathy. Am. J. Hum. Genet. 2019, 105, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Malek, S.; Cowell, J.K.; Ren, M. Transformation of Human CD34+ Hematopoietic Progenitor Cells with DEK-NUP214 Induces AML in an Immunocompromized Mouse Model. Oncogene 2016, 35, 5686–5691. [Google Scholar] [CrossRef] [PubMed]
- Sandén, C.; Ageberg, M.; Petersson, J.; Lennartsson, A.; Gullberg, U. Forced Expression of the DEK-NUP214 Fusion Protein Promotes Proliferation Dependent on Upregulation of MTOR. BMC Cancer 2013, 13, 440. [Google Scholar] [CrossRef]
- Oancea, C.; Rüster, B.; Henschler, R.; Puccetti, E.; Ruthardt, M. The t(6;9) Associated DEK/CAN Fusion Protein Targets a Population of Long-Term Repopulating Hematopoietic Stem Cells for Leukemogenic Transformation. Leukemia 2010, 24, 1910–1919. [Google Scholar] [CrossRef]
- Park, S.; Osmers, U.; Raman, G.; Schwantes, R.H.; Diaz, M.O.; Bushweller, J.H. The PHD3 Domain of MLL Acts as a CYP33-Regulated Switch between MLL-Mediated Activation and Repression. Biochemistry 2010, 49, 6576–6586. [Google Scholar] [CrossRef]
- Dou, Y.; Milne, T.A.; Tackett, A.J.; Smith, E.R.; Fukuda, A.; Wysocka, J.; Allis, C.D.; Chait, B.T.; Hess, J.L.; Roeder, R.G. Physical Association and Coordinate Function of the H3 K4 Methyltransferase MLL1 and the H4 K16 Acetyltransferase MOF. Cell 2005, 121, 873–885. [Google Scholar] [CrossRef]
- Slany, R.K. When Epigenetics Kills: MLL Fusion Proteins in Leukemia. Hematol. Oncol. 2005, 23, 1–9. [Google Scholar] [CrossRef]
- von Neuhoff, C.; Reinhardt, D.; Sander, A.; Zimmermann, M.; Bradtke, J.; Betts, D.R.; Zemanova, Z.; Stary, J.; Bourquin, J.-P.; Haas, O.A.; et al. Prognostic Impact of Specific Chromosomal Aberrations in a Large Group of Pediatric Patients with Acute Myeloid Leukemia Treated Uniformly According to Trial AML-BFM 98. J. Clin. Oncol. 2010, 28, 2682–2689. [Google Scholar] [CrossRef] [PubMed]
- on Behalf of the Acute Leukemia Working Party EBMT; Pigneux, A.; Labopin, M.; Maertens, J.; Cordonnier, C.; Volin, L.; Socié, G.; Blaise, D.; Craddock, C.; Milpied, N.; et al. Outcome of Allogeneic Hematopoietic Stem-Cell Transplantation for Adult Patients with AML and 11q23/MLL Rearrangement (MLL-r AML). Leukemia 2015, 29, 2375–2381. [Google Scholar] [CrossRef] [PubMed]
- Dafflon, C.; Craig, V.J.; Méreau, H.; Gräsel, J.; Schacher Engstler, B.; Hoffman, G.; Nigsch, F.; Gaulis, S.; Barys, L.; Ito, M.; et al. Complementary Activities of DOT1L and Menin Inhibitors in MLL-Rearranged Leukemia. Leukemia 2017, 31, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Somervaille, T.C.P.; Smith, K.S.; Rozenblatt-Rosen, O.; Meyerson, M.; Cleary, M.L. The Menin Tumor Suppressor Protein Is an Essential Oncogenic Cofactor for MLL-Associated Leukemogenesis. Cell 2005, 123, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, S.A.; Staunton, J.E.; Silverman, L.B.; Pieters, R.; den Boer, M.L.; Minden, M.D.; Sallan, S.E.; Lander, E.S.; Golub, T.R.; Korsmeyer, S.J. MLL Translocations Specify a Distinct Gene Expression Profile That Distinguishes a Unique Leukemia. Nat. Genet. 2002, 30, 41–47. [Google Scholar] [CrossRef]
- Milne, T.A.; Martin, M.E.; Brock, H.W.; Slany, R.K.; Hess, J.L. Leukemogenic MLL Fusion Proteins Bind across a Broad Region of the Hox A9 Locus, Promoting Transcription and Multiple Histone Modifications. Cancer Res. 2005, 65, 11367–11374. [Google Scholar] [CrossRef]
- Yokoyama, A.; Cleary, M.L. Menin Critically Links MLL Proteins with LEDGF on Cancer-Associated Target Genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef]
- Yuan, Z.M.; Huang, Y.; Ishiko, T.; Kharbanda, S.; Weichselbaum, R.; Kufe, D. Regulation of DNA Damage-Induced Apoptosis by the c-Abl Tyrosine Kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 1437–1440. [Google Scholar] [CrossRef]
- Yuan, Z.M.; Huang, Y.; Ishiko, T.; Nakada, S.; Utsugisawa, T.; Kharbanda, S.; Wang, R.; Sung, P.; Shinohara, A.; Weichselbaum, R.; et al. Regulation of Rad51 Function by C-Abl in Response to DNA Damage. J. Biol. Chem. 1998, 273, 3799–3802. [Google Scholar] [CrossRef]
- Verschraegen, C.F.; Kantarjian, H.M.; Hirsch-Ginsberg, C.; Lee, M.S.; O’Brien, S.; Rios, M.B.; Stass, S.A.; Keating, M.; Talpaz, M. The BreakpoInt. Cluster Region Site in Patients with Philadelphia Chromosome-Positive Chronic Myelogenous Leukemia. Clinical, Laboratory, and Prognostic Correlations. Cancer 1995, 76, 992–997. [Google Scholar] [CrossRef]
- Sattler, M.; Salgia, R. Activation of Hematopoietic Growth Factor Signal Transduction Pathways by the Human Oncogene BCR/ABL. Cytokine Growth Factor Rev. 1997, 8, 63–79. [Google Scholar] [CrossRef]
- Skorski, T.; Kanakaraj, P.; Nieborowska-Skorska, M.; Ratajczak, M.Z.; Wen, S.-C.; Zon, G.; Gewirtz, A.M.; Perussia, B.; Calabretta, B. Phosphatidylinositol-3 Kinase Activity Is Regulated by BCR/ABL and Is Required for the Growth of Philadelphia Chromosome-Positive Cells. Blood 1995, 86, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Uemura, N.; Okuda, K.; Li, J.L.; Pisick, E.; Sattler, M.; de Jong, R.; Druker, B.; Heisterkamp, N.; Chen, L.B. CRKL Links P210BCR/ABL with Paxillin in Chronic Myelogenous Leukemia Cells. J. Biol. Chem. 1995, 270, 29145–29150. [Google Scholar] [CrossRef] [PubMed]
- de Jong, R.; ten Hoeve, J.; Heisterkamp, N.; Groffen, J. Crkl Is Complexed with Tyrosine-Phosphorylated Cbl in Ph-Positive Leukemia. J. Biol. Chem. 1995, 270, 21468–21471. [Google Scholar] [CrossRef] [PubMed]
- Shuai, K.; Halpern, J.; ten Hoeve, J.; Rao, X.; Sawyers, C.L. Constitutive Activation of STAT5 by the BCR-ABL Oncogene in Chronic Myelogenous Leukemia. Oncogene 1996, 13, 247–254. [Google Scholar] [PubMed]
- Gishizky, M.L.; Cortez, D.; Pendergast, A.M. Mutant Forms of Growth Factor-Binding Protein-2 Reverse BCR-ABL-Induced Transformation. Proc. Natl. Acad. Sci. USA 1995, 92, 10889–10893. [Google Scholar] [CrossRef]
- Gorre, M.E.; Ellwood-Yen, K.; Chiosis, G.; Rosen, N.; Sawyers, C.L. BCR-ABL PoInt. Mutants Isolated from Patients with Imatinib Mesylate–Resistant Chronic Myeloid Leukemia Remain Sensitive to Inhibitors of the BCR-ABL Chaperone Heat Shock Protein 90. Blood 2002, 100, 3041–3044. [Google Scholar] [CrossRef]
- Bagatell, R.; Whitesell, L. Altered Hsp90 Function in Cancer: A Unique Therapeutic Opportunity. Mol. Cancer 2004, 3, 1021–1030. [Google Scholar]
- Keung, Y.-K.; Beaty, M.; Powell, B.L.; Molnar, I.; Buss, D.; Pettenati, M. Philadelphia Chromosome Positive Myelodysplastic Syndrome and Acute Myeloid Leukemia-Retrospective Study and Review of Literature. Leuk. Res. 2004, 28, 579–586. [Google Scholar] [CrossRef]
- Berger, R. Differences between Blastic Chronic Myeloid Leukemia and Ph-Positive Acute Leukemia. Leuk. Lymphoma 1993, 11 (Suppl. S1), 235–237. [Google Scholar] [CrossRef]
- Neuendorff, N.R.; Burmeister, T.; Dörken, B.; Westermann, J. BCR-ABL-Positive Acute Myeloid Leukemia: A New Entity? Analysis of Clinical and Molecular Features. Ann. Hematol. 2016, 95, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Morishita, K.; Parganas, E.; William, C.L.; Whittaker, M.H.; Drabkin, H.; Oval, J.; Taetle, R.; Valentine, M.B.; Ihle, J.N. Activation of EVI1 Gene Expression in Human Acute Myelogenous Leukemias by Translocations Spanning 300-400 Kilobases on Chromosome Band 3q26. Proc. Natl. Acad. Sci. USA 1992, 89, 3937–3941. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, M.; Mitani, K.; Irie, K.; Matsuyama, T.; Takahashi, T.; Chiba, S.; Yazaki, Y.; Matsumoto, K.; Hirai, H. The Oncoprotein Evi-1 Represses TGF-Beta Signalling by Inhibiting Smad3. Nature 1998, 394, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, M.; Mitani, K.; Yamagata, T.; Takahashi, T.; Izutsu, K.; Ogawa, S.; Moriguchi, T.; Nishida, E.; Yazaki, Y.; Hirai, H. The Evi-1 Oncoprotein Inhibits c-Jun N-Terminal Kinase and Prevents Stress-Induced Cell Death. EMBO J. 2000, 19, 2958–2968. [Google Scholar] [CrossRef]
- Katayama, S.; Suzuki, M.; Yamaoka, A.; Keleku-Lukwete, N.; Katsuoka, F.; Otsuki, A.; Kure, S.; Engel, J.D.; Yamamoto, M. GATA2 Haploinsufficiency Accelerates EVI1-Driven Leukemogenesis. Blood 2017, 130, 908–919. [Google Scholar] [CrossRef]
- Minegishi, N.; Suzuki, N.; Yokomizo, T.; Pan, X.; Fujimoto, T.; Takahashi, S.; Hara, T.; Miyajima, A.; Nishikawa, S.-I.; Yamamoto, M. Expression and Domain-Specific Function of GATA-2 during Differentiation of the Hematopoietic Precursor Cells in Midgestation Mouse Embryos. Blood 2003, 102, 896–905. [Google Scholar] [CrossRef]
- Hsu, A.P.; Johnson, K.D.; Falcone, E.L.; Sanalkumar, R.; Sanchez, L.; Hickstein, D.D.; Cuellar-Rodriguez, J.; Lemieux, J.E.; Zerbe, C.S.; Bresnick, E.H.; et al. GATA2 Haploinsufficiency Caused by Mutations in a Conserved Intronic Element Leads to MonoMAC Syndrome. Blood 2013, 121, 3830–3837, S1–7. [Google Scholar] [CrossRef]
- Dickinson, R.E.; Griffin, H.; Bigley, V.; Reynard, L.N.; Hussain, R.; Haniffa, M.; Lakey, J.H.; Rahman, T.; Wang, X.-N.; McGovern, N.; et al. Exome Sequencing Identifies GATA-2 Mutation as the Cause of Dendritic Cell, Monocyte, B and NK Lymphoid Deficiency. Blood 2011, 118, 2656–2658. [Google Scholar] [CrossRef]
- Galera, P.; Hsu, A.P.; Wang, W.; Droll, S.; Chen, R.; Schwartz, J.R.; Klco, J.M.; Arai, S.; Maese, L.; Zerbe, C.; et al. Donor-Derived MDS/AML in Families with Germline GATA2 Mutation. Blood 2018, 132, 1994–1998. [Google Scholar] [CrossRef]
- Lugthart, S.; Gröschel, S.; Beverloo, H.B.; Kayser, S.; Valk, P.J.M.; van Zelderen-Bhola, S.L.; Jan Ossenkoppele, G.; Vellenga, E.; van den Berg-de Ruiter, E.; Schanz, U.; et al. Clinical, Molecular, and Prognostic Significance of WHO Type Inv(3)(Q21q26.2)/t(3;3)(Q21;Q26.2) and Various Other 3q Abnormalities in Acute Myeloid Leukemia. JCO 2010, 28, 3890–3898. [Google Scholar] [CrossRef]
- Barjesteh van Waalwijk van Doorn-Khosrovani, S.; Erpelinck, C.; van Putten, W.L.J.; Valk, P.J.M.; van der Poel-van de Luytgaarde, S.; Hack, R.; Slater, R.; Smit, E.M.E.; Beverloo, H.B.; Verhoef, G.; et al. High EVI1 Expression Predicts Poor Survival in Acute Myeloid Leukemia: A Study of 319 de Novo AML Patients. Blood 2003, 101, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Lugthart, S.; van Drunen, E.; van Norden, Y.; van Hoven, A.; Erpelinck, C.A.J.; Valk, P.J.M.; Beverloo, H.B.; Löwenberg, B.; Delwel, R. High EVI1 Levels Predict Adverse Outcome in Acute Myeloid Leukemia: Prevalence of EVI1 Overexpression and Chromosome 3q26 Abnormalities Underestimated. Blood 2008, 111, 4329–4337. [Google Scholar] [CrossRef] [PubMed]
- Mrózek, K. Cytogenetic, Molecular Genetic, and Clinical Characteristics of Acute Myeloid Leukemia with a Complex Karyotype. Semin. Oncol. 2008, 35, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Stölzel, F.; Mohr, B.; Kramer, M.; Oelschlägel, U.; Bochtler, T.; Berdel, W.E.; Kaufmann, M.; Baldus, C.D.; Schäfer-Eckart, K.; Stuhlmann, R.; et al. Karyotype Complexity and Prognosis in Acute Myeloid Leukemia. Blood Cancer J. 2016, 6, e386. [Google Scholar] [CrossRef] [PubMed]
- Hannum, C.; Culpepper, J.; Campbell, D.; McClanahan, T.; Zurawski, S.; Bazan, J.F.; Kastelein, R.; Hudak, S.; Wagner, J.; Mattson, J. Ligand for FLT3/FLK2 Receptor Tyrosine Kinase Regulates Growth of Haematopoietic Stem Cells and Is Encoded by Variant RNAs. Nature 1994, 368, 643–648. [Google Scholar] [CrossRef]
- Zhang, S.; Mantel, C.; Broxmeyer, H.E. Flt3 Signaling Involves Tyrosyl-Phosphorylation of SHP-2 and SHIP and Their Association with Grb2 and Shc in Baf3/Flt3 Cells. J. Leukoc. Biol. 1999, 65, 372–380. [Google Scholar] [CrossRef]
- Brandts, C.H.; Sargin, B.; Rode, M.; Biermann, C.; Lindtner, B.; Schwäble, J.; Buerger, H.; Müller-Tidow, C.; Choudhary, C.; McMahon, M.; et al. Constitutive Activation of Akt by Flt3 Internal Tandem Duplications Is Necessary for Increased Survival, Proliferation, and Myeloid Transformation. Cancer Res. 2005, 65, 9643–9650. [Google Scholar] [CrossRef]
- Ekim, B.; Magnuson, B.; Acosta-Jaquez, H.A.; Keller, J.A.; Feener, E.P.; Fingar, D.C. MTOR Kinase Domain Phosphorylation Promotes MTORC1 Signaling, Cell Growth, and Cell Cycle Progression. Mol. Cell Biol. 2011, 31, 2787–2801. [Google Scholar] [CrossRef]
- Bagrintseva, K.; Geisenhof, S.; Kern, R.; Eichenlaub, S.; Reindl, C.; Ellwart, J.W.; Hiddemann, W.; Spiekermann, K. FLT3-ITD-TKD Dual Mutants Associated with AML Confer Resistance to FLT3 PTK Inhibitors and Cytotoxic Agents by Overexpression of Bcl-x(L). Blood 2005, 105, 3679–3685. [Google Scholar] [CrossRef]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The MTOR-Regulated Phosphoproteome Reveals a Mechanism of MTORC1-Mediated Inhibition of Growth Factor Signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef]
- Mizuki, M.; Fenski, R.; Halfter, H.; Matsumura, I.; Schmidt, R.; Müller, C.; Grüning, W.; Kratz-Albers, K.; Serve, S.; Steur, C.; et al. Flt3 Mutations from Patients with Acute Myeloid Leukemia Induce Transformation of 32D Cells Mediated by the Ras and STAT5 Pathways. Blood 2000, 96, 3907–3914. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Ye, B.H.; Dalla-Favera, R. Antigen Receptor Signaling Induces MAP Kinase-Mediated Phosphorylation and Degradation of the BCL-6 Transcription Factor. Genes Dev. 1998, 12, 1953–1961. [Google Scholar] [CrossRef] [PubMed]
- Domina, A.M.; Vrana, J.A.; Gregory, M.A.; Hann, S.R.; Craig, R.W. MCL1 Is Phosphorylated in the PEST Region and Stabilized upon ERK Activation in Viable Cells, and at Additional Sites with Cytotoxic Okadaic Acid or Taxol. Oncogene 2004, 23, 5301–5315. [Google Scholar] [CrossRef] [PubMed]
- Maggi, L.B.; Kuchenruether, M.; Dadey, D.Y.A.; Schwope, R.M.; Grisendi, S.; Townsend, R.R.; Pandolfi, P.P.; Weber, J.D. Nucleophosmin Serves as a Rate-Limiting Nuclear Export Chaperone for the Mammalian Ribosome. Mol. Cell Biol. 2008, 28, 7050–7065. [Google Scholar] [CrossRef]
- Swaminathan, V.; Kishore, A.H.; Febitha, K.K.; Kundu, T.K. Human Histone Chaperone Nucleophosmin Enhances Acetylation-Dependent Chromatin Transcription. Mol. Cell Biol. 2005, 25, 7534–7545. [Google Scholar] [CrossRef]
- Wang, H.-F.; Takenaka, K.; Nakanishi, A.; Miki, Y. BRCA2 and Nucleophosmin Coregulate Centrosome Amplification and Form a Complex with the Rho Effector Kinase ROCK2. Cancer Res. 2011, 71, 68–77. [Google Scholar] [CrossRef]
- Liu, X.; Liu, D.; Qian, D.; Dai, J.; An, Y.; Jiang, S.; Stanley, B.; Yang, J.; Wang, B.; Liu, X.; et al. Nucleophosmin (NPM1/B23) Interacts with Activating Transcription Factor 5 (ATF5) Protein and Promotes Proteasome- and Caspase-Dependent ATF5 Degradation in Hepatocellular Carcinoma Cells. J. Biol. Chem. 2012, 287, 19599–19609. [Google Scholar] [CrossRef]
- Korgaonkar, C.; Hagen, J.; Tompkins, V.; Frazier, A.A.; Allamargot, C.; Quelle, F.W.; Quelle, D.E. Nucleophosmin (B23) Targets ARF to Nucleoli and Inhibits Its Function. Mol. Cell Biol. 2005, 25, 1258–1271. [Google Scholar] [CrossRef]
- Colombo, E.; Marine, J.-C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin Regulates the Stability and Transcriptional Activity of P53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Cho, Y.-E.; Park, J.-H. The Nucleolar Protein GLTSCR2 Is an UpstreAm. Negative Regulator of the Oncogenic Nucleophosmin-MYC Axis. Am. J. Pathol. 2015, 185, 2061–2068. [Google Scholar] [CrossRef]
- Falini, B.; Bolli, N.; Shan, J.; Martelli, M.P.; Liso, A.; Pucciarini, A.; Bigerna, B.; Pasqualucci, L.; Mannucci, R.; Rosati, R.; et al. Both Carboxy-Terminus NES Motif and Mutated Tryptophan(s) Are Crucial for Aberrant Nuclear Export of Nucleophosmin Leukemic Mutants in NPMc+ AML. Blood 2006, 107, 4514–4523. [Google Scholar] [CrossRef] [PubMed]
- Heath, E.M.; Chan, S.M.; Minden, M.D.; Murphy, T.; Shlush, L.I.; Schimmer, A.D. Biological and Clinical Consequences of NPM1 Mutations in AML. Leukemia 2017, 31, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.J.; Han, Y.; Jia, N.; Chen, P.; Minden, M.D. NPM1c Impedes CTCF Functions through Cytoplasmic Mislocalization in Acute Myeloid Leukemia. Leukemia 2020, 34, 1278–1290. [Google Scholar] [CrossRef]
- Gu, X.; Ebrahem, Q.; Mahfouz, R.Z.; Hasipek, M.; Enane, F.; Radivoyevitch, T.; Rapin, N.; Przychodzen, B.; Hu, Z.; Balusu, R.; et al. Leukemogenic Nucleophosmin Mutation Disrupts the Transcription Factor Hub That Regulates Granulomonocytic Fates. J. Clin. Investig. 2018, 128, 4260–4279. [Google Scholar] [CrossRef]
- Alcalay, M.; Tiacci, E.; Bergomas, R.; Bigerna, B.; Venturini, E.; Minardi, S.P.; Meani, N.; Diverio, D.; Bernard, L.; Tizzoni, L.; et al. Acute Myeloid Leukemia Bearing Cytoplasmic Nucleophosmin (NPMc+ AML) Shows a Distinct Gene Expression Profile Characterized by up-Regulation of Genes Involved in Stem-Cell Maintenance. Blood 2005, 106, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic Nucleophosmin in Acute Myelogenous Leukemia with a Normal Karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Kennedy, A.; Zhou, X.; Radtke, I.; Phillips, L.A.; Shurtleff, S.A.; Downing, J.R. Pediatric Acute Myeloid Leukemia with NPM1 Mutations Is Characterized by a Gene Expression Profile with Dysregulated HOX Gene Expression Distinct from MLL-Rearranged Leukemias. Leukemia 2007, 21, 2000–2009. [Google Scholar] [CrossRef]
- Collins, C.T.; Hess, J.L. Deregulation of the HOXA9/MEIS1 Axis in Acute Leukemia. Curr. Opin. Hematol. 2016, 23, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Gale, R.E.; Green, C.; Allen, C.; Mead, A.J.; Burnett, A.K.; Hills, R.K.; Linch, D.C. The Impact of FLT3 Internal Tandem Duplication Mutant Level, Number, Size, and Interaction with NPM1 Mutations in a Large Cohort of Young Adult Patients with Acute Myeloid Leukemia. Blood 2008, 111, 2776–2784. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M.; Yamaguchi, H.; Najima, Y.; Usuki, K.; Ueki, T.; Oh, I.; Mori, S.; Kawata, E.; Uoshima, N.; Kobayashi, Y.; et al. Prognostic Impact of Low Allelic Ratio FLT3-ITD and NPM1 Mutation in Acute Myeloid Leukemia. Blood Adv. 2018, 2, 2744–2754. [Google Scholar] [CrossRef]
- Döhner, K.; Thiede, C.; Jahn, N.; Panina, E.; Gambietz, A.; Larson, R.A.; Prior, T.W.; Marcucci, G.; Jones, D.; Krauter, J.; et al. Impact of NPM1/FLT3-ITD Genotypes Defined by the 2017 European LeukemiaNet in Patients with Acute Myeloid Leukemia. Blood 2020, 135, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Anderson, K.; Jacobsen, S.E.W.; Nishikawa, S.-I.; Nerlov, C. Cdk6 Blocks Myeloid Differentiation by Interfering with Runx1 DNA Binding and Runx1-C/EBPalpha Interaction. EMBO J. 2007, 26, 2361–2370. [Google Scholar] [CrossRef]
- Morita, K.; Noura, M.; Tokushige, C.; Maeda, S.; Kiyose, H.; Kashiwazaki, G.; Taniguchi, J.; Bando, T.; Yoshida, K.; Ozaki, T.; et al. Autonomous Feedback Loop of RUNX1-P53-CBFB in Acute Myeloid Leukemia Cells. Sci. Rep. 2017, 7, 16604. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-J.; Wang, Z.; Wang, L.; Jiang, Y.; Kost, N.; Soong, T.D.; Chen, W.-Y.; Tang, Z.; Nakadai, T.; Elemento, O.; et al. A Stable Transcription Factor Complex Nucleated by Oligomeric AML1-ETO Controls Leukaemogenesis. Nature 2013, 500, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Nagata, Y.; Kihara, R.; Ishikawa, Y.; Asou, N.; Ohtake, S.; Miyawaki, S.; Sakura, T.; Ozawa, Y.; Usui, N.; et al. Prognostic Analysis According to the 2017 ELN Risk Stratification by Genetics in Adult Acute Myeloid Leukemia Patients Treated in the Japan Adult Leukemia Study Group (JALSG) AML201 Study. Leuk. Res. 2018, 66, 20–27. [Google Scholar] [CrossRef]
- Gaidzik, V.I.; Teleanu, V.; Papaemmanuil, E.; Weber, D.; Paschka, P.; Hahn, J.; Wallrabenstein, T.; Kolbinger, B.; Köhne, C.H.; Horst, H.A.; et al. RUNX1 Mutations in Acute Myeloid Leukemia Are Associated with Distinct Clinico-Pathologic and Genetic Features. Leukemia 2016, 30, 2160–2168. [Google Scholar] [CrossRef]
- Bera, R.; Chiu, M.-C.; Huang, Y.-J.; Lin, T.-H.; Kuo, M.-C.; Shih, L.-Y. RUNX1 Mutations Promote Leukemogenesis of Myeloid Malignancies in ASXL1-Mutated Leukemia. J. Hematol. Oncol. 2019, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.C.; Leeanansaksiri, W.; Ji, M.; Klarmann, K.D.; Renn, K.; Gooya, J.; Smith, D.; McNiece, I.; Lugthart, S.; Valk, P.J.M.; et al. Id1 Immortalizes Hematopoietic Progenitors in Vitro and Promotes a Myeloproliferative Disease in Vivo. Oncogene 2008, 27, 5612–5623. [Google Scholar] [CrossRef]
- Gu, X.; Hu, Z.; Ebrahem, Q.; Crabb, J.S.; Mahfouz, R.Z.; Radivoyevitch, T.; Crabb, J.W.; Saunthararajah, Y. Runx1 Regulation of Pu.1 Corepressor/Coactivator Exchange Identifies Specific Molecular Targets for Leukemia Differentiation Therapy. J. Biol. Chem. 2014, 289, 14881–14895. [Google Scholar] [CrossRef]
- Kweon, S.-M.; Chen, Y.; Moon, E.; Kvederaviciutė, K.; Klimasauskas, S.; Feldman, D.E. An Adversarial DNA N6-Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol. Cell 2019, 74, 1138–1147.e6. [Google Scholar] [CrossRef]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856.e5. [Google Scholar] [CrossRef] [PubMed]
- Uni, M.; Masamoto, Y.; Sato, T.; Kamikubo, Y.; Arai, S.; Hara, E.; Kurokawa, M. Modeling ASXL1 Mutation Revealed Impaired Hematopoiesis Caused by Derepression of P16Ink4a through Aberrant PRC1-Mediated Histone Modification. Leukemia 2019, 33, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Xu, S.; Zhang, Y.; Xu, J.; Huang, Z.; Liu, W.; Wang, S.; Yen, K.; Zhang, W. Asxl1 C-Terminal Mutation Perturbs Neutrophil Differentiation in Zebrafish. Leukemia 2021, 35, 2299–2310. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, R.; Wu, X. The Functions and Mechanisms of PR-DUB in Malignancy. Front. Mol. Biosci. 2021, 8, 657150. [Google Scholar] [CrossRef] [PubMed]
- Asada, S.; Fujino, T.; Goyama, S.; Kitamura, T. The Role of ASXL1 in Hematopoiesis and Myeloid Malignancies. Cell Mol. Life Sci. 2019, 76, 2511–2523. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Gao, J.; Adli, M.; Dey, A.; Trimarchi, T.; Chung, Y.R.; Kuscu, C.; Hricik, T.; Ndiaye-Lobry, D.; Lafave, L.M.; et al. Deletion of Asxl1 Results in Myelodysplasia and Severe Developmental Defects in Vivo. J. Exp. Med. 2013, 210, 2641–2659. [Google Scholar] [CrossRef]
- Zhang, P.; He, F.; Bai, J.; Yamamoto, S.; Chen, S.; Zhang, L.; Sheng, M.; Zhang, L.; Guo, Y.; Man, N.; et al. Chromatin Regulator Asxl1 Loss and Nf1 Haploinsufficiency Cooperate to Accelerate Myeloid Malignancy. J. Clin. Investig. 2018, 128, 5383–5398. [Google Scholar] [CrossRef]
- Schnittger, S.; Eder, C.; Jeromin, S.; Alpermann, T.; Fasan, A.; Grossmann, V.; Kohlmann, A.; Illig, T.; Klopp, N.; Wichmann, H.-E.; et al. ASXL1 Exon 12 Mutations Are Frequent in AML with Intermediate Risk Karyotype and Are Independently Associated with an Adverse Outcome. Leukemia 2013, 27, 82–91. [Google Scholar] [CrossRef]
- Wu, M.; Bellas, R.E.; Shen, J.; Sonenshein, G.E. Roles of the Tumor Suppressor P53 and the Cyclin-Dependent Kinase Inhibitor P21WAF1/CIP1 in Receptor-Mediated Apoptosis of WEHI 231 B Lymphoma Cells. J. Exp. Med. 1998, 187, 1671–1679. [Google Scholar] [CrossRef]
- Chipuk, J.E.; Kuwana, T.; Bouchier-Hayes, L.; Droin, N.M.; Newmeyer, D.D.; Schuler, M.; Green, D.R. Direct Activation of Bax by P53 Mediates Mitochondrial Membrane Permeabilization and Apoptosis. Science 2004, 303, 1010–1014. [Google Scholar] [CrossRef]
- Thomas, A.; Giesler, T.; White, E. P53 Mediates Bcl-2 Phosphorylation and Apoptosis via Activation of the Cdc42/JNK1 Pathway. Oncogene 2000, 19, 5259–5269. [Google Scholar] [CrossRef]
- Kuwana, T.; Newmeyer, D.D. Bcl-2-Family Proteins and the Role of Mitochondria in Apoptosis. Curr. Opin. Cell Biol. 2003, 15, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Letai, A.; Bassik, M.C.; Walensky, L.D.; Sorcinelli, M.D.; Weiler, S.; Korsmeyer, S.J. Distinct BH3 Domains Either Sensitize or Activate Mitochondrial Apoptosis, Serving as Prototype Cancer Therapeutics. Cancer Cell 2002, 2, 183–192. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Hu, C.; Qutub, A.; Qiu, Y.H.; Zhang, X.; Post, S.M.; Zhang, N.; Coombes, K.; Kornblau, S.M. P53 Pathway Dysfunction Is Highly Prevalent in Acute Myeloid Leukemia Independent of TP53 Mutational Status. Leukemia 2017, 31, 1296–1305. [Google Scholar] [CrossRef]
- Nahi, H.; Lehmann, S.; Bengtzen, S.; Jansson, M.; Möllgård, L.; Paul, C.; Merup, M. Chromosomal Aberrations in 17p Predict in Vitro Drug Resistance and Short Overall Survival in Acute Myeloid Leukemia. Leuk. Lymphoma 2008, 49, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, D.H.; Andersen, M.K.; Pedersen-Bjergaard, J. Mutations With Loss of Heterozygosity of P53 Are Common in Therapy-Related Myelodysplasia and Acute Myeloid Leukemia After Exposure to Alkylating Agents and Significantly Associated With Deletion or Loss of 5q, a Complex Karyotype, and a Poor Prognosis. JCO 2001, 19, 1405–1413. [Google Scholar] [CrossRef]
- Barbosa, K.; Li, S.; Adams, P.D.; Deshpande, A.J. The Role of TP53 in Acute Myeloid Leukemia: Challenges and Opportunities. Genes Chromosomes Cancer 2019, 58, 875–888. [Google Scholar] [CrossRef]
- Sallman, D.A.; McLemore, A.F.; Aldrich, A.L.; Komrokji, R.S.; McGraw, K.L.; Dhawan, A.; Geyer, S.; Hou, H.-A.; Eksioglu, E.A.; Sullivan, A.; et al. TP53 Mutations in Myelodysplastic Syndromes and Secondary AML Confer an Immunosuppressive Phenotype. Blood 2020, 136, 2812–2823. [Google Scholar] [CrossRef]
- Ohtake, S.; Miyawaki, S.; Fujita, H.; Kiyoi, H.; Shinagawa, K.; Usui, N.; Okumura, H.; Miyamura, K.; Nakaseko, C.; Miyazaki, Y.; et al. Randomized Study of Induction Therapy Comparing Standard-Dose Idarubicin with High-Dose Daunorubicin in Adult Patients with Previously Untreated Acute Myeloid Leukemia: The JALSG AML201 Study. Blood 2011, 117, 2358–2365. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, S.; Ohtake, S.; Fujisawa, S.; Kiyoi, H.; Shinagawa, K.; Usui, N.; Sakura, T.; Miyamura, K.; Nakaseko, C.; Miyazaki, Y.; et al. A Randomized Comparison of 4 Courses of Standard-Dose Multiagent Chemotherapy versus 3 Courses of High-Dose Cytarabine Alone in Postremission Therapy for Acute Myeloid Leukemia in Adults: The JALSG AML201 Study. Blood 2011, 117, 2366–2372. [Google Scholar] [CrossRef]
- Miyamoto, K.; Fukushima, K.; Chi, S.; Shibayama, H.; Hosono, N.; Yamauchi, T.; Katagiri, S.; Gotoh, A.; Morishita, T.; Yanada, M.; et al. Interim Analysis of Hematologic Malignancies (HM)-Screen-Japan 01: A Mutation Profiling Multicenter Study of Patients with AML. Blood 2020, 136, 2–3. [Google Scholar] [CrossRef]
- Hosono, N.; Yamauchi, T.; Chi, S.; Fukushima, K.; Shibayama, H.; Katagiri, S.; Gotoh, A.; Eguchi, M.; Morishita, T.; Ogasawara, R.; et al. Hematologic Malignancies (HM)-Screen-Japan 01: A Mutation Profiling Multicenter Study on Patients with Acute Myeloid Leukemia. Blood 2021, 138, 4457. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Döhner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Weber, D.; Fiedler, W.; Salih, H.R.; Wulf, G.; Salwender, H.; Schroeder, T.; Kindler, T.; Lübbert, M.; Wolf, D.; et al. Midostaurin Added to Chemotherapy and Continued Single-Agent Maintenance Therapy in Acute Myeloid Leukemia with FLT3-ITD. Blood 2019, 133, 840–851. [Google Scholar] [CrossRef]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3 -Mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Khaled, S.; Martinelli, G.; Perl, A.E.; Ganguly, S.; Russell, N.; Krämer, A.; Dombret, H.; Hogge, D.; Jonas, B.A.; et al. Quizartinib versus Salvage Chemotherapy in Relapsed or Refractory FLT3-ITD Acute Myeloid Leukaemia (QuANTUM-R): A Multicentre, Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet Oncol. 2019, 20, 984–997. [Google Scholar] [CrossRef]
- Wang, E.S.; Stone, R.M.; Tallman, M.S.; Walter, R.B.; Eckardt, J.R.; Collins, R. Crenolanib, a Type I FLT3 TKI, Can Be Safely Combined with Cytarabine and Anthracycline Induction Chemotherapy and Results in High Response Rates in Patients with Newly Diagnosed FLT3 Mutant Acute Myeloid Leukemia (AML). Blood 2016, 128, 1071. [Google Scholar] [CrossRef]
- Iyer, S.P.; Jethava, Y.; Karanes, C.; Eckardt, J.R.; Collins, R. Safety Study of Salvage Chemotherapy High-Dose Ara-C/Mitoxantrone (HAM) and Type I FLT3-TKI Crenolanib in First Relapsed/Primary Refractory AML. Blood 2016, 128, 3983. [Google Scholar] [CrossRef]
- Ohanian, M.; Kantarjian, H.M.; Borthakur, G.; Kadia, T.M.; Konopleva, M.; Garcia-Manero, G.; Estrov, Z.; Ferrajoli, A.; Takahashi, K.; Jabbour, E.J.; et al. Efficacy of a Type I FLT3 Inhibitor, Crenolanib, with Idarubicin and High-Dose Ara-C in Multiply Relapsed/Refractory FLT3+ AML. Blood 2016, 128, 2744. [Google Scholar] [CrossRef]
- Pratz, K.W.; Cherry, M.; Altman, J.K.; Cooper, B.; Cruz, J.C.; Jurcic, J.G.; Levis, M.J.; Lin, T.L.; Perl, A.E.; Podoltsev, N.A.; et al. Updated Results from a Phase 1 Study of Gilteritinib in Combination with Induction and Consolidation Chemotherapy in Subjects with Newly Diagnosed Acute Myeloid Leukemia (AML). Blood 2018, 132, 564. [Google Scholar] [CrossRef]
- Luger, S.M.; Sun, Z.; Loghavi, S.; Lazarus, H.M.; Rowe, J.M.; Tallman, M.S.; Pratz, K.W.; Litzow, M. Phase II Randomized Trial of Gilteritinib Vs Midostaurin in Newly Diagnosed FLT3 Mutated Acute Myeloid Leukemia (AML). Blood 2019, 134, 1309. [Google Scholar] [CrossRef]
- Daiichi Sankyo, Inc. A Phase 3, Double-Blind, Placebo-Controlled Study of Quizartinib Administered in Combination With Induction and Consolidation Chemotherapy, and Administered as Continuation Therapy in Subjects 18 to 75 Years Old With Newly Diagnosed FLT3-ITD (+) Acute Myeloid Leukemia (QuANTUM First). Available online: https://clinicaltrials.gov/ct2/show/NCT02668653 (accessed on 28 June 2020).
- Press Releases—Daiichi Sankyo, US. Available online: https://daiichisankyo.us/press-releases/-/article/364091/11880925 (accessed on 27 March 2022).
- Martelli, A.M.; Evangelisti, C.; Chiarini, F.; McCubrey, J.A. The Phosphatidylinositol 3-Kinase/Akt/MTOR Signaling Network as a Therapeutic Target in Acute Myelogenous Leukemia Patients. Oncotarget 2010, 1, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Faderl, S.; Pal, A.; Bornmann, W.; Albitar, M.; Maxwell, D.; Van, Q.; Peng, Z.; Harris, D.; Liu, Z.; Hazan-Halevy, I.; et al. Kit Inhibitor APcK110 Induces Apoptosis and Inhibits Proliferation of Acute Myeloid Leukemia Cells. Cancer Res. 2009, 69, 3910–3917. [Google Scholar] [CrossRef]
- Birkenkamp, K.U.; Geugien, M.; Schepers, H.; Westra, J.; Lemmink, H.H.; Vellenga, E. Constitutive NF-KappaB DNA-Binding Activity in AML Is Frequently Mediated by a Ras/PI3-K/PKB-Dependent Pathway. Leukemia 2004, 18, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Sujobert, P.; Bardet, V.; Cornillet-Lefebvre, P.; Hayflick, J.S.; Prie, N.; Verdier, F.; Vanhaesebroeck, B.; Muller, O.; Pesce, F.; Ifrah, N.; et al. Essential Role for the P110delta Isoform in Phosphoinositide 3-Kinase Activation and Cell Proliferation in Acute Myeloid Leukemia. Blood 2005, 106, 1063–1066. [Google Scholar] [CrossRef]
- Rizzieri, D.A.; Feldman, E.; Dipersio, J.F.; Gabrail, N.; Stock, W.; Strair, R.; Rivera, V.M.; Albitar, M.; Bedrosian, C.L.; Giles, F.J. A Phase 2 Clinical Trial of Deforolimus (AP23573, MK-8669), a Novel Mammalian Target of Rapamycin Inhibitor, in Patients with Relapsed or Refractory Hematologic Malignancies. Clin. Cancer Res. 2008, 14, 2756–2762. [Google Scholar] [CrossRef]
- Callera, F.; Lopes, C.O.; Rosa, E.S.; Mulin, C.C. Lack of Antileukemic Activity of Rapamycin in Elderly Patients with Acute Myeloid Leukemia Evolving from a Myelodysplastic Syndrome. Leuk. Res. 2008, 32, 1633–1634. [Google Scholar] [CrossRef]
- Chapuis, N.; Tamburini, J.; Green, A.S.; Vignon, C.; Bardet, V.; Neyret, A.; Pannetier, M.; Willems, L.; Park, S.; Macone, A.; et al. Dual Inhibition of PI3K and MTORC1/2 Signaling by NVP-BEZ235 as a New Therapeutic Strategy for Acute Myeloid Leukemia. Clin. Cancer Res. 2010, 16, 5424–5435. [Google Scholar] [CrossRef]
- Kampa-Schittenhelm, K.M.; Heinrich, M.C.; Akmut, F.; Rasp, K.H.; Illing, B.; Döhner, H.; Döhner, K.; Schittenhelm, M.M. Cell Cycle-Dependent Activity of the Novel Dual PI3K-MTORC1/2 Inhibitor NVP-BGT226 in Acute Leukemia. Mol. Cancer 2013, 12, 46. [Google Scholar] [CrossRef]
- Park, S.; Chapuis, N.; Bardet, V.; Tamburini, J.; Gallay, N.; Willems, L.; Knight, Z.A.; Shokat, K.M.; Azar, N.; Viguié, F.; et al. PI-103, a Dual Inhibitor of Class IA Phosphatidylinositide 3-Kinase and MTOR, Has Antileukemic Activity in AML. Leukemia 2008, 22, 1698–1706. [Google Scholar] [CrossRef]
- Ikezoe, T.; Kojima, S.; Furihata, M.; Yang, J.; Nishioka, C.; Takeuchi, A.; Isaka, M.; Koeffler, H.P.; Yokoyama, A. Expression of P-JAK2 Predicts Clinical Outcome and Is a Potential Molecular Target of Acute Myelogenous Leukemia. Int. J. Cancer 2011, 129, 2512–2521. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, S.; Bar-Natan, M.; Mascarenhas, J.O. JAKs to STATs: A Tantalizing Therapeutic Target in Acute Myeloid Leukemia. Blood Rev. 2020, 40, 100634. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Tsay, W.; Tang, J.-L.; Shen, H.-L.; Lin, S.-W.; Huang, S.-Y.; Yao, M.; Chen, Y.-C.; Shen, M.-C.; Wang, C.-H.; et al. SOCS1 Methylation in Patients with Newly Diagnosed Acute Myeloid Leukemia. Genes Chromosomes Cancer 2003, 37, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Pemmaraju, N.; Kantarjian, H.; Kadia, T.; Cortes, J.; Borthakur, G.; Newberry, K.; Garcia-Manero, G.; Ravandi, F.; Jabbour, E.; Dellasala, S.; et al. A Phase I/II Study of the Janus Kinase (JAK)1 and 2 Inhibitor Ruxolitinib in Patients with Relapsed or Refractory Acute Myeloid Leukemia. Clin. Lymphoma Myeloma Leuk. 2015, 15, 171–176. [Google Scholar] [CrossRef]
- Rampal, R.K.; Mascarenhas, J.; Kosiorek, H.E.; Bhave, R.; Hexner, E.O.; Wang, E.S.; Gerds, A.T.; Heaney, M.L.; Abboud, C.N.; Kremyanskaya, M.; et al. Efficacy of Combined Ruxolitinib and Decitabine in Patients with Accelerated and Blast-Phase Myeloproliferative Neoplasms: Results of a Phase II Study (MPN-RC 109 Trial). Blood 2018, 132, 3027. [Google Scholar] [CrossRef]
- Levis, M.; Ravandi, F.; Wang, E.S.; Baer, M.R.; Perl, A.; Coutre, S.; Erba, H.; Stuart, R.K.; Baccarani, M.; Cripe, L.D.; et al. Results from a Randomized Trial of Salvage Chemotherapy Followed by Lestaurtinib for Patients with FLT3 Mutant AML in First Relapse. Blood 2011, 117, 3294–3301. [Google Scholar] [CrossRef]
- Verstovsek, S.; Odenike, O.; Singer, J.W.; Granston, T.; Al-Fayoumi, S.; Deeg, H.J. Phase 1/2 Study of Pacritinib, a next Generation JAK2/FLT3 Inhibitor, in Myelofibrosis or Other Myeloid Malignancies. J. Hematol. Oncol. 2016, 9, 137. [Google Scholar] [CrossRef]
- Ricciardi, M.R.; McQueen, T.; Chism, D.; Milella, M.; Estey, E.; Kaldjian, E.; Sebolt-Leopold, J.; Konopleva, M.; Andreeff, M. Quantitative Single Cell Determination of ERK Phosphorylation and Regulation in Relapsed and Refractory Primary Acute Myeloid Leukemia. Leukemia 2005, 19, 1543–1549. [Google Scholar] [CrossRef]
- Jain, N.; Curran, E.; Iyengar, N.M.; Diaz-Flores, E.; Kunnavakkam, R.; Popplewell, L.; Kirschbaum, M.H.; Karrison, T.; Erba, H.P.; Green, M.; et al. Phase II Study of the Oral MEK Inhibitor Selumetinib in Advanced Acute Myelogenous Leukemia: A University of Chicago Phase II Consortium Trial. Clin. Cancer Res. 2014, 20, 490–498. [Google Scholar] [CrossRef]
- Borthakur, G.; Popplewell, L.; Boyiadzis, M.; Foran, J.; Platzbecker, U.; Vey, N.; Walter, R.B.; Olin, R.; Raza, A.; Giagounidis, A.; et al. Activity of the Oral Mitogen-Activated Protein Kinase Kinase Inhibitor Trametinib in RAS-Mutant Relapsed or Refractory Myeloid Malignancies. Cancer 2016, 122, 1871–1879. [Google Scholar] [CrossRef]
- Maiti, A.; Naqvi, K.; Kadia, T.M.; Borthakur, G.; Takahashi, K.; Bose, P.; Daver, N.G.; Patel, A.; Alvarado, Y.; Ohanian, M.; et al. Phase II Trial of MEK Inhibitor Binimetinib (MEK162) in RAS-Mutant Acute Myeloid Leukemia. Clin. Lymphoma Myeloma Leuk. 2019, 19, 142–148.e1. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-B.; Henry, J.R.; Kaufman, M.D.; Lu, W.-P.; Smith, B.D.; Vogeti, S.; Rutkoski, T.J.; Wise, S.; Chun, L.; Zhang, Y.; et al. Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-Tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell 2015, 28, 384–398. [Google Scholar] [CrossRef]
- Müller, E.; Bauer, S.; Stühmer, T.; Mottok, A.; Scholz, C.-J.; Steinbrunn, T.; Brünnert, D.; Brandl, A.; Schraud, H.; Kreßmann, S.; et al. Pan-Raf Co-Operates with PI3K-Dependent Signalling and Critically Contributes to Myeloma Cell Survival Independently of Mutated RAS. Leukemia 2017, 31, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Tambe, M.; Karjalainen, E.; Vähä-Koskela, M.; Bulanova, D.; Gjertsen, B.T.; Kontro, M.; Porkka, K.; Heckman, C.A.; Wennerberg, K. Pan-RAF Inhibition Induces Apoptosis in Acute Myeloid Leukemia Cells and Synergizes with BCL2 Inhibition. Leukemia 2020, 34, 3186–3196. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Evans, K.; Gadrey, J.Y.; Eschle, B.K.; Hatton, C.; Uckelmann, H.J.; Ross, K.N.; Perner, F.; Olsen, S.N.; Pritchard, T.; et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 2019, 36, 660–673.e11. [Google Scholar] [CrossRef]
- Klossowski, S.; Miao, H.; Kempinska, K.; Wu, T.; Purohit, T.; Kim, E.; Linhares, B.M.; Chen, D.; Jih, G.; Perkey, E.; et al. Menin Inhibitor MI-3454 Induces Remission in MLL1-Rearranged and NPM1-Mutated Models of Leukemia. J. Clin. Investig. 2020, 130, 981–997. [Google Scholar] [CrossRef]
- Fiskus, W.; Boettcher, S.; Daver, N.; Mill, C.P.; Sasaki, K.; Birdwell, C.E.; Davis, J.A.; Takahashi, K.; Kadia, T.M.; DiNardo, C.D.; et al. Effective Menin Inhibitor-Based Combinations against AML with MLL Rearrangement or NPM1 Mutation (NPM1c). Blood Cancer J. 2022, 12, 5. [Google Scholar] [CrossRef]
- Dzama, M.M.; Steiner, M.; Rausch, J.; Sasca, D.; Schönfeld, J.; Kunz, K.; Taubert, M.C.; McGeehan, G.M.; Chen, C.-W.; Mupo, A.; et al. Synergistic Targeting of FLT3 Mutations in AML via Combined Menin-MLL and FLT3 Inhibition. Blood 2020, 136, 2442–2456. [Google Scholar] [CrossRef]
- Stein, E. Safety and Efficacy of Menin Inhibition in Patients (Pts) with MLL-Rearranged and NPM1 Mutant Acute Leukemia: A Phase (Ph) 1, First-in-Human Study of SNDX-5613 (AUGMENT 101). Blood 2021, 138, 699. [Google Scholar] [CrossRef]
- Bykov, V.J.N.; Issaeva, N.; Zache, N.; Shilov, A.; Hultcrantz, M.; Bergman, J.; Selivanova, G.; Wiman, K.G. Reactivation of Mutant P53 and Induction of Apoptosis in Human Tumor Cells by Maleimide Analogs *. J. Biol. Chem. 2005, 280, 30384–30391. [Google Scholar] [CrossRef]
- Bykov, V.J.N.; Zache, N.; Stridh, H.; Westman, J.; Bergman, J.; Selivanova, G.; Wiman, K.G. PRIMA-1MET Synergizes with Cisplatin to Induce Tumor Cell Apoptosis. Oncogene 2005, 24, 3484–3491. [Google Scholar] [CrossRef]
- Lambert, J.M.R.; Gorzov, P.; Veprintsev, D.B.; Söderqvist, M.; Segerbäck, D.; Bergman, J.; Fersht, A.R.; Hainaut, P.; Wiman, K.G.; Bykov, V.J.N. PRIMA-1 Reactivates Mutant P53 by Covalent Binding to the Core Domain. Cancer Cell 2009, 15, 376–388. [Google Scholar] [CrossRef]
- Sallman, D.A.; DeZern, A.E.; Garcia-Manero, G.; Steensma, D.P.; Roboz, G.J.; Sekeres, M.A.; Cluzeau, T.; Sweet, K.L.; McLemore, A.; McGraw, K.L.; et al. Eprenetapopt (APR-246) and Azacitidine in TP53-Mutant Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1584–1594. [Google Scholar] [CrossRef] [PubMed]
- Aprea Therapeutics. A Phase III Multicenter, Randomized, Open Label Study of APR-246 in Combination with Azacitidine Versus Azacitidine Alone for the Treatment of (Tumor Protein) TP53 Mutant Myelodysplastic Syndromes. Available online: https://clinicaltrials.gov/ct2/show/NCT03745716 (accessed on 7 October 2021).
- Aprea Therapeutics. Phase 1 Study to Evaluate Safety and Efficacy of APR-548 in Combination with Azacitidine for the Treatment of TP53-Mutant Myelodysplastic Syndromes. Available online: https://clinicaltrials.gov/ct2/show/NCT04638309 (accessed on 4 November 2021).
- Moll, U.M.; Petrenko, O. The MDM2-P53 Interaction. Mol. Cancer Res. 2003, 1, 1001–1008. [Google Scholar]
- Lehmann, C.; Friess, T.; Birzele, F.; Kiialainen, A.; Dangl, M. Superior Anti-Tumor Activity of the MDM2 Antagonist Idasanutlin and the Bcl-2 Inhibitor Venetoclax in P53 Wild-Type Acute Myeloid Leukemia Models. J. Hematol. Oncol. 2016, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Andreeff, M.; Kelly, K.R.; Yee, K.; Assouline, S.; Strair, R.; Popplewell, L.; Bowen, D.; Martinelli, G.; Drummond, M.W.; Vyas, P.; et al. Results of the Phase 1 Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin. Cancer Res. 2016, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Grob, T.; Al Hinai, A.S.A.; Sanders, M.A.; Kavelaars, F.G.; Rijken, M.; Gradowska, P.L.; Biemond, B.J.; Breems, D.A.; Maertens, J.; van Marwijk Kooy, M.; et al. Molecular Characterization of Mutant TP53 Acute Myeloid Leukemia and High-Risk Myelodysplastic Syndrome. Blood 2022, 139, 2347–2354. [Google Scholar] [CrossRef] [PubMed]
- Traina, F.; Visconte, V.; Elson, P.; Tabarroki, A.; Jankowska, A.M.; Hasrouni, E.; Sugimoto, Y.; Szpurka, H.; Makishima, H.; O’Keefe, C.L.; et al. Impact of Molecular Mutations on Treatment Response to DNMT Inhibitors in Myelodysplasia and Related Neoplasms. Leukemia 2014, 28, 78–87. [Google Scholar] [CrossRef] [PubMed]
- R. Gorbacheva Memorial Institute of Children Oncology, Hematology and Transplantation, First Pavlov State Medical University of St. Petersburg, St. Petersburg, Russia; Tcvetkov, N.U.; Epifanovskaya, O.S.; Rudnitskaya, Y.V.; Morozova, E.V.; Moiseev, I.S.; Afanasyev, B.V. Meta-Analysis of Studies with Genome Sequencing in Myelodysplastic Syndrome Treated with Hypomethylating Agents. CTT 2018, 7, 44–51. [Google Scholar] [CrossRef][Green Version]
- Barclay, A.N.; Brown, M.H. The SIRP Family of Receptors and Immune Regulation. Nat. Rev. Immunol. 2006, 6, 457–464. [Google Scholar] [CrossRef]
- Latour, S.; Tanaka, H.; Demeure, C.; Mateo, V.; Rubio, M.; Brown, E.J.; Maliszewski, C.; Lindberg, F.P.; Oldenborg, A.; Ullrich, A.; et al. Bidirectional Negative Regulation of Human T and Dendritic Cells by CD47 and Its Cognate Receptor Signal-Regulator Protein-α: Down-Regulation of IL-12 Responsiveness and Inhibition of Dendritic Cell Activation. J. Immunol. 2001, 167, 2547–2554. [Google Scholar] [CrossRef] [PubMed]
- Oldenborg, P.-A.; Gresham, H.D.; Lindberg, F.P. Cd47-Signal Regulatory Protein α (Sirpα) Regulates Fcγ and Complement Receptor–Mediated Phagocytosis. J. Exp. Med. 2001, 193, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Majeti, R.; Chao, M.P.; Alizadeh, A.A.; Pang, W.W.; Jaiswal, S.; Gibbs, K.D.; van Rooijen, N.; Weissman, I.L. CD47 Is an Adverse Prognostic Factor and Therapeutic Antibody Target on Human Acute Myeloid Leukemia Stem Cells. Cell 2009, 138, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D. The First-in-Class Anti-CD47 Antibody Magrolimab Combined with Azacitidine Is Well-Tolerated and Effective in AML Patients: Phase 1b Results. Blood 2020, 21, S213. [Google Scholar]
- Gilead Sciences. A Phase 2 Multi-Arm Study of Magrolimab Combinations in Patients with Myeloid Malignancies. Available online: https://clinicaltrials.gov/ct2/show/NCT04778410 (accessed on 23 March 2022).

| Risk Category | Genetic Abnormality |
|---|---|
| Favorable | t(8;21)(q22;q22.1); RUNX1-RUNX1T1 |
| inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB-MYH11 | |
| Mutated NPM1 without FLT3-ITD or with FLT3-ITD low | |
| Biallelic-mutated CEBPA | |
| Intermediate | Mutated NPM1 and FLT3-ITD high |
| Wild-type NPM1 without FLT3-ITD or with FLT3-ITD low (without adverse-risk genetic lesions) | |
| t(9;11)(p21.3;q23.3); MLLT3-KMT2A | |
| Cytogenetic abnormalities not classified as favorable or adverse | |
| Adverse | t(6;9)(p23;q34.1); DEK-NUP214 |
| t(v;11q23.3); KMT2A rearranged | |
| t(9;22)(q34.1;q11.2); BCR-ABL1 | |
| inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2,MECOM(EVI1) | |
| −5 or del(5q); −7; −17/abn(17p) | |
| Complex karyotype, monosomal karyotype | |
| Wild-type NPM1 and FLT3-ITD high | |
| Mutated RUNX1 | |
| Mutated ASXL1 | |
| Mutated TP53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, D.; Chi, S.; Uchiyama, S.; Nakamura, H.; Guo, Y.-M.; Yamauchi, N.; Yuda, J.; Minami, Y. Molecular Classification and Overcoming Therapy Resistance for Acute Myeloid Leukemia with Adverse Genetic Factors. Int. J. Mol. Sci. 2022, 23, 5950. https://doi.org/10.3390/ijms23115950
Ikeda D, Chi S, Uchiyama S, Nakamura H, Guo Y-M, Yamauchi N, Yuda J, Minami Y. Molecular Classification and Overcoming Therapy Resistance for Acute Myeloid Leukemia with Adverse Genetic Factors. International Journal of Molecular Sciences. 2022; 23(11):5950. https://doi.org/10.3390/ijms23115950
Chicago/Turabian StyleIkeda, Daisuke, SungGi Chi, Satoshi Uchiyama, Hirotaka Nakamura, Yong-Mei Guo, Nobuhiko Yamauchi, Junichiro Yuda, and Yosuke Minami. 2022. "Molecular Classification and Overcoming Therapy Resistance for Acute Myeloid Leukemia with Adverse Genetic Factors" International Journal of Molecular Sciences 23, no. 11: 5950. https://doi.org/10.3390/ijms23115950
APA StyleIkeda, D., Chi, S., Uchiyama, S., Nakamura, H., Guo, Y.-M., Yamauchi, N., Yuda, J., & Minami, Y. (2022). Molecular Classification and Overcoming Therapy Resistance for Acute Myeloid Leukemia with Adverse Genetic Factors. International Journal of Molecular Sciences, 23(11), 5950. https://doi.org/10.3390/ijms23115950






