How Cell Geometry and Cellular Patterning Influence Tissue Stiffness
Abstract
:1. Introduction
2. Results and Discussion
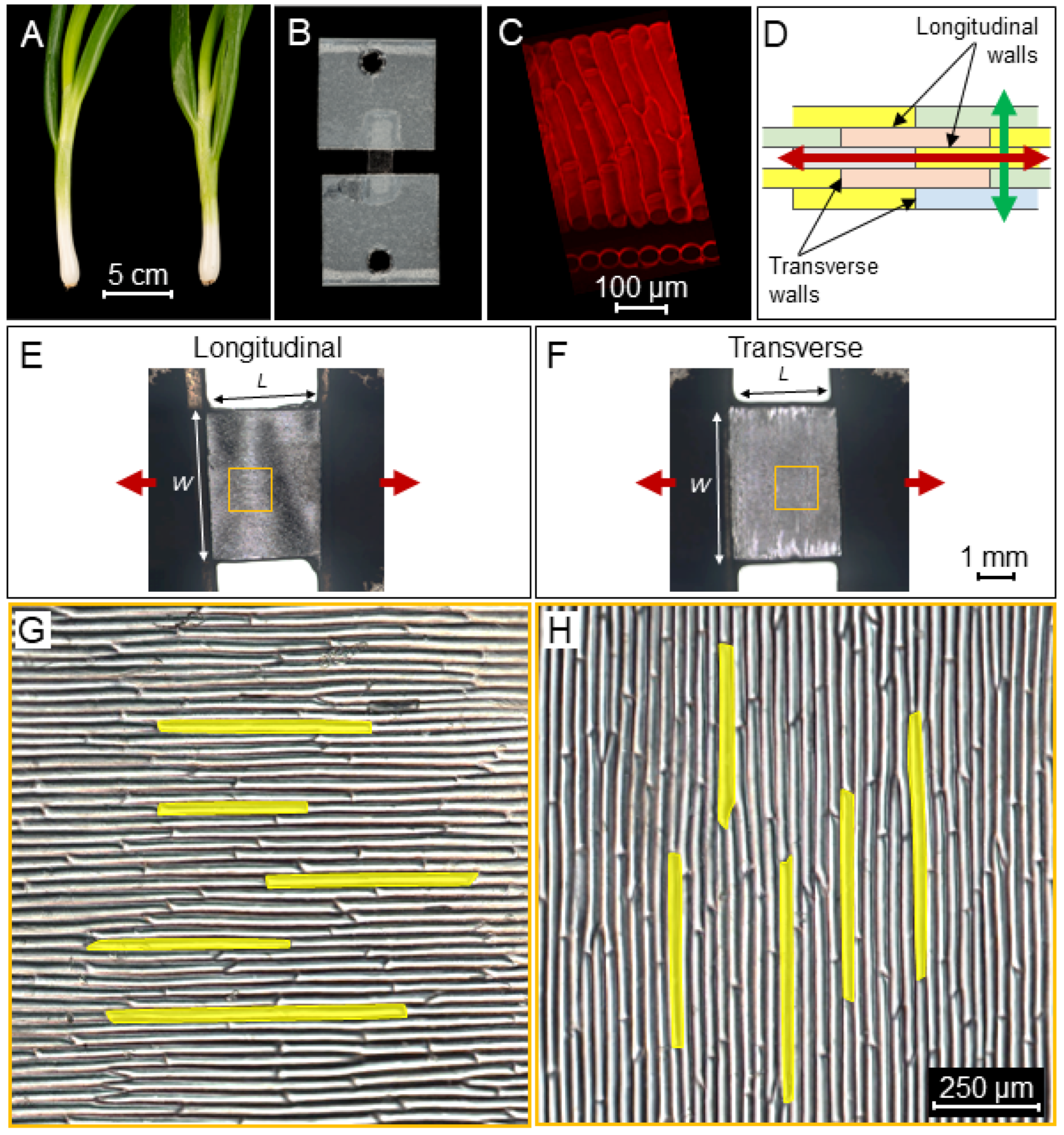
2.1. Tissue Mechanical Stiffness Is Anisotropic
2.2. A Simulation Model of the Extensometer Experiments
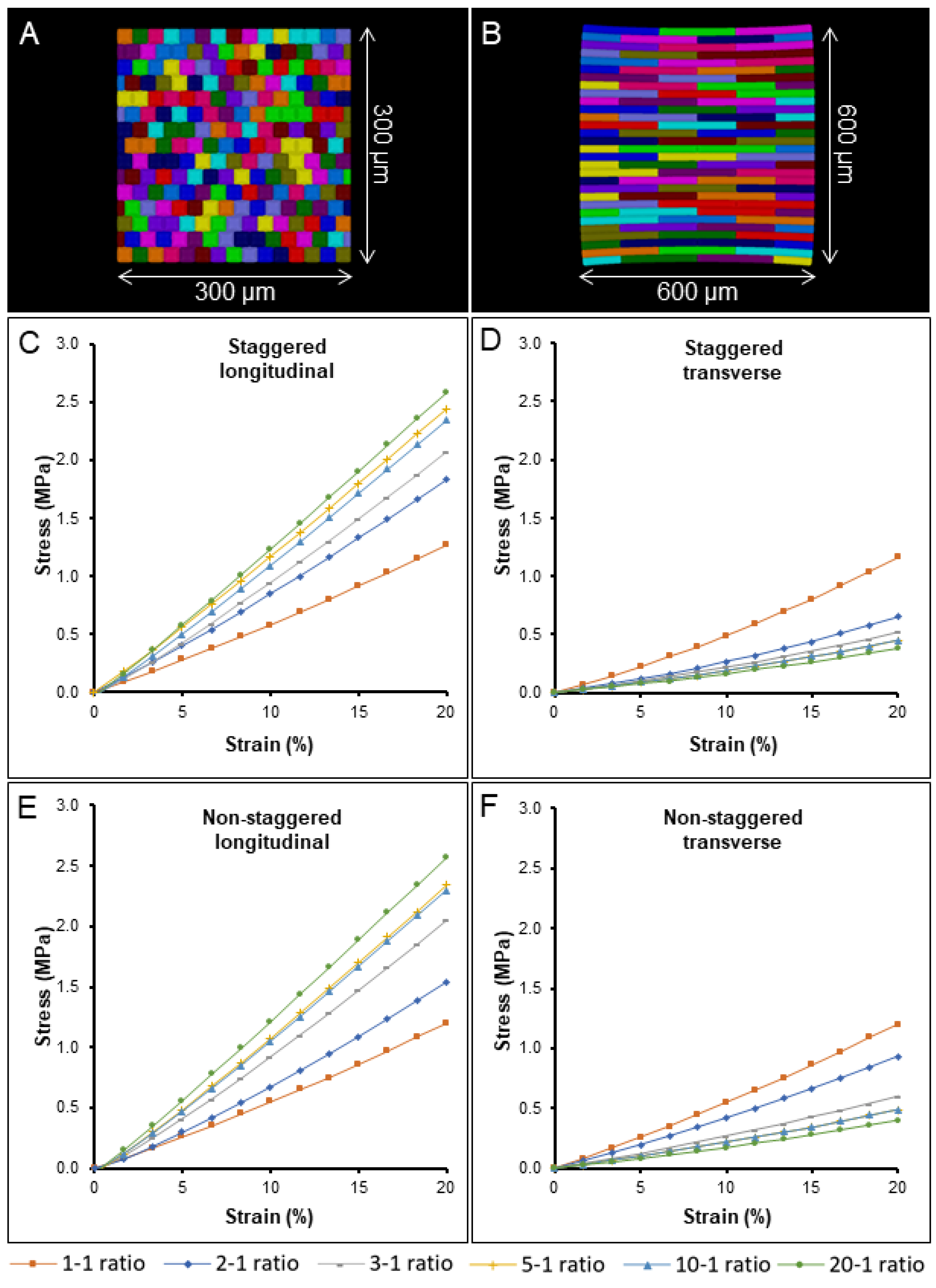
2.3. The Impact of Cell Aspect Ratio and Staggering on Stiffness and Nonlinearity
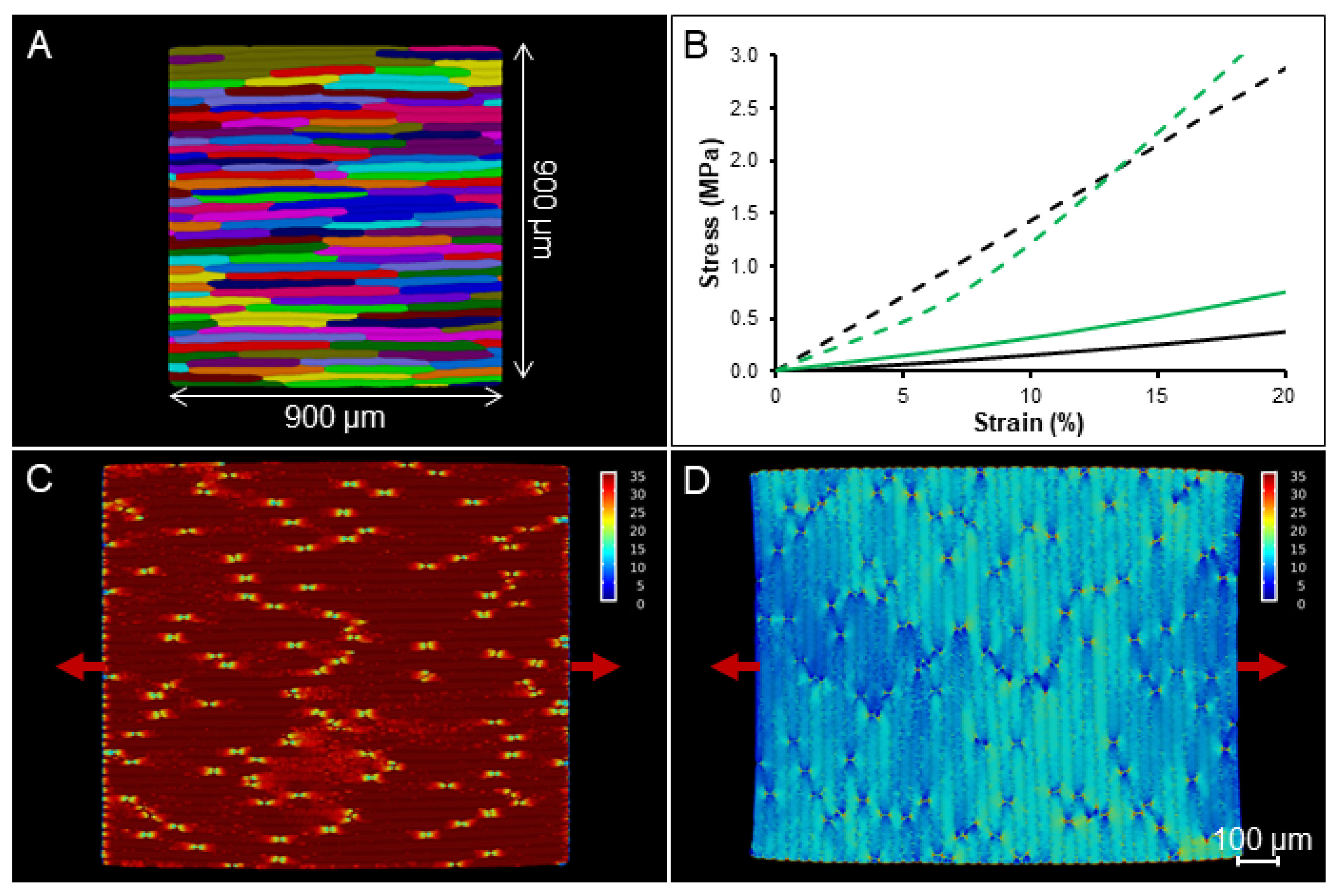
2.4. Simulation on a Template with Realistic Cell Shapes
2.5. Estimating Cell-Wall Anisotropy
3. Materials and Methods
3.1. Plant Material and Sample Preparation
3.2. Microscopy and Image Processing
3.3. Microextensometer Setup
3.4. Stretching the Samples with the Extensometer
3.5. Mechanical Modeling
3.5.1. Idealized Cell Templates
3.5.2. Generation of Realistic Cellular Templates
3.5.3. Simulations of Extensometer Experiments
3.5.4. Model Parameters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, D.J. Diffuse Growth of Plant Cell Walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockhart, J.A. An analysis of irreversible plant cell elongation. J. Theor. Biol. 1965, 8, 264–275. [Google Scholar] [CrossRef]
- Cosgrove, D. Biophysical control of plant cell growth. Annu. Rev. Plant Physiol. 1986, 37, 377–405. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Wall relaxation and the driving forces for cell expansive growth. Plant Physiol. 1987, 84, 561–564. [Google Scholar] [CrossRef] [Green Version]
- Cavalier, D.M.; Lerouxel, O.; Neumetzler, L.; Yamauchi, K.; Reinecke, A.; Freshour, G.; Zabotina, O.A.; Hahn, M.G.; Burgert, I.; Pauly, M.; et al. Disrupting Two Arabidopsis thaliana Xylosyltransferase Genes Results in Plants Deficient in Xyloglucan, a Major Primary Cell Wall Component. Plant Cell 2008, 20, 1519–1537. [Google Scholar] [CrossRef] [Green Version]
- Kuchen, E.E.; Fox, S.; Barbier De Reuille, P.; Kennaway, R.; Bensmihen, S.; Avondo, J.; Calder, G.M.; Southam, P.; Robinson, S.; Bangham, A.; et al. Generation of Leaf Shape Through Early Patterns of Growth and Tissue Polarity. Science 2012, 335, 1092–1096. [Google Scholar] [CrossRef] [Green Version]
- Bassel, G.W.; Stamm, P.; Mosca, G.; Barbier De Reuille, P.; Gibbs, D.J.; Winter, R.; Janka, A.; Holdsworth, M.J.; Smith, R.S. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc. Natl. Acad. Sci. USA 2014, 111, 8685–8690. [Google Scholar] [CrossRef] [Green Version]
- Rebocho, A.B.; Southam, P.; Kennaway, J.R.; Bangham, J.A.; Coen, E. Generation of shape complexity through tissue conflict resolution. eLife 2017, 6, e20156. [Google Scholar] [CrossRef] [Green Version]
- Mosca, G.; Sapala, A.; Strauss, S.; Routier-Kierzkowska, A.L.; Smith, R.S. On the micro-indentation of plant cells in a tissue context. Phys. Biol. 2017, 14, 015003. [Google Scholar] [CrossRef]
- Sapala, A.; Runions, A.; Routier-Kierzkowska, A.L.; Das Gupta, M.; Hong, L.; Hofhuis, H.; Verger, S.; Mosca, G.; Li, C.B.; Hay, A.; et al. Why plants make puzzle cells, and how their shape emerges. eLife 2018, 7, e32794. [Google Scholar] [CrossRef]
- Kierzkowski, D.; Routier-Kierzkowska, A.L. Cellular basis of growth in plants: Geometry matters. Curr. Opin. Plant Biol. 2019, 47, 56–63. [Google Scholar] [CrossRef]
- Bidhendi, A.J.; Geitmann, A. Methods to quantify primary plant cell wall mechanics. J. Exp. Bot. 2019, 70, 3615–3648. [Google Scholar] [CrossRef] [Green Version]
- Hong, L.; Dumond, M.; Tsugawa, S.; Sapala, A.; Routier-Kierzkowska, A.-L.; Zhou, Y.; Chen, C.; Kiss, A.; Zhu, M.; Hamant, O.; et al. Variable Cell Growth Yields Reproducible Organ Development through Spatiotemporal Averaging. Dev. Cell 2016, 38, 15–32. [Google Scholar] [CrossRef] [Green Version]
- Kierzkowski, D.; Nakayama, N.; Routier-Kierzkowska, A.L.; Weber, A.; Bayer, E.; Schorderet, M.; Reinhardt, D.; Kuhlemeier, C.; Smith, R.S. Elastic Domains Regulate Growth and Organogenesis in the Plant Shoot Apical Meristem. Science 2012, 335, 1096–1099. [Google Scholar] [CrossRef] [Green Version]
- Hofhuis, H.; Moulton, D.; Lessinnes, T.; Routier-Kierzkowska, A.L.; Bomphrey, R.J.; Mosca, G.; Reinhardt, H.; Sarchet, P.; Gan, X.; Tsiantis, M.; et al. Morphomechanical Innovation Drives Explosive Seed Dispersal. Cell 2016, 166, 222–233. [Google Scholar] [CrossRef] [Green Version]
- Milani, P.; Gholamirad, M.; Traas, J.; Arnéodo, A.; Boudaoud, A.; Argoul, F.; Hamant, O. In vivo analysis of local wall stiffness at the shoot apical meristem in Arabidopsis using atomic force microscopy. Plant J. 2011, 67, 1116–1123. [Google Scholar] [CrossRef]
- Radotić, K.; Roduit, C.; Simonović, J.; Hornitschek, P.; Fankhauser, C.; Mutavdžić, D.; Steinbach, G.; Dietler, G.; Kasas, S. Atomic Force Microscopy Stiffness Tomography on Living Arabidopsis thaliana Cells Reveals the Mechanical Properties of Surface and Deep Cell-Wall Layers during Growth. Biophys. J. 2012, 103, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Majda, M. Atomic Force Microscopy to Study Cell Wall Mechanics in Plants. Methods Mol. Biol. 2021, 2200, 349–369. [Google Scholar] [CrossRef]
- Majda, M.; Grones, P.; Sintorn, I.M.; Vain, T.; Milani, P.; Krupinski, P.; Zagorska-Marek, B.; Viotti, C.; Jonsson, H.; Mellerowicz, E.J.; et al. Mechanochemical Polarization of Contiguous Cell Walls Shapes Plant Pavement Cells. Dev. Cell 2017, 43, 290–304. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.A.; le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-Induced Changes in Cell Wall Mechanics Underlie Organ Initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef] [Green Version]
- Beauzamy, L.; Derr, J.; Boudaoud, A. Quantifying hydrostatic pressure in plant cells by using indentation with an atomic force microscope. Biophys. J. 2015, 108, 2448–2456. [Google Scholar] [CrossRef] [Green Version]
- Routier-Kierzkowska, A.-L.; Weber, A.; Kochova, P.; Felekis, D.; Nelson, B.J.; Kuhlemeier, C.; Smith, R.S. Cellular Force Microscopy for in Vivo Measurements of Plant Tissue Mechanics. Plant Physiol. 2012, 158, 1514–1522. [Google Scholar] [CrossRef] [Green Version]
- Majda, M.; Sapala, A.; Routier-Kierzkowska, A.L.; Smith, R.S. Cellular Force Microscopy to Measure Mechanical Forces in Plant Cells. Methods Mol. Biol. 2019, 1992, 215–230. [Google Scholar] [CrossRef]
- Scarcelli, G.; Polacheck, W.J.; Nia, H.T.; Patel, K.; Grodzinsky, A.J.; Kamm, R.D.; Yun, S.H. Noncontact three-dimensional mapping of intracellular hydromechanical properties by Brillouin microscopy. Nat. Methods 2015, 12, 1132–1134. [Google Scholar] [CrossRef] [Green Version]
- Elsayad, K.; Werner, S.; Gallemi, M.; Kong, J.; Sanchez Guajardo, E.R.; Zhang, L.; Jaillais, Y.; Greb, T.; Belkhadir, Y. Mapping the subcellular mechanical properties of live cells in tissues with fluorescence emission-Brillouin imaging. Sci. Signal. 2016, 9, rs5. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Wall extensibility: Its nature, measurement and relationship to plant cell growth. New Phytol. 1993, 124, 1–23. [Google Scholar] [CrossRef]
- Phyo, P.; Wang, T.; Kiemle, S.N.; O’Neill, H.; Pingali, S.V.; Hong, M.; Cosgrove, D.J. Gradients in Wall Mechanics and Polysaccharides along Growing Inflorescence Stems. Plant Physiol. 2017, 175, 1593–1607. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Measuring in vitro extensibility of growing plant cell walls. Methods Mol. Biol. 2011, 715, 291–303. [Google Scholar] [CrossRef]
- Burgert, I. Exploring the micromechanical design of plant cell walls. Am. J. Bot. 2006, 93, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.; Huflejt, M.; Barbier De Reuille, P.; Braybrook, S.A.; Schorderet, M.; Reinhardt, D.; Kuhlemeier, C. An Automated Confocal Micro-Extensometer Enables in Vivo Quantification of Mechanical Properties with Cellular Resolution. Plant Cell 2017, 29, 2959–2973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schluck, T.; Nienhaus, U.; Aegerter-Wilmsen, T.; Aegerter, C.M. Mechanical Control of Organ Size in the Development of the Drosophila Wing Disc. PLoS ONE 2013, 8, e76171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomos, A.D.; Leigh, R.A. THE PRESSURE PROBE: A Versatile Tool in Plant Cell Physiology. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1999, 50, 447–472. [Google Scholar] [CrossRef]
- Sapala, A.; Smith, R.S. Osmotic Treatment for Quantifying Cell Wall Elasticity in the Sepal of Arabidopsis thaliana. Methods Mol. Biol. 2020, 2094, 101–112. [Google Scholar] [CrossRef]
- Natonik-Białoń, S.; Borowska-Wykręt, D.; Mosca, G.; Grelowski, M.; Wrzalik, R.; Smith, R.S.; Kwiatkowska, D. Deformation of a cell monolayer due to osmotic treatment: A case study of onion scale epidermis. Botany 2020, 98, 21–36. [Google Scholar] [CrossRef]
- Kim, K.; Yi, H.; Zamil, M.S.; Haque, M.A.; Puri, V.M. Multiscale stress-strain characterization of onion outer epidermal tissue in wet and dry states. Am. J. Bot. 2015, 102, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Tang, H.; Vavylonis, D.; Cosgrove, D.J. Disentangling loosening from softening: Insights into primary cell wall structure. Plant J. 2019, 100, 1101–1117. [Google Scholar] [CrossRef] [Green Version]
- Vanstreels, E.; Alamar, M.C.; Verlinden, B.E.; Enninghorst, A.; Loodts, J.K.A.; Tijskens, E.; Ramon, H.; Nicolaï, B.M. Micromechanical behaviour of onion epidermal tissue. Postharvest Biol. Technol. 2005, 37, 163–173. [Google Scholar] [CrossRef]
- Zamil, M.S.; Yi, H.; Haque, M.A.; Puri, V.M. Characterizing microscale biological samples under tensile loading: Stress–strain behavior of cell wall fragment of onion outer epidermis. Am. J. Bot. 2013, 100, 1105–1115. [Google Scholar] [CrossRef]
- Suslov, D.; Verbelen, J.P. Cellulose orientation determines mechanical anisotropy in onion epidermis cell walls. J. Exp. Bot. 2006, 57, 2183–2192. [Google Scholar] [CrossRef] [Green Version]
- Bidhendi, A.J.; Li, H.; Geitmann, A. Modeling the nonlinear elastic behavior of plant epidermis. Botany 2020, 98, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Barbier De Reuille, P.; Routier-Kierzkowska, A.-L.; Kierzkowski, D.; Bassel, G.W.; Schüpbach, T.; Tauriello, G.; Bajpai, N.; Strauss, S.; Weber, A.; Kiss, A.; et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 2015, 4, e05864. [Google Scholar] [CrossRef]
- Strauss, S.; Runions, A.; Lane, B.; Eschweiler, D.; Bajpai, N.; Trozzi, N.; Routier-Kierzkowska, A.-L.; Yoshida, S.; Rodrigues da Silveira, S.; Vijayan, A.; et al. MorphoGraphX 2.0: Providing context for biological image analysis with positional information. bioRxiv 2021, 8, 12. [Google Scholar] [CrossRef]
- Baskin, T.I. Anisotropic expansion of the plant cell wall. Annu. Rev. Cell. Dev. Biol. 2005, 21, 203–222. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Zienkiewicz, O.C.; Taylor, R.L.; Zhu, J.Z. The Finite Element Method: Its Basis and Fundamentals; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Holzapfel, G.A. Nonlinear solid mechanics: A continuum approach for engineering science. Meccanica 2002, 37, 489–490. [Google Scholar] [CrossRef]
- Bonet, J.; Burton, A.J. A simple orthotropic, transversely isotropic hyperelastic constitutive equation for large strain computations. Comput. Methods Appl. Mech. Eng. 1998, 162, 151–164. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Wang, X.; Durachko, D.M.; Zhang, S.; Cosgrove, D.J. Molecular insights into the complex mechanics of plant epidermal cell walls. Science 2021, 372, 706–711. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majda, M.; Trozzi, N.; Mosca, G.; Smith, R.S. How Cell Geometry and Cellular Patterning Influence Tissue Stiffness. Int. J. Mol. Sci. 2022, 23, 5651. https://doi.org/10.3390/ijms23105651
Majda M, Trozzi N, Mosca G, Smith RS. How Cell Geometry and Cellular Patterning Influence Tissue Stiffness. International Journal of Molecular Sciences. 2022; 23(10):5651. https://doi.org/10.3390/ijms23105651
Chicago/Turabian StyleMajda, Mateusz, Nicola Trozzi, Gabriella Mosca, and Richard S. Smith. 2022. "How Cell Geometry and Cellular Patterning Influence Tissue Stiffness" International Journal of Molecular Sciences 23, no. 10: 5651. https://doi.org/10.3390/ijms23105651
APA StyleMajda, M., Trozzi, N., Mosca, G., & Smith, R. S. (2022). How Cell Geometry and Cellular Patterning Influence Tissue Stiffness. International Journal of Molecular Sciences, 23(10), 5651. https://doi.org/10.3390/ijms23105651





