Bioprospecting for Thermozymes and Characterization of a Novel Lipolytic Thermozyme Belonging to the SGNH/GDSL Family of Hydrolases
Abstract
:1. Introduction
2. Results
2.1. Water Sampling
2.2. Library Construction
2.3. Functional Screening
2.3.1. Screening for Plant Cell Wall Degrading Enzymes
2.3.2. Screening for Proteases
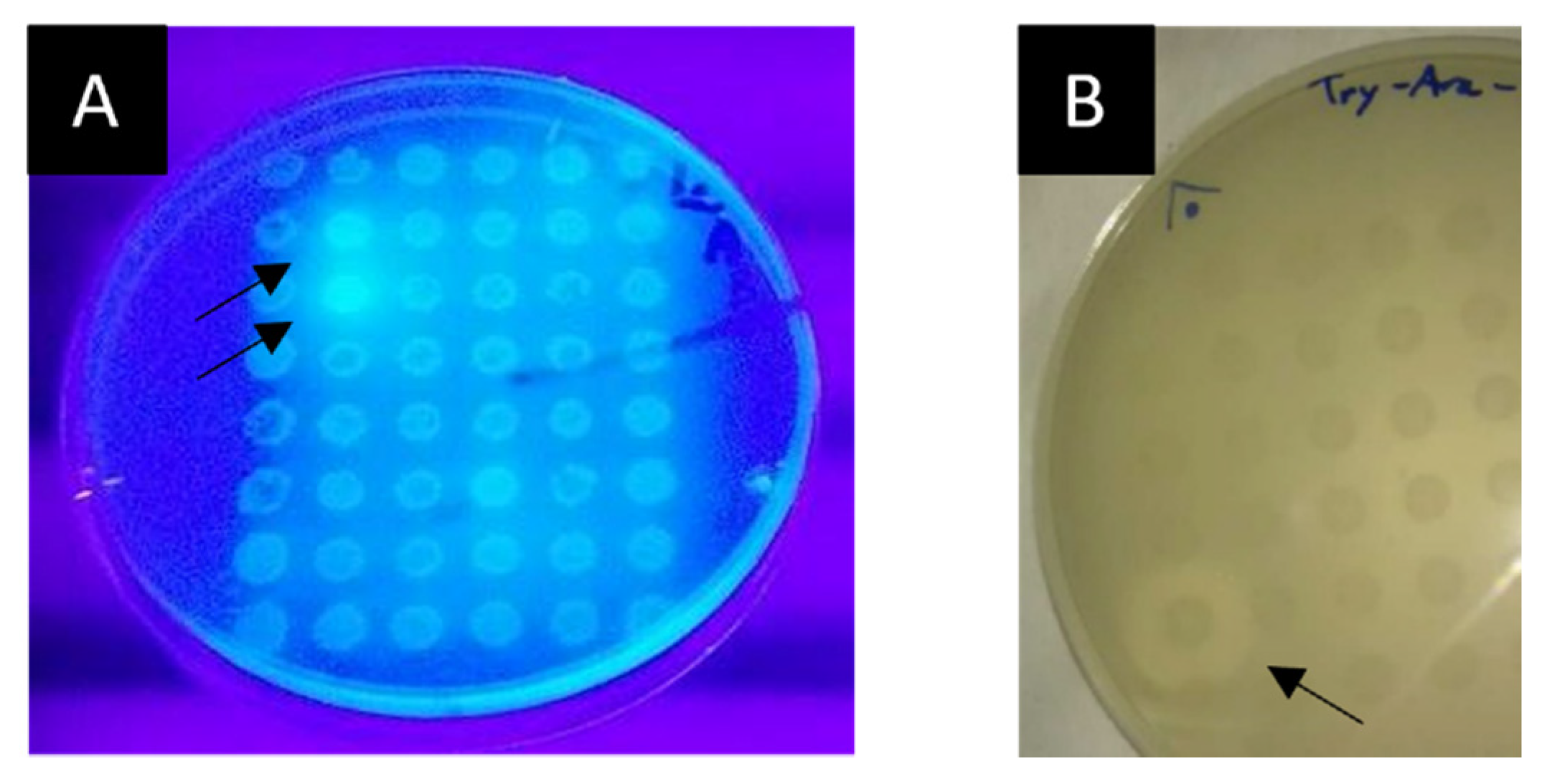
2.3.3. Screening for Lipolytic and Feruloyl Esterase Activities
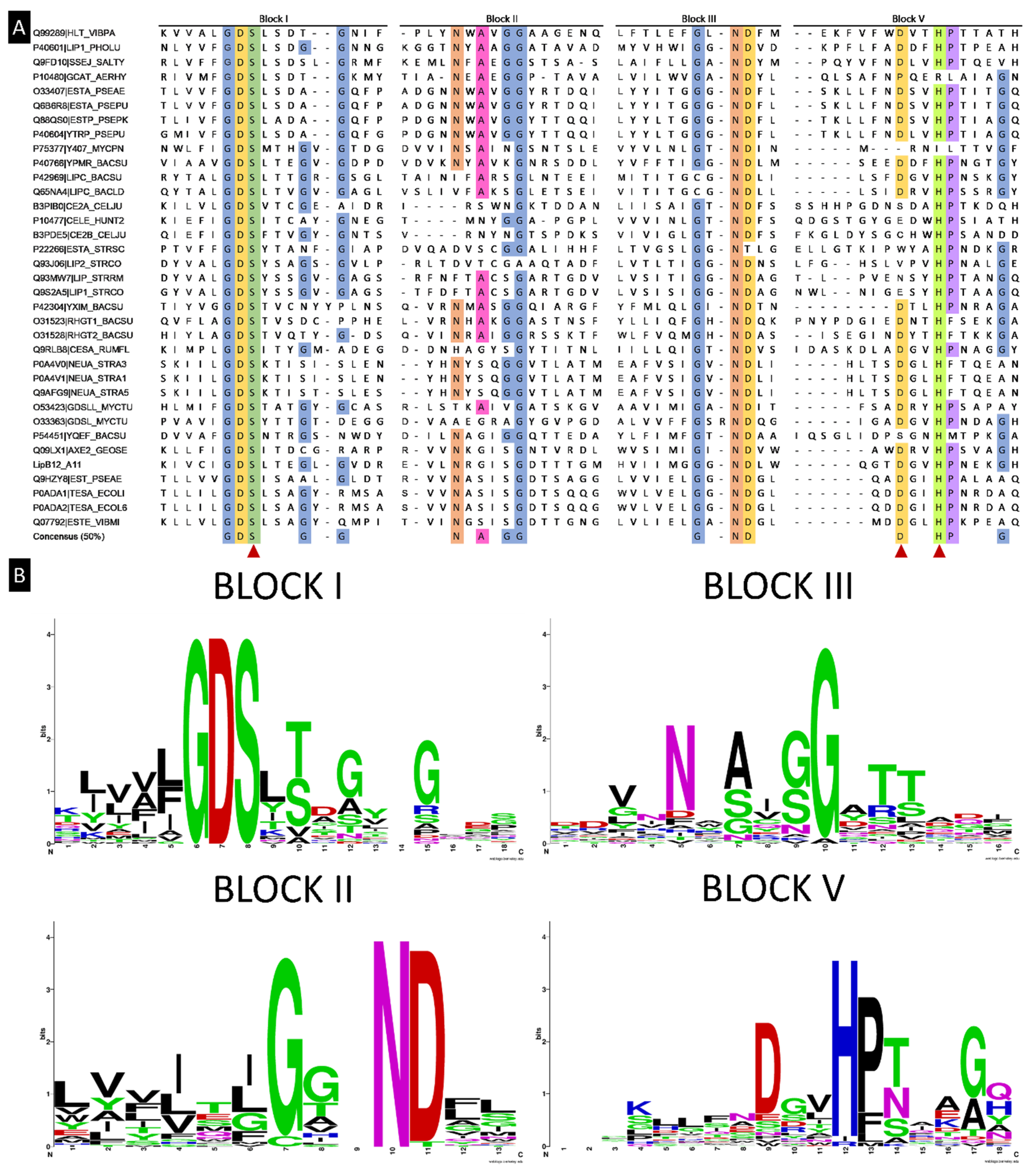
2.4. Subcloning of Selected Positive Lypolitic Activity-Conferring Genes and Sequence Analysis
2.5. Alignment of LipB12_A11 with NCBI Databases for DNA and Protein Sequences
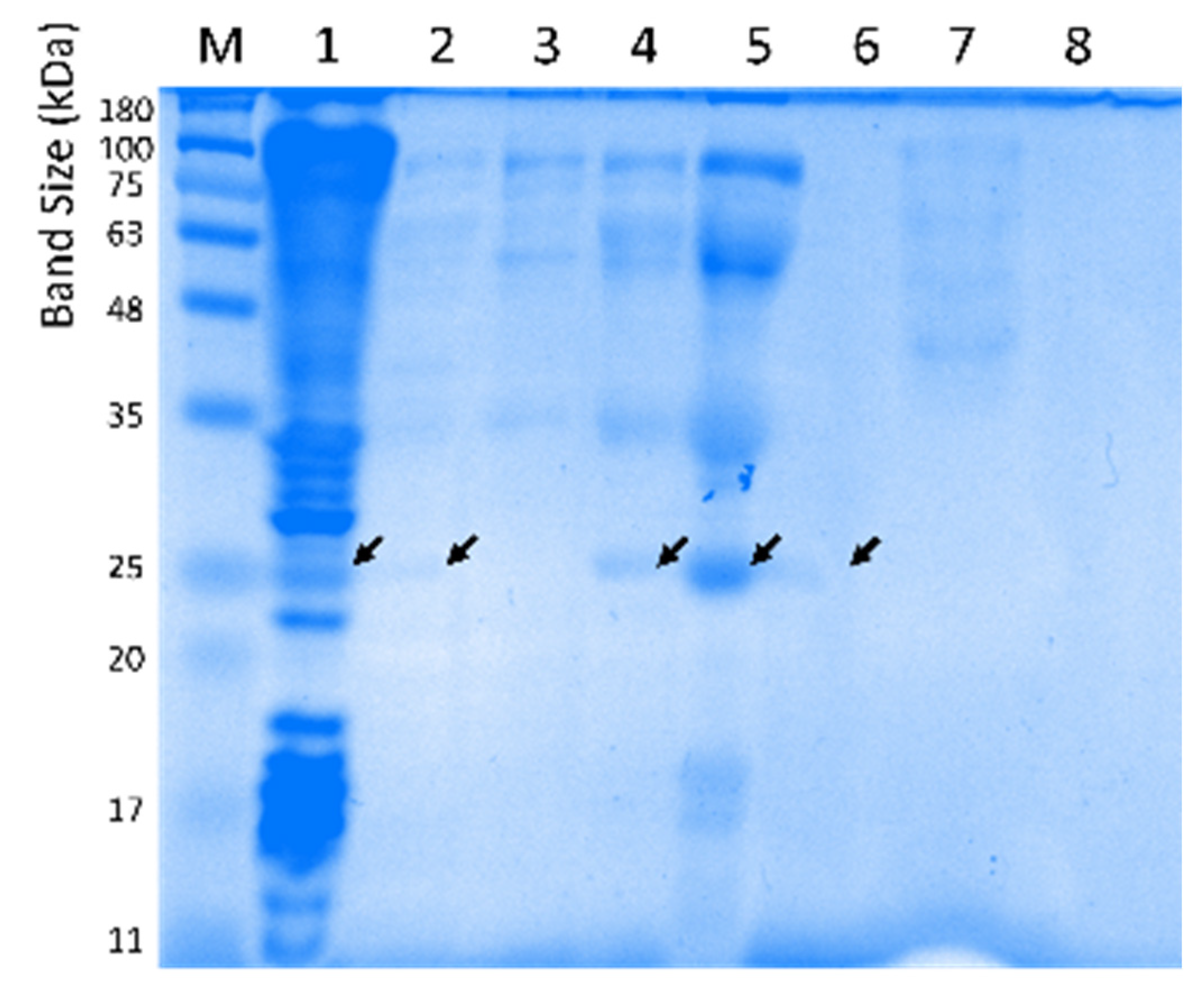
2.6. Purification and Biochemical Characterization of Lipolytic Activity Enzyme LipB12_A11
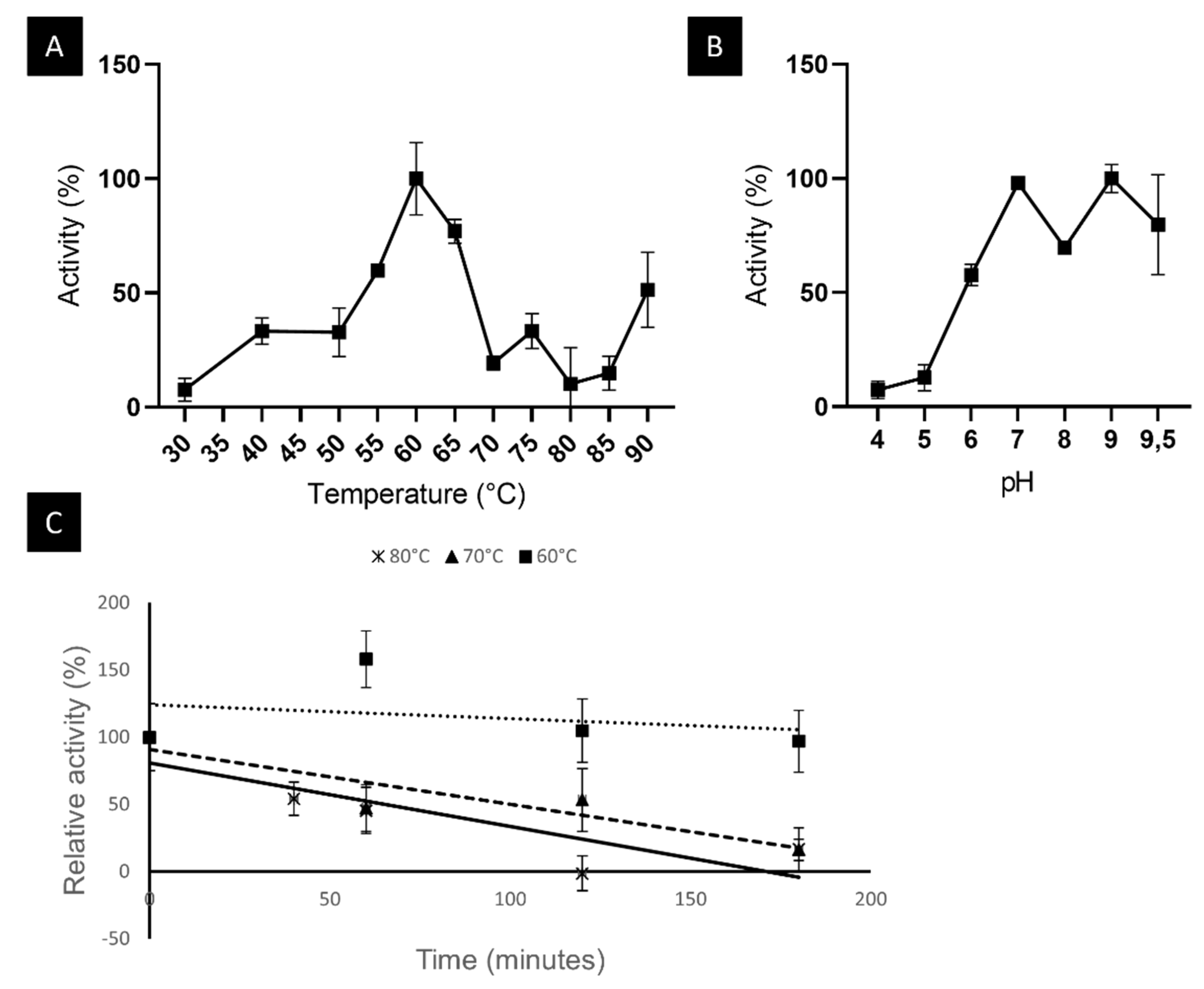
2.6.1. LipB12_A11 Temperature and pH Optima and Thermostability
2.6.2. LipB12_A11 Substrate Preference
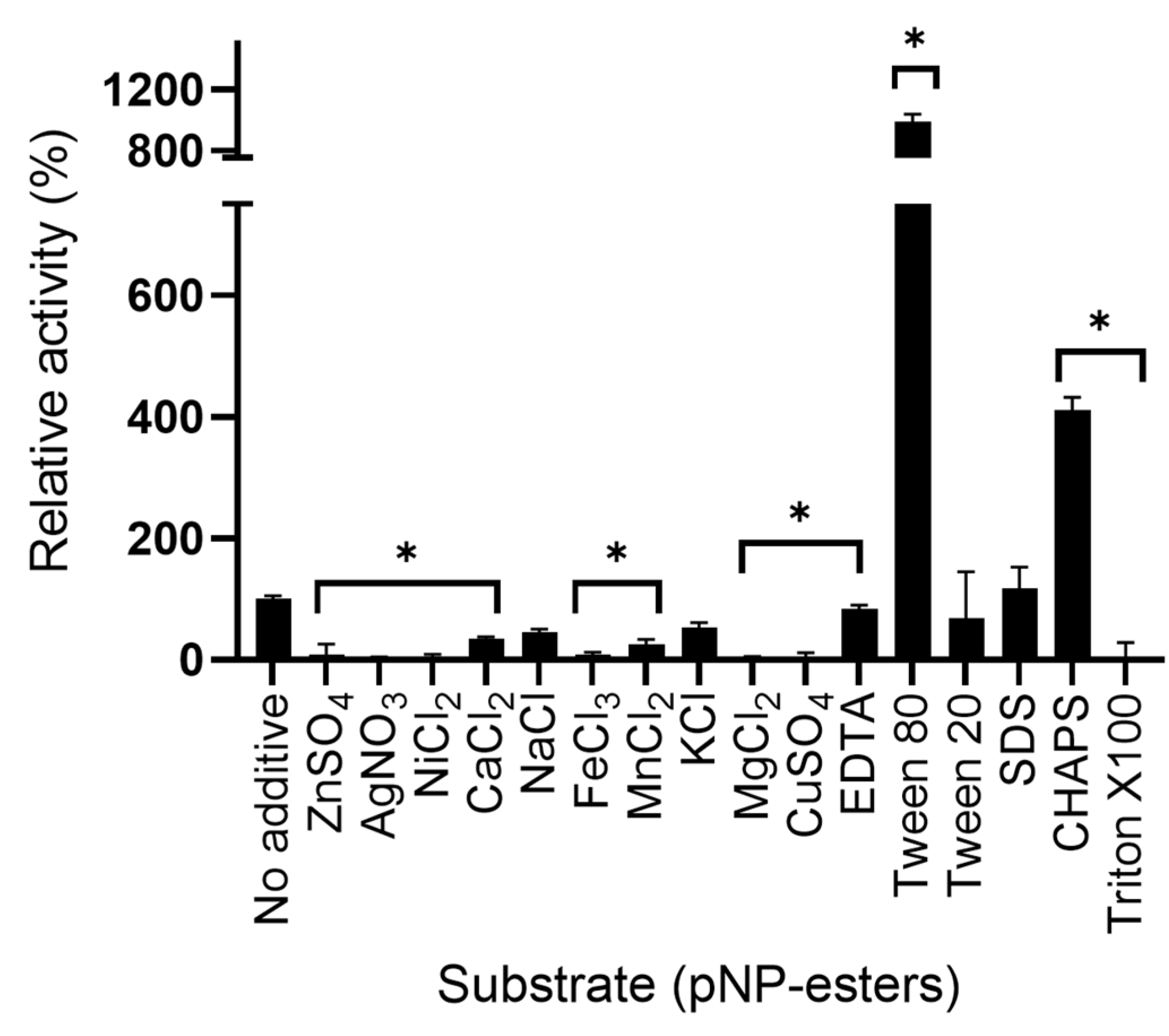
2.6.3. Effect of Additives and Detergents on LipB12_A11 Enzymatic Activity
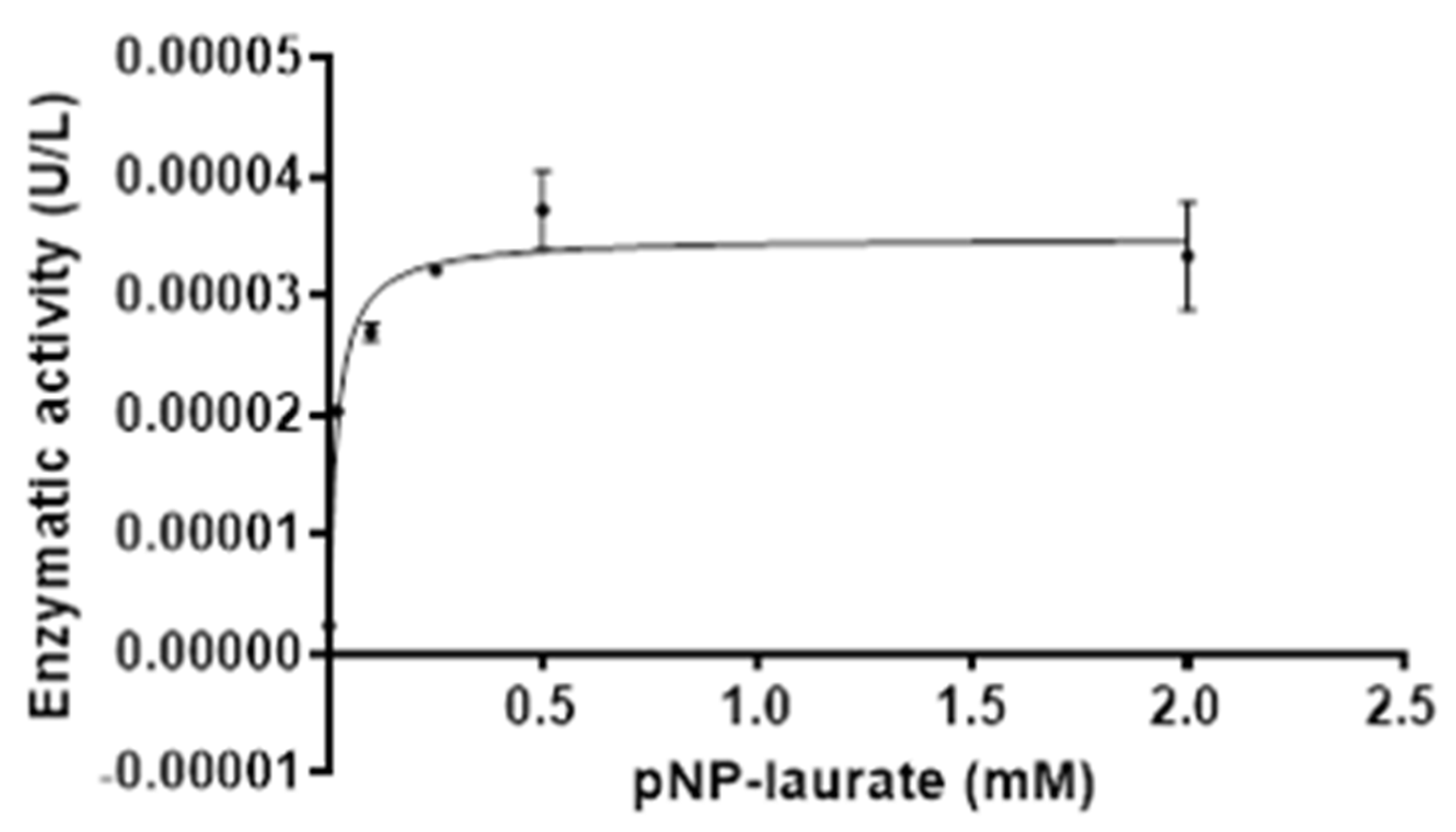
2.6.4. LipB12_A11 Enzyme Kinetics
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. DNA Extraction
4.3. Metagenomic Libraries Construction
4.3.1. End Repair and Size Selection of Metagenomic DNA
4.3.2. Vector
4.3.3. Ligation and Packaging of Metagenomic DNA
4.4. Functional Screening Methods
4.4.1. Cellulase Screening
4.4.2. Xylanase Screening
4.4.3. β-Xylosidase Screening
4.4.4. Lipolytic Activity Screening
4.4.5. Feruloyl Esterase Screening
4.4.6. β-Glucosidase Screening
4.4.7. Protease Screening
4.5. Subcloning of a DNA Fragment Containing a Lipolytic Activity-Conferring Gene
4.6. Sequencing and Primer Walking of a DNA Fragment Containing a Lipolytic Activity-Conferring Gene
4.7. Sequence Analysis
4.8. Purification
4.9. Activity Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Felczykowska, A.; Krajewska, A.; Zielińska, S.; Łoś, J.M.; Bloch, S.K.; Nejman-Faleńczyk, B. The Most Widespread Problems in the Function-Based Microbial Metagenomics. Acta Biochim. Pol. 2015, 62, 161–166. [Google Scholar] [CrossRef]
- Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Becerra, M.; Rodríguez-Belmonte, E.; González-Siso, M.-I. Advances of Functional Metagenomics in Harnessing Thermozymes. In Metagenomics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 289–307. ISBN 9780128134030. [Google Scholar]
- Ferrer, M.; Golyshina, O.; Beloqui, A.; Golyshin, P.N. Mining Enzymes from Extreme Environments. Curr. Opin. Microbiol. 2007, 10, 207–214. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Kadnikov, V.V.; Ravin, N.V. Metagenomics: A Paradigm Shift in Microbiology. In Metagenomics; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1–13. ISBN 9780128134030. [Google Scholar]
- Armstrong, Z.; Rahfeld, P.; Withers, S.G. Discovery of New Glycosidases from Metagenomic Libraries. In Methods in Enzymology; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 597, pp. 3–23. [Google Scholar]
- DeCastro, M.-E.; Rodríguez-Belmonte, E.; González-Siso, M.-I. Metagenomics of Thermophiles with a Focus on Discovery of Novel Thermozymes. Front. Microbiol. 2016, 7, 1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guazzaroni, M.; Silva-Rocha, R.; Ward, R.J. Synthetic Biology Approaches to Improve Biocatalyst Identification in Metagenomic Library Screening. Microb. Biotechnol. 2015, 8, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Taupp, M.; Mewis, K.; Hallam, S.J. The Art and Design of Functional Metagenomic Screens. Curr. Opin. Biotechnol. 2011, 22, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel Enzymes from Non-Culturable Microbes. FEMS Microbiol. Lett. 2017, 364, fnx211. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the Success of Enzyme Bioprospecting through Metagenomics: Current Status and Future Trends. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar] [CrossRef]
- Ekkers, D.M.; Cretoiu, M.S.; Kielak, A.M.; van Elsas, J.D. The Great Screen Anomaly—A New Frontier in Product Discovery through Functional Metagenomics. Appl. Microbiol. Biotechnol. 2012, 93, 1005–1020. [Google Scholar] [CrossRef] [Green Version]
- an der Helm, E.; Genee, H.J.; Sommer, M.O.A. The Evolving Interface between Synthetic Biology and Functional Metagenomics. Nat. Chem. Biol. 2018, 14, 752–759. [Google Scholar] [CrossRef]
- Uchiyama, T.; Miyazaki, K. Functional Metagenomics for Enzyme Discovery: Challenges to Efficient Screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef]
- Craig, J.W.; Chang, F.-Y.; Kim, J.H.; Obiajulu, S.C.; Brady, S.F. Expanding Small-Molecule Functional Metagenomics through Parallel Screening of Broad-Host-Range Cosmid Environmental DNA Libraries in Diverse Proteobacteria. Appl. Environ. Microbiol. 2010, 76, 1633–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, P.; Eck, J. Metagenomics and Industrial Applications. Nat. Rev. Microbiol. 2005, 3, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Gabor, E.M.; Alkema, W.B.L.; Janssen, D.B. Quantifying the Accessibility of the Metagenome by Random Expression Cloning Techniques. Environ. Microbiol. 2004, 6, 879–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, J.-P.; Reymond, J.-L. Recent Advances in Enzyme Assays. Trends Biotechnol. 2004, 22, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Demirjian, D.C.; Morıs-Varas, F.; Cassidy, C.S. Enzymes from Extremophiles. Curr. Opin. Chem. Biol. 2001, 5, 144–151. [Google Scholar] [CrossRef]
- Haki, G.; Rakshit, S. Developments in Industrially Important Thermostable Enzymes: A Review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Bruins, M.E.; Janssen, A.E.M.; Boom, R.M. Thermozymes and Their Applications: A Review of Recent Literature and Patents. Appl. Biochem. Biotechnol. 2001, 90, 155–186. [Google Scholar] [CrossRef]
- Kumar, S.; Dangi, A.K.; Shukla, P.; Baishya, D.; Khare, S.K. Thermozymes: Adaptive Strategies and Tools for Their Biotechnological Applications. Bioresour. Technol. 2019, 278, 372–382. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Jiang, X.-R. Next Generation Industrial Biotechnology Based on Extremophilic Bacteria. Curr. Opin. Biotechnol. 2018, 50, 94–100. [Google Scholar] [CrossRef]
- Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Rodríguez-Belmonte, E.; Becerra, M.; María-Isabel, G.-S. Microbiomes of Extreme Environments. In Microbiomes of Extreme Environments: Biodiversity and Biotechnological Applications; Yadav, A.N., Rastegari, A.A., Yadav, N., Eds.; CRC Press: Boca Raton, CA, USA, 2021; pp. 87–105. ISBN 9780429328633. [Google Scholar]
- Prayogo, F.A.; Budiharjo, A.; Kusumaningrum, H.P.; Wijanarka, W.; Suprihadi, A.; Nurhayati, N. Metagenomic Applications in Exploration and Development of Novel Enzymes from Nature: A Review. J. Genet. Eng. Biotechnol. 2020, 18, 39. [Google Scholar] [CrossRef]
- Escuder-Rodríguez, J.-J.; López-López, O.; Becerra, M.; Cerdán, M.-E.; González-Siso, M.-I. Extremophilic Esterases for Bioprocessing of Lignocellulosic Feedstocks. In Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy; Springer International Publishing: Cham, Switzerland, 2017; pp. 205–223. ISBN 9783319546841. [Google Scholar]
- Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Cerdán, M.-E.; Rodríguez-Belmonte, E.; Becerra, M.; González-Siso, M.-I. Cellulases from Thermophiles Found by Metagenomics. Microorganisms 2018, 6, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juturu, V.; Wu, J.C. Insight into Microbial Hemicellulases Other than Xylanases: A Review. J. Chem. Technol. Biotechnol. 2013, 88, 353–363. [Google Scholar] [CrossRef]
- DeCastro, M.-E.; Escuder-Rodriguez, J.-J.; Cerdan, M.-E.; Becerra, M.; Rodriguez-Belmonte, E.; Gonzalez-Siso, M.-I. Heat-Loving β-Galactosidases from Cultured and Uncultured Microorganisms. Curr. Protein Pept. Sci. 2018, 19, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Barcelos, M.C.S.; Ramos, C.L.; Kuddus, M.; Rodriguez-Couto, S.; Srivastava, N.; Ramteke, P.W.; Mishra, P.K.; Molina, G. Enzymatic Potential for the Valorization of Agro-Industrial by-Products. Biotechnol. Lett. 2020, 42, 1799–1827. [Google Scholar] [CrossRef]
- Elleuche, S.; Schröder, C.; Sahm, K.; Antranikian, G. Extremozymes—Biocatalysts with Unique Properties from Extremophilic Microorganisms. Curr. Opin. Biotechnol. 2014, 29, 116–123. [Google Scholar] [CrossRef]
- Wohlgemuth, R.; Littlechild, J.; Monti, D.; Schnorr, K.; van Rossum, T.; Siebers, B.; Menzel, P.; Kublanov, I.V.; Rike, A.G.; Skretas, G.; et al. Discovering Novel Hydrolases from Hot Environments. Biotechnol. Adv. 2018, 36, 2077–2100. [Google Scholar] [CrossRef] [Green Version]
- Dadwal, A.; Sharma, S.; Satyanarayana, T. Progress in Ameliorating Beneficial Characteristics of Microbial Cellulases by Genetic Engineering Approaches for Cellulose Saccharification. Front. Microbiol. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Sadhu, S.; Maiti, T.K. Cellulase Production by Bacteria: A Review. Br. Microbiol. Res. J. 2013, 3, 235–258. [Google Scholar] [CrossRef] [Green Version]
- Urbieta, M.S.; Donati, E.R.; Chan, K.-G.; Shahar, S.; Sin, L.L.; Goh, K.M. Thermophiles in the Genomic Era: Biodiversity, Science, and Applications. Biotechnol. Adv. 2015, 33, 633–647. [Google Scholar] [CrossRef]
- Delgado-Outeiriño, I.; Araujo-Nespereira, P.; Cid-Fernández, J.A.; Mejuto, J.C.; Martínez-Carballo, E.; Simal-Gándara, J. Behaviour of Thermal Waters through Granite Rocks Based on Residence Time and Inorganic Pattern. J. Hydrol. 2009, 373, 329–336. [Google Scholar] [CrossRef]
- López, D.L.; Araujo, P.A.; Outeiriño, I.D.; Cid, J.A.; Astray, G. Geochemical Signatures of the Groundwaters from Ourense Thermal Springs, Galicia, Spain. Sustain. Water Resour. Manag. 2019, 5, 103–116. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.-C.; Liaw, Y.-C.; Huang, T.-H.; Shaw, J.-F. GDSL Family of Serine Esterases/Lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Alalouf, O.; Balazs, Y.; Volkinshtein, M.; Grimpel, Y.; Shoham, G.; Shoham, Y. A New Family of Carbohydrate Esterases Is Represented by a GDSL Hydrolase/Acetylxylan Esterase from Geobacillus Stearothermophilus. J. Biol. Chem. 2011, 286, 41993–42001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.; Hazlewood, G.P.; Barker, P.J.; Gilbert, H.J. Conserved Reiterated Domains in Clostridium Thermocellum Endoglucanases Are Not Essential for Catalytic Activity. Gene 1988, 69, 29–38. [Google Scholar] [CrossRef]
- Montanier, C.; Money, V.A.; Pires, V.M.R.; Flint, J.E.; Pinheiro, B.A.; Goyal, A.; Prates, J.A.M.; Izumi, A.; Stålbrand, H.; Morland, C.; et al. The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions. PLoS Biol. 2009, 7, e1000071. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhang, Y.; Shen, T.; Xie, Y.; Mao, Y.; Ji, C. Cloning, Expression and Biochemical Characterization of a Novel, Moderately Thermostable GDSL Family Esterase from Geobacillus Thermodenitrificans T2. J. Biosci. Bioeng. 2013, 115, 133–137. [Google Scholar] [CrossRef]
- Soni, S.; Sathe, S.S.; Odaneth, A.A.; Lali, A.M.; Chandrayan, S.K. SGNH Hydrolase-Type Esterase Domain Containing Cbes-AcXE2: A Novel and Thermostable Acetyl Xylan Esterase from Caldicellulosiruptor Bescii. Extremophiles 2017, 21, 687–697. [Google Scholar] [CrossRef]
- Jo, E.; Kim, J.; Lee, A.; Moon, K.; Cha, J. Identification and Characterization of a Novel Thermostable GDSL-Type Lipase from Geobacillus Thermocatenulatus. J. Microbiol. Biotechnol. 2021, 31, 483–491. [Google Scholar] [CrossRef]
- Chahiniana, H.; Sarda, L. Distinction Between Esterases and Lipases: Comparative Biochemical Properties of Sequence-Related Carboxylesterases. Protein Pept. Lett. 2009, 16, 1149–1161. [Google Scholar] [CrossRef]
- Fojan, P. What Distinguishes an Esterase from a Lipase: A Novel Structural Approach. Biochimie 2000, 82, 1033–1041. [Google Scholar] [CrossRef]
- Google Maps Imagery ©2021 TerraMetrics. Map Data ©2021 GeoBasis-DE/BKG (©2009) CNES/Airbus, Landsat/Copernicus, Maxar Technologies. Map Data ©2021 Instituto Geográfico Nacional Spain. Available online: https://www.google.com/maps/ (accessed on 5 August 2021).
- Serial Cloner. Available online: http://serialbasics.free.fr/Serial_Cloner.html (accessed on 28 July 2021).
- Berlemont, R.; Pipers, D.; Delsaute, M.; Angiono, F.; Feller, G.; Galleni, M.; Power, P. Exploring the Antarctic Soil Metagenome as a Source of Novel Cold-Adapted Enzymes and Genetic Mobile Elements. Rev. Argent. De Microbiol. 2011, 43, 94–103. [Google Scholar] [CrossRef]
- Pandey, S.; Singh, S.; Yadav, A.N.; Nain, L.; Saxena, A.K. Phylogenetic Diversity and Characterization of Novel and Efficient Cellulase Producing Bacterial Isolates from Various Extreme Environments. Biosci Biotechnol Biochem 2013, 77, 1474–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A Rapid and Easy Method for the Detection of Microbial Cellulases on Agar Plates Using Gram’s Iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Knapik, K.; Becerra, M.; González-Siso, M.-I. Microbial Diversity Analysis and Screening for Novel Xylanase Enzymes from the Sediment of the Lobios Hot Spring in Spain. Sci. Rep. 2019, 9, 11195. [Google Scholar] [CrossRef]
- Anderson, I.; Abt, B.; Lykidis, A.; Klenk, H.-P.; Kyrpides, N.; Ivanova, N. Genomics of Aerobic Cellulose Utilization Systems in Actinobacteria. PLoS ONE 2012, 7, e39331. [Google Scholar] [CrossRef] [Green Version]
- Nyyssönen, M.; Tran, H.M.; Karaoz, U.; Weihe, C.; Hadi, M.Z.; Martiny, J.B.H.; Martiny, A.C.; Brodie, E.L. Coupled High-Throughput Functional Screening and next Generation Sequencing for Identification of Plant Polymer Decomposing Enzymes in Metagenomic Libraries. Front. Microbiol. 2013, 4, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Blank, S.; Schrödern, C.; Schirrmacher, G.; Reisinger, C.A.G. Biochemical Characterization of a Recombinant Xylanase from Thermus Brockianus, Suitable for Biofuel Production. JSM Biotechnol. Bioeng. 2014, 2, 1027. [Google Scholar]
- Pinheiro, G.L.; Correa, R.F.; Cunha, R.S.; Cardoso, A.M.; Chaia, C.; Clementino, M.M.; Garcia, E.S.; de Souza, W.; Frasés, S. Isolation of Aerobic Cultivable Cellulolytic Bacteria from Different Regions of the Gastrointestinal Tract of Giant Land Snail Achatina Fulica. Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ohlhoff, C.W.; Kirby, B.M.; van Zyl, L.; Mutepfa, D.L.R.; Casanueva, A.; Huddy, R.J.; Bauer, R.; Cowan, D.A.; Tuffin, M. An Unusual Feruloyl Esterase Belonging to Family VIII Esterases and Displaying a Broad Substrate Range. J. Mol. Catal. B Enzym. 2015, 118, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Hårdeman, F.; Sjöling, S. Metagenomic Approach for the Isolation of a Novel Low-Temperature-Active Lipase from Uncultured Bacteria of Marine Sediment. FEMS Microbiol. Ecol. 2007, 59, 524–534. [Google Scholar] [CrossRef] [Green Version]
- Leis, B.; Angelov, A.; Mientus, M.; Li, H.; Pham, V.T.T.; Lauinger, B.; Bongen, P.; Pietruszka, J.; Gonçalves, L.G.; Santos, H.; et al. Identification of Novel Esterase-Active Enzymes from Hot Environments by Use of the Host Bacterium Thermus Thermophilus. Front. Microbiol. 2015, 6, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvin, J.; Kennedy, J.; Lejon, D.P.H.; Kiran, G.S.; Dobson, A.D.W. Isolation Identification and Biochemical Characterization of a Novel Halo-Tolerant Lipase from the Metagenome of the Marine Sponge Haliclona Simulans. Microb. Cell Factories 2012, 11, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, S.L.; Li, G.; Hu, X.P.; Liu, Y.H. Molecular Cloning, Overexpression and Characterization of a Novel Feruloyl Esterase from a Soil Metagenomic Library. J. Mol. Microbiol. Biotechnol. 2011, 20, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.; Svendsen, A.; Vind, J.; Lassen, S.F.; Hjort, C.; Borch, K.; Patkar, S.A. Studies on Ferulic Acid Esterase Activity in Fungal Lipases and Cutinases. Colloids Surf. B Biointerfaces 2002, 26, 47–55. [Google Scholar] [CrossRef]
- Biosoft, P. Net Primer. Available online: http://www.premierbiosoft.com/netprimer/index.html (accessed on 18 June 2021).
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger LCC The PyMOL Molecular Graphics System, Version 1.2r3pre. Available online: https://pymol.org/2/#page-top (accessed on 21 June 2021).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–608. ISBN 9781588293435. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Fuciños, P.; Abadín, C.M.; Sanromán, A.; Longo, M.A.; Pastrana, L.; Rúa, M.L. Identification of Extracellular Lipases/Esterases Produced by Thermus Thermophilus HB27: Partial Purification and Preliminary Biochemical Characterisation. J. Biotechnol. 2005, 117, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Miguel-Ruano, V.; Rivera, I.; Rajkovic, J.; Knapik, K.; Torrado, A.; Otero, J.M.; Beneventi, E.; Becerra, M.; Sánchez-Costa, M.; Hidalgo, A.; et al. Biochemical and Structural Characterization of a Novel Thermophilic Esterase EstD11 Provide Catalytic Insights for the HSL Family. Comput. Struct. Biotechnol. J. 2021, 19, 1214–1232. [Google Scholar] [CrossRef] [PubMed]


| Enzymatic Activity | Substrate | Method | As Burgas | Muiño da Veiga |
|---|---|---|---|---|
| Endoglucanase | CMC | Halo | 0 | 0 |
| AZCL-HE-Celulose | Colorimetric | 0 | 0 | |
| EnzChek cellulase | Fluorimetric | 2 | 4 | |
| Exoglucanase | AVICEL | Halo | 0 | 0 |
| β-glucosidase | pNP-glucopyranoside | Colorimetric | 5 | 1 |
| Xylanase | AZCL-Xylan | Colorimetric | 0 | 0 |
| β-xylosidase | MUX | Fluorimetric | 2 | 0 |
| Protease | EnzChek Casein | Fluorimetric | 4 | 6 |
| Lipolytic | Tributyrin | Halo | 1 | 8 |
| Feruloyl esterase | MUTMAC | Fluorimetric | 0 | 0 |
| Purification Step | Volume (mL) | Total Activity (U/µL) | Total Protein (mg) | Specific Activity (U/mg) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|---|
| Crude extract | 16 | 0.000201 | 17.01 | 1.1807 × 10−8 | 100 | 1 |
| Differential thermal precipitation | 8 | 0.000155 | 10.88 | 1.4287 × 10−8 | 77.42 | 1.21 |
| Ammonium sulphate precipitation | 8 | 0.000149 | 4.33 | 3.4641 × 10−8 | 74.69 | 2.93 |
| Molecular exclusion chromatography | 3 | 0.000145 | 2.70 | 5.383 × 10−8 | 72.49 | 4.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Saavedra-Bouza, A.; González-Siso, M.-I.; Becerra, M. Bioprospecting for Thermozymes and Characterization of a Novel Lipolytic Thermozyme Belonging to the SGNH/GDSL Family of Hydrolases. Int. J. Mol. Sci. 2022, 23, 5733. https://doi.org/10.3390/ijms23105733
Escuder-Rodríguez J-J, DeCastro M-E, Saavedra-Bouza A, González-Siso M-I, Becerra M. Bioprospecting for Thermozymes and Characterization of a Novel Lipolytic Thermozyme Belonging to the SGNH/GDSL Family of Hydrolases. International Journal of Molecular Sciences. 2022; 23(10):5733. https://doi.org/10.3390/ijms23105733
Chicago/Turabian StyleEscuder-Rodríguez, Juan-José, María-Eugenia DeCastro, Almudena Saavedra-Bouza, María-Isabel González-Siso, and Manuel Becerra. 2022. "Bioprospecting for Thermozymes and Characterization of a Novel Lipolytic Thermozyme Belonging to the SGNH/GDSL Family of Hydrolases" International Journal of Molecular Sciences 23, no. 10: 5733. https://doi.org/10.3390/ijms23105733
APA StyleEscuder-Rodríguez, J.-J., DeCastro, M.-E., Saavedra-Bouza, A., González-Siso, M.-I., & Becerra, M. (2022). Bioprospecting for Thermozymes and Characterization of a Novel Lipolytic Thermozyme Belonging to the SGNH/GDSL Family of Hydrolases. International Journal of Molecular Sciences, 23(10), 5733. https://doi.org/10.3390/ijms23105733








