Genome-Wide Identification of Auxin Response Factors in Peanut (Arachis hypogaea L.) and Functional Analysis in Root Morphology
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of Peanut ARF Genes
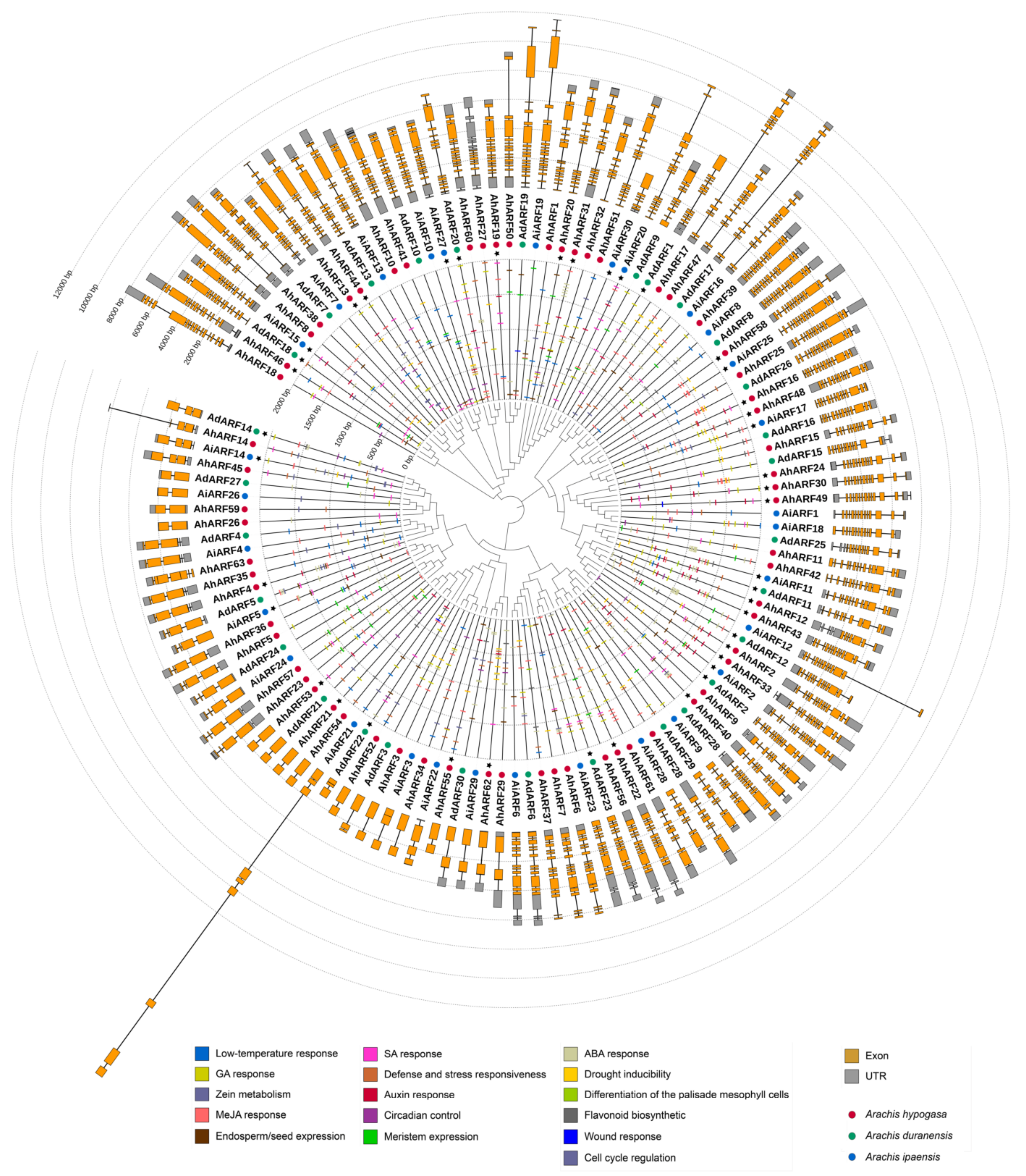
2.2. Phylogenetic Analysis of Peanut ARFs
2.3. Gene Duplication on the Expansion of ARF Genes
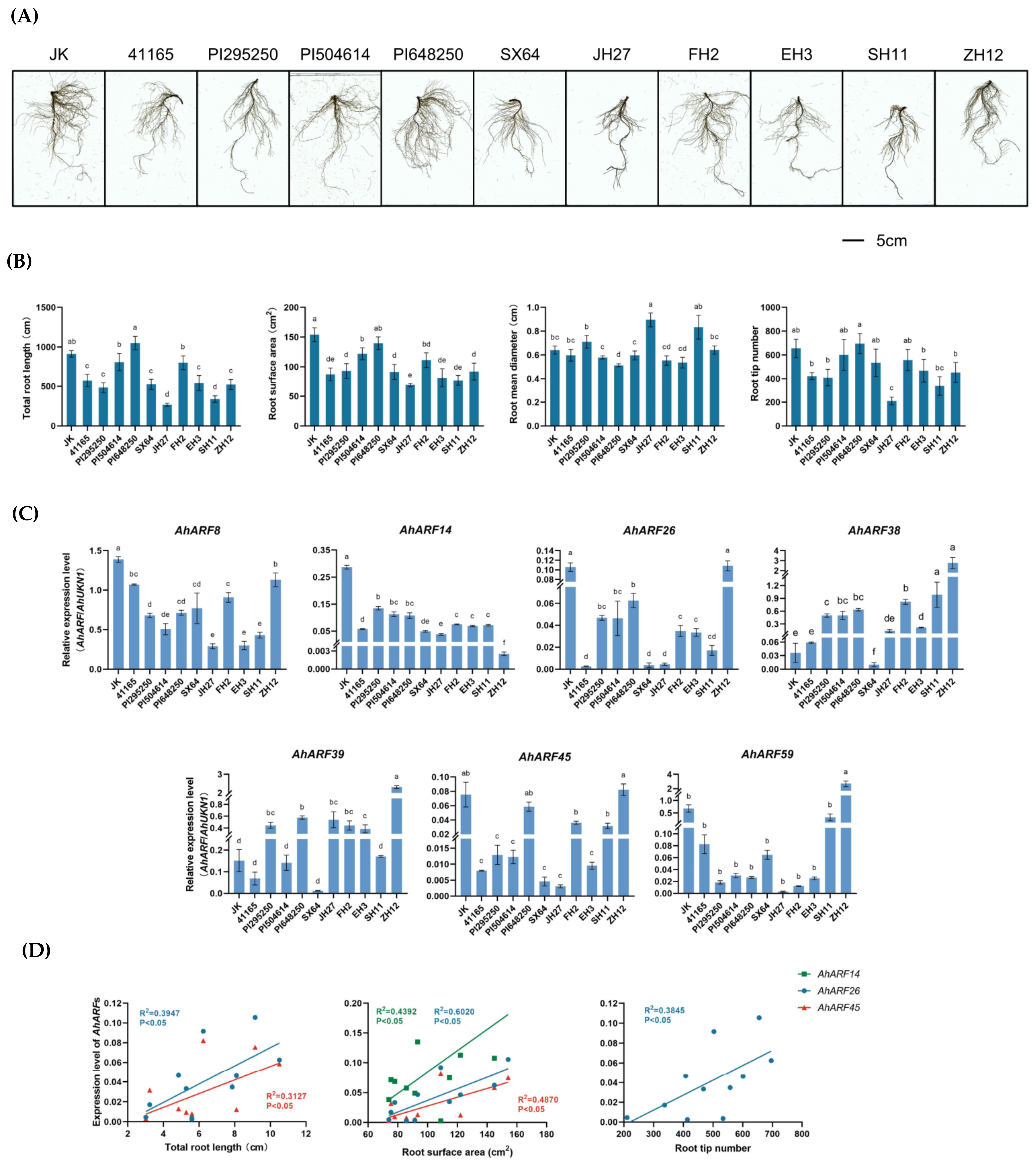
2.4. Expression Profiles of AhARFs
2.5. Characterization of Downstream Target Genes of AhARFs
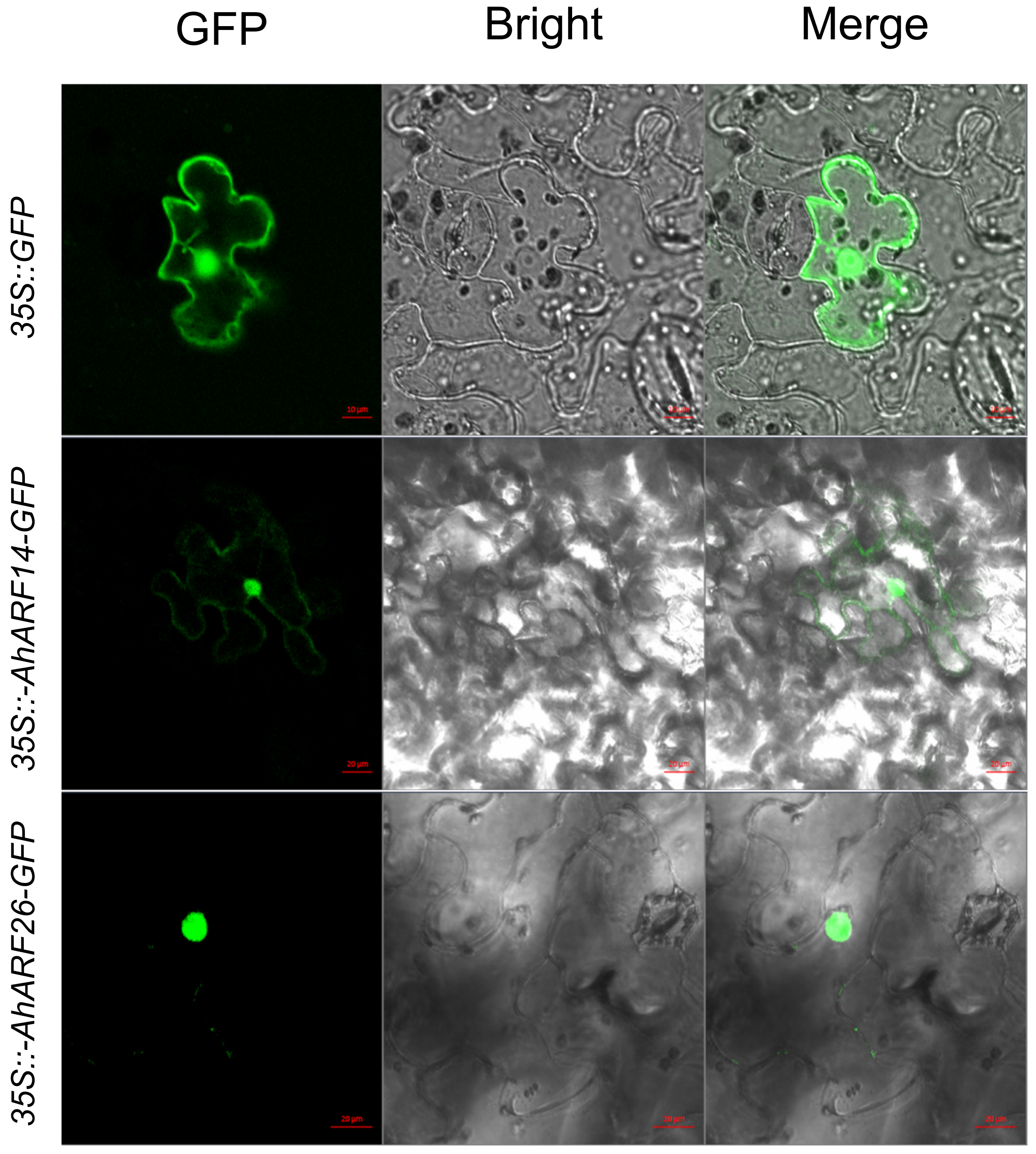
2.6. The Possible Regulatory Roles of AhARFs in Peanut Root Morphology
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Genome-Wide Identification of AhARFs, AdARFs, and AiARFs
4.3. Chromosome Distribution, Gene Structure, Phylogenetic Analysis, and cis-Elements Analysis
4.4. Syntenic and Genome Duplication Analysis of Peanut ARFs
4.5. Genome-Wide Search for Downstream Targets of AhARFs
4.6. Expression Analysis of AhARFs
4.7. Root Morphology Analysis
4.8. Statistical Analysis
4.9. Subcellular Localization Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The Systems Biology of Auxin in Developing Embryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Gallei, M.; Luschnig, C.; Friml, J. Auxin signalling in growth: Schrodinger’s cat out of the bag. Curr. Opin. Plant Biol. 2020, 53, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, Z.; Xu, Y.; Guo, C.; Zhang, L.; Wu, C.; Yang, G.; Huang, J.; Yan, K.; Shu, H.; et al. Function identification of MdTIR1 in apple root growth benefited from the predicted MdPPI network. J. Integr. Plant Biol. 2020, 63, 723–739. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boer, D.R.; Freire-Rios, A.; van den Berg, W.A.; Saaki, T.; Manfield, I.W.; Kepinski, S.; Lopez-Vidrieo, I.; Franco-Zorrilla, J.M.; de Vries, S.C.; Solano, R.; et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marletaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Swarup, R.; Bhosale, R. Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.I. Chemical Biology in Auxin Research. Cold Spring Harb. Perspect. Biol. 2021, 13, a040105. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [Green Version]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [Green Version]
- Wilmoth, J.C.; Wang, S.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. Cell Mol. Biol. 2005, 43, 118–130. [Google Scholar] [CrossRef]
- Zhang, F.; Tao, W.; Sun, R.; Wang, J.; Li, C.; Kong, X.; Tian, H.; Ding, Z. PRH1 mediates ARF7-LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana. PLoS Genet. 2020, 16, e1008044. [Google Scholar] [CrossRef]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-Directed Regulation of the Auxin Response Factors GmARF8a and GmARF8b Is Required for Soybean Nodulation and Lateral Root Development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.; Liu, R.; Gu, W.; Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. Int. J. Exp. Plant Biol. 2017, 256, 103–111. [Google Scholar] [CrossRef]
- Stigliani, A.; Martin-Arevalillo, R.; Lucas, J.; Bessy, A.; Vinos-Poyo, T.; Mironova, V.; Vernoux, T.; Dumas, R.; Parcy, F. Capturing Auxin Response Factors Syntax Using DNA Binding Models. Mol. Plant 2019, 12, 822–832. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretzsohn, M.C.; Gouvea, E.G.; Inglis, P.W.; Leal-Bertioli, S.C.; Valls, J.F.; Bertioli, D.J. A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann. Bot. 2013, 111, 113–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhu, S.; Zhang, K.; Wan, Y.; Liu, F.; Sun, Q.; Li, Y. Establishment and evaluation of a peanut association panel and analysis of key nutritional traits. J. Integr. Plant Biol. 2018, 60, 195–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Liu, Y.; Luo, L.; Zhang, X.; Li, G.; Wan, Y.; Liu, F. Root traits of peanut cultivars with different drought resistant under drought stress at flowering and pegging phase. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2021, 71, 363–376. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, C.; Zheng, H.; Sun, M.; Zhang, F.; Zhang, M.; Cui, F.; Lv, D.; Liu, L.; Guo, S.; et al. Antagonistic Interaction between Auxin and SA Signaling Pathways Regulates Bacterial Infection through Lateral Root in Arabidopsis. Cell Rep. 2020, 32, 108060. [Google Scholar] [CrossRef] [PubMed]
- Attia, K.A.; Abdelkhalik, A.F.; Ammar, M.H.; Wei, C.; Yang, J.; Lightfoot, D.A.; El-Sayed, W.M.; El-Shemy, H.A. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 2009, 11 (Suppl. 1), i29–i34. [Google Scholar] [PubMed]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef]
- Huang, K.L.; Ma, G.J.; Zhang, M.L.; Xiong, H.; Wu, H.; Zhao, C.Z.; Liu, C.S.; Jia, H.X.; Chen, L.; Kjorven, J.O.; et al. The ARF7 and ARF19 Transcription Factors Positively Regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis Roots. Plant Physiol. 2018, 178, 413–427. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, S.; Sun, C.; Xu, Y.; Chen, Y.; Yu, C.; Qian, Q.; Jiang, D.A.; Qi, Y. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 2014, 201, 91–103. [Google Scholar] [CrossRef]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Breitel, D.A.; Chappell-Maor, L.; Meir, S.; Panizel, I.; Puig, C.P.; Hao, Y.; Yifhar, T.; Yasuor, H.; Zouine, M.; Bouzayen, M.; et al. AUXIN RESPONSE FACTOR 2 Intersects Hormonal Signals in the Regulation of Tomato Fruit Ripening. PLoS Genet. 2016, 12, e1005903. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. Cell Mol. Biol. 2014, 80, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Xu, X.; Gong, Z.; Tang, Y.; Wu, M.; Yan, F.; Zhang, X.; Zhang, Q.; Yang, F.; Hu, X.; et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang de, A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [Green Version]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 Regulates Floral Meristem Determinacy by Repressing Cytokinin Biosynthesis and Signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef] [Green Version]
- Goetz, M.; Vivian-Smith, A.; Johnson, S.D.; Koltunow, A.M. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 2006, 18, 1873–1886. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.F.; Wang, B.; Feng, Y.F.; Xue, J.S.; Qian, X.X.; Liu, S.Q.; Zhou, J.; Yu, Y.H.; Yang, N.Y.; Xu, P.; et al. AUXIN RESPONSE FACTOR17 Directly Regulates MYB108 for Anther Dehiscence. Plant Physiol. 2019, 181, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.; Zhu, Y.; Wu, M.F.; Simonini, S.; Kuhn, A.; Armenta-Medina, A.; Jin, R.; Ostergaard, L.; Gillmor, C.S.; Wagner, D. Auxin Response Factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat. Commun. 2019, 10, 886. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yan, F.; Tang, Y.; Yuan, Y.; Deng, W.; Li, Z. Auxin Response Gene SlARF3 Plays Multiple Roles in Tomato Development and is Involved in the Formation of Epidermal Cells and Trichomes. Plant Cell Physiol. 2015, 56, 2110–2124. [Google Scholar] [CrossRef] [Green Version]
- Pashkovskiy, P.P.; Kartashov, A.V.; Zlobin, I.E.; Pogosyan, S.I.; Kuznetsov, V.V. Blue light alters miR167 expression and microRNA-targeted auxin response factor genes in Arabidopsis thaliana plants. Plant Physiol. Biochem. PPB 2016, 104, 146–154. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; He, Y.; Qin, Q.; Chen, C.; Wei, Z.; Tan, X.; Xie, K.; Zhang, R.; Hong, G.; et al. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA virumses for infection. Proc. Natl. Acad. Sci. USA 2020, 117, 9112–9121. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsogon, A.; Smouni, A.; et al. Down Regulation and Loss of Auxin Response Factor 4 Function Using CRISPR/Cas9 Alters Plant Growth, Stomatal Function and Improves Tomato Tolerance to Salinity and Osmotic Stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef] [Green Version]
- Kelley, D.R.; Arreola, A.; Gallagher, T.L.; Gasser, C.S. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 2012, 139, 1105–1109. [Google Scholar] [CrossRef] [Green Version]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.H.; Liu, Y.B.; Zhou, C.; Li, X.M.; Zhang, X.S. The microRNA167 controls somatic embryogenesis in Arabidopsis through regulating its target genes ARF6 and ARF8. Plant Cell, Tissue and Organ Culture 2016, 124, 405–417. [Google Scholar] [CrossRef]
- Luo, L.; Zeng, J.; Wu, H.; Tian, Z.; Zhao, Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant 2018, 11, 899–913. [Google Scholar] [CrossRef] [Green Version]
- Ghelli, R.; Brunetti, P.; Napoli, N.; De Paolis, A.; Cecchetti, V.; Tsuge, T.; Serino, G.; Matsui, M.; Mele, G.; Rinaldi, G.; et al. A Newly Identified Flower-Specific Splice Variant of AUXIN RESPONSE FACTOR8 Regulates Stamen Elongation and Endothecium Lignification in Arabidopsis. Plant Cell 2018, 30, 620–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.; Yue, R.; Sun, T.; Zhang, L.; Yang, Y.; Wang, H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. Int. J. Exp. Plant Biol. 2015, 231, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Chen, Z.; Wei, Y.; Qi, Y.; Wu, C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 2020, 18, 2015–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalve, S.; Sizani, B.L.; Markakis, M.N.; Helsmoortel, C.; Vandeweyer, G.; Laukens, K.; Sommen, M.; Naulaerts, S.; Vissenberg, K.; Prinsen, E.; et al. Osmotic stress inhibits leaf growth of Arabidopsis thaliana by enhancing ARF-mediated auxin responses. New Phytol. 2020, 226, 1766–1780. [Google Scholar] [CrossRef]
- Finet, C.; Fourquin, C.; Vinauger, M.; Berne-Dedieu, A.; Chambrier, P.; Paindavoine, S.; Scutt, C.P. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. Cell Mol. Biol. 2010, 63, 952–959. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Wang, L.; Sun, W.; Zhang, Y.; Zhou, C.; Su, Y.H.; Li, W.; Sun, T.T.; Zhao, X.Y.; Li, X.G.; et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 2013, 161, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Orosa-Puente, B.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef] [Green Version]
- Nziengui, H.; Lasok, H.; Kochersperger, P.; Ruperti, B.; Rebeille, F.; Palme, K.; Ditengou, F.A. Root Gravitropism Is Regulated by a Crosstalk between para-Aminobenzoic Acid, Ethylene, and Auxin. Plant Physiol. 2018, 178, 1370–1389. [Google Scholar] [CrossRef] [Green Version]
- Sagar, M.; Chervin, C.; Mila, I.; Hao, Y.; Roustan, J.P.; Benichou, M.; Gibon, Y.; Biais, B.; Maury, P.; Latche, A.; et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013, 161, 1362–1374. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Mei, L.; Wu, M.; Wei, W.; Shan, W.; Gong, Z.; Zhang, Q.; Yang, F.; Yan, F.; Zhang, Q.; et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef] [Green Version]
- De Jong, M.; Wolters-Arts, M.; Schimmel, B.C.; Stultiens, C.L.; de Groot, P.F.; Powers, S.J.; Tikunov, Y.M.; Bovy, A.G.; Mariani, C.; Vriezen, W.H.; et al. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 2015, 66, 3405–3416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three Auxin Response Factors Promote Hypocotyl Elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, C.; Willmann, M.R.; Wu, G.; Yoshikawa, M.; de la Luz Gutierrez-Nava, M.; Poethig, S.R. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roodbarkelari, F.; Du, F.; Truernit, E.; Laux, T. ZLL/AGO10 maintains shoot meristem stem cells during Arabidopsis embryogenesis by down-regulating ARF2-mediated auxin response. BMC Biol. 2015, 13, 74. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yu, R.; Wang, J.; Lin, Z.; Han, X.; Deng, Z.; Fan, L.; He, H.; Deng, X.W.; Chen, H. The Asymmetric Expression of SAUR Genes Mediated by ARF7/19 Promotes the Gravitropism and Phototropism of Plant Hypocotyls. Cell Rep. 2020, 31, 107529. [Google Scholar] [CrossRef]
- Peng, J.; Berbel, A.; Madueno, F.; Chen, R. AUXIN RESPONSE FACTOR3 Regulates Compound Leaf Patterning by Directly Repressing PALMATE-LIKE PENTAFOLIATA1 Expression in Medicago truncatula. Front. Plant Sci. 2017, 8, 1630. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Li, P.; Ma, Q.; Qu, C.; Zhu, S.; Zhao, K.; Ma, X.; Li, Z.; Zhang, X.; Gong, F.; Yin, D. Genome-wide identification and expression analysis of auxin response factors in peanut (Arachis hypogaea L.). PeerJ 2021, 9, e12319. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, G.; Ding, H.; Ci, D.; Dai, L.; Zhang, Z. Influence of salt stress on the rhizosphere soil bacterial community structure and growth performance of groundnut (Arachis hypogaea L.). Int. Microbiol. 2020, 23, 453–465. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, D.; Dai, L.; Ding, H.; Ci, D.; Qin, F.; Zhang, G.; Zhang, Z. Influence of Salt Stress on Growth of Spermosphere Bacterial Communities in Different Peanut (Arachis hypogaea L.) Cultivars. Int. J. Mol. Sci. 2020, 21, 2131. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.-c.; Lü, J.-l.; Xu, X.-p.; Lin, X.-m.; Luiz, M.R.; Qiu, S.-j.; Ciampitti, I.; He, P. Peanut yield, nutrient uptake and nutrient requirements in different regions of China. J. Integr. Agric. 2021, 20, 2502–2511. [Google Scholar] [CrossRef]
- Dai, L.; Zhang, G.; Yu, Z.; Ding, H.; Xu, Y.; Zhang, Z. Effect of Drought Stress and Developmental Stages on Microbial Community Structure and Diversity in Peanut Rhizosphere Soil. Int. J. Mol. Sci. 2019, 20, 2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ci, D.; Tang, Z.; Ding, H.; Cui, L.; Zhang, G.; Li, S.; Dai, L.; Qin, F.; Zhang, Z.; Yang, J.; et al. The synergy effect of arbuscular mycorrhizal fungi symbiosis and exogenous calcium on bacterial community composition and growth performance of peanut (Arachis hypogaea L.) in saline alkali soil. J. Microbiol. 2021, 59, 51–63. [Google Scholar] [CrossRef] [PubMed]
- De Dorlodot, S.; Forster, B.; Pages, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef]
- Ding, T.; Zhang, F.; Wang, J.; Wang, F.; Liu, J.; Xie, C.; Hu, Y.; Shani, E.; Kong, X.; Ding, Z.; et al. Cell-type action specificity of auxin on Arabidopsis root growth. Plant J. 2021, 106, 928–941. [Google Scholar] [CrossRef]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol. Plant 2021, 14, 285–297. [Google Scholar] [CrossRef]
- Jing, H.; Strader, L.C. Interplay of Auxin and Cytokinin in Lateral Root Development. Int. J. Mol. Sci. 2019, 20, 486. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Fletcher, R.S.; Mullen, J.L.; Heiliger, A.; McKay, J.K. QTL analysis of root morphology, flowering time, and yield reveals trade-offs in response to drought in Brassica napus. J. Exp. Bot. 2015, 66, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Soriano, J.M.; Alvaro, F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 2019, 9, 10537. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Kumawat, G.; Yan, Y.; Fan, B.; Xu, D. Mapping and validation of a major QTL for primary root length of soybean seedlings grown in hydroponic conditions. BMC Genom. 2021, 22, 132. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, W.; Zhang, N.; Chen, M.; Zheng, S.; Zhao, C.; Han, J.; Liu, J.; Zhang, X.; Song, L.; et al. Identification of QTL regions for seedling root traits and their effect on nitrogen use efficiency in wheat (Triticum aestivum L.). TAG Theor. Appl. Genet. Theor. Angew. Genet. 2018, 131, 2677–2698. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, S.; Zhang, H.; Li, F.; Li, G.; Fan, C.; Sun, R.; Zhang, S. QTL Mapping and Candidate Gene Identification of Swollen Root Formation in Turnip. Int. J. Mol. Sci. 2021, 22, 653. [Google Scholar] [CrossRef]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-wide identification of the ARF (auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Pan, C.L.; Yao, S.C.; Xiong, W.J.; Luo, S.Z.; Wang, Y.L.; Wang, A.Q.; Xiao, D.; Zhan, J.; He, L.F. Nitric Oxide Inhibits Al-Induced Programmed Cell Death in Root Tips of Peanut (Arachis hypogaea L.) by Affecting Physiological Properties of Antioxidants Systems and Cell Wall. Front. Physiol. 2017, 8, 1037. [Google Scholar] [CrossRef] [Green Version]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Noble, W.S. Searching for statistically significant regulatory modules. Bioinformatics 2003, 19, ii16–ii25. [Google Scholar] [CrossRef] [Green Version]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Boer, D.R.; et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Zhang, X.; Luo, L.; Yang, H.; Li, P.; Zhang, K.; Liu, F.; Wan, Y. Characterization of glycerol-3-phosphate acyltransferase 9 (AhGPAT9) genes, their allelic polymorphism and association with oil content in peanut (Arachis hypogaea L.). Sci. Rep. 2020, 10, 14648. [Google Scholar] [CrossRef] [PubMed]




| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Divergence Time (Mya) |
|---|---|---|---|---|---|
| AhARF3 | AhARF34 | 0.0377 | 0.0517 | 0.7288 | 6.37 |
| AhARF1 | AhARF31 | 0.0116 | 0.0166 | 0.6989 | 2.04 |
| AhARF18 | AhARF46 | 0.0349 | 0.0571 | 0.6102 | 7.03 |
| AhARF17 | AhARF47 | 0.0083 | 0.0169 | 0.4911 | 2.08 |
| AhARF26 | AhARF59 | 0.0141 | 0.0304 | 0.4641 | 3.74 |
| AhARF6 | AhARF37 | 0.0295 | 0.0644 | 0.4585 | 7.93 |
| AhARF28 | AhARF61 | 0.0045 | 0.0107 | 0.4234 | 1.32 |
| AhARF12 | AhARF43 | 0.0132 | 0.0320 | 0.4110 | 3.94 |
| AhARF16 | AhARF48 | 0.0165 | 0.0416 | 0.3973 | 5.12 |
| AhARF15 | AhARF49 | 0.0097 | 0.0323 | 0.3015 | 3.98 |
| AhARF25 | AhARF58 | 0.0138 | 0.0461 | 0.2984 | 5.68 |
| AhARF40 | AhARF61 | 0.1741 | 0.6958 | 0.2502 | 85.69 |
| AhARF28 | AhARF40 | 0.1844 | 0.7419 | 0.2486 | 91.36 |
| AhARF37 | AhARF56 | 0.1638 | 0.6923 | 0.2367 | 85.25 |
| AhARF19 | AhARF50 | 0.0049 | 0.0206 | 0.2362 | 2.54 |
| AhARF36 | AhARF57 | 0.1618 | 0.7533 | 0.2148 | 92.77 |
| AhARF14 | AhARF45 | 0.0077 | 0.0442 | 0.1741 | 5.44 |
| AhARF5 | AhARF36 | 0.0025 | 0.0150 | 0.1645 | 1.85 |
| AhARF39 | AhARF58 | 0.1335 | 0.8681 | 0.1538 | 106.91 |
| AhARF11 | AhARF42 | 0.0023 | 0.0188 | 0.1234 | 2.32 |
| AhARF2 | AhARF33 | 0.0025 | 0.0311 | 0.0805 | 3.83 |
| AhARF22 | AhARF56 | 0.0022 | 0.0273 | 0.0793 | 3.36 |
| AhARF8 | AhARF38 | 0.0034 | 0.0525 | 0.0647 | 6.47 |
| AhARF9 | AhARF40 | 0.0013 | 0.0204 | 0.0617 | 2.51 |
| AhARF20 | AhARF51 | 0.0005 | 0.0329 | 0.0154 | 4.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Wan, Q.; Yu, Z.; Zhang, K.; Zhang, X.; Zhu, S.; Wan, Y.; Ding, Z.; Liu, F. Genome-Wide Identification of Auxin Response Factors in Peanut (Arachis hypogaea L.) and Functional Analysis in Root Morphology. Int. J. Mol. Sci. 2022, 23, 5309. https://doi.org/10.3390/ijms23105309
Luo L, Wan Q, Yu Z, Zhang K, Zhang X, Zhu S, Wan Y, Ding Z, Liu F. Genome-Wide Identification of Auxin Response Factors in Peanut (Arachis hypogaea L.) and Functional Analysis in Root Morphology. International Journal of Molecular Sciences. 2022; 23(10):5309. https://doi.org/10.3390/ijms23105309
Chicago/Turabian StyleLuo, Lu, Qian Wan, Zipeng Yu, Kun Zhang, Xiurong Zhang, Suqing Zhu, Yongshan Wan, Zhaojun Ding, and Fengzhen Liu. 2022. "Genome-Wide Identification of Auxin Response Factors in Peanut (Arachis hypogaea L.) and Functional Analysis in Root Morphology" International Journal of Molecular Sciences 23, no. 10: 5309. https://doi.org/10.3390/ijms23105309
APA StyleLuo, L., Wan, Q., Yu, Z., Zhang, K., Zhang, X., Zhu, S., Wan, Y., Ding, Z., & Liu, F. (2022). Genome-Wide Identification of Auxin Response Factors in Peanut (Arachis hypogaea L.) and Functional Analysis in Root Morphology. International Journal of Molecular Sciences, 23(10), 5309. https://doi.org/10.3390/ijms23105309







