The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum)
Abstract
:1. Introduction
2. Results
2.1. Molecular Cloning and Sequence Analysis of LlHsfA4
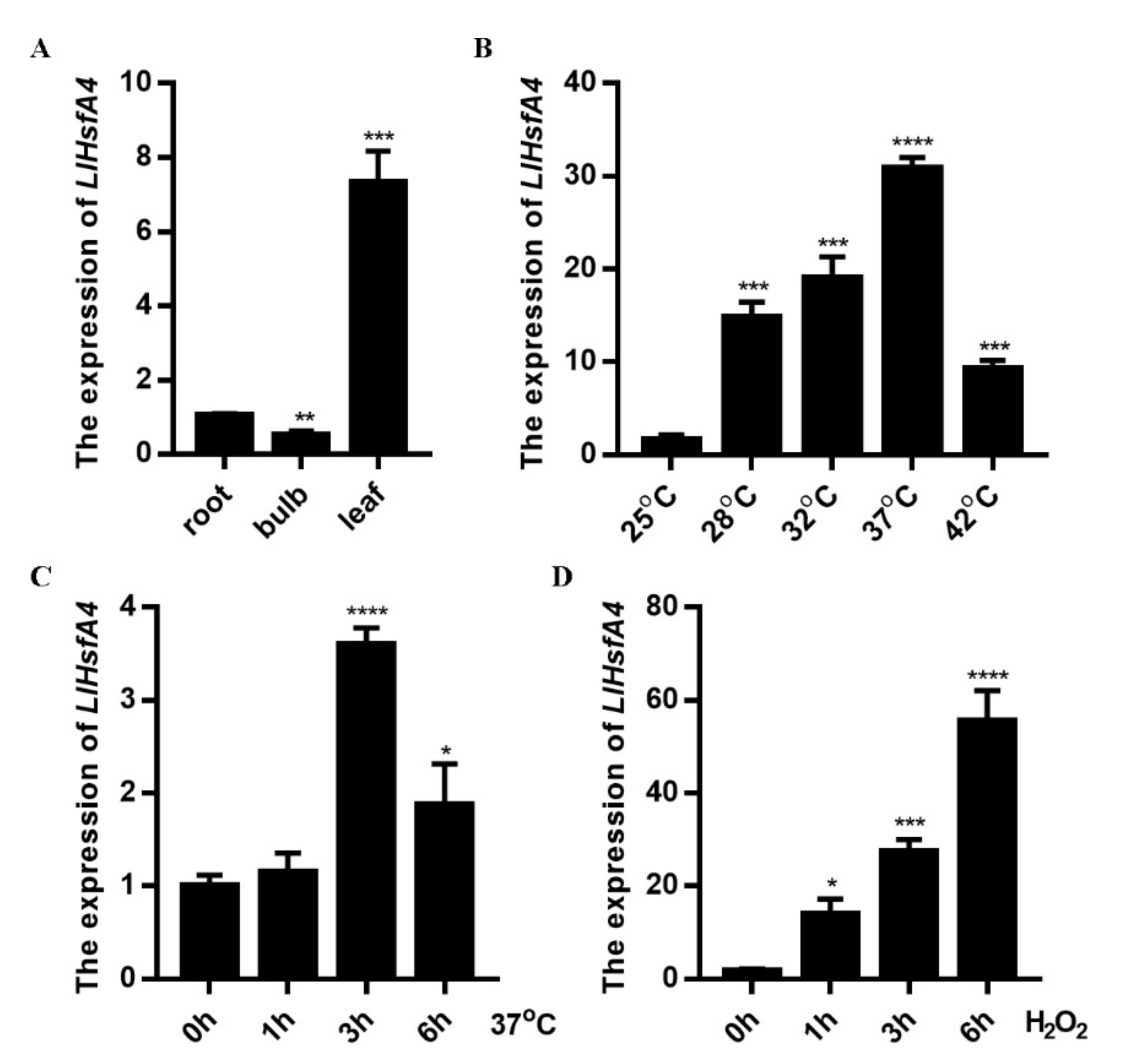
2.2. Expression Analysis of LlHsfA4
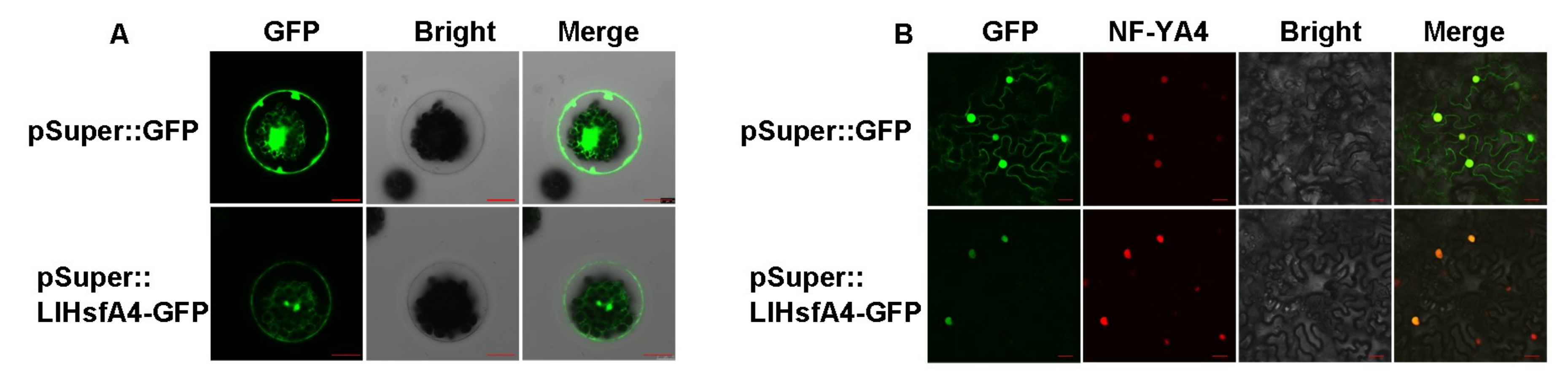
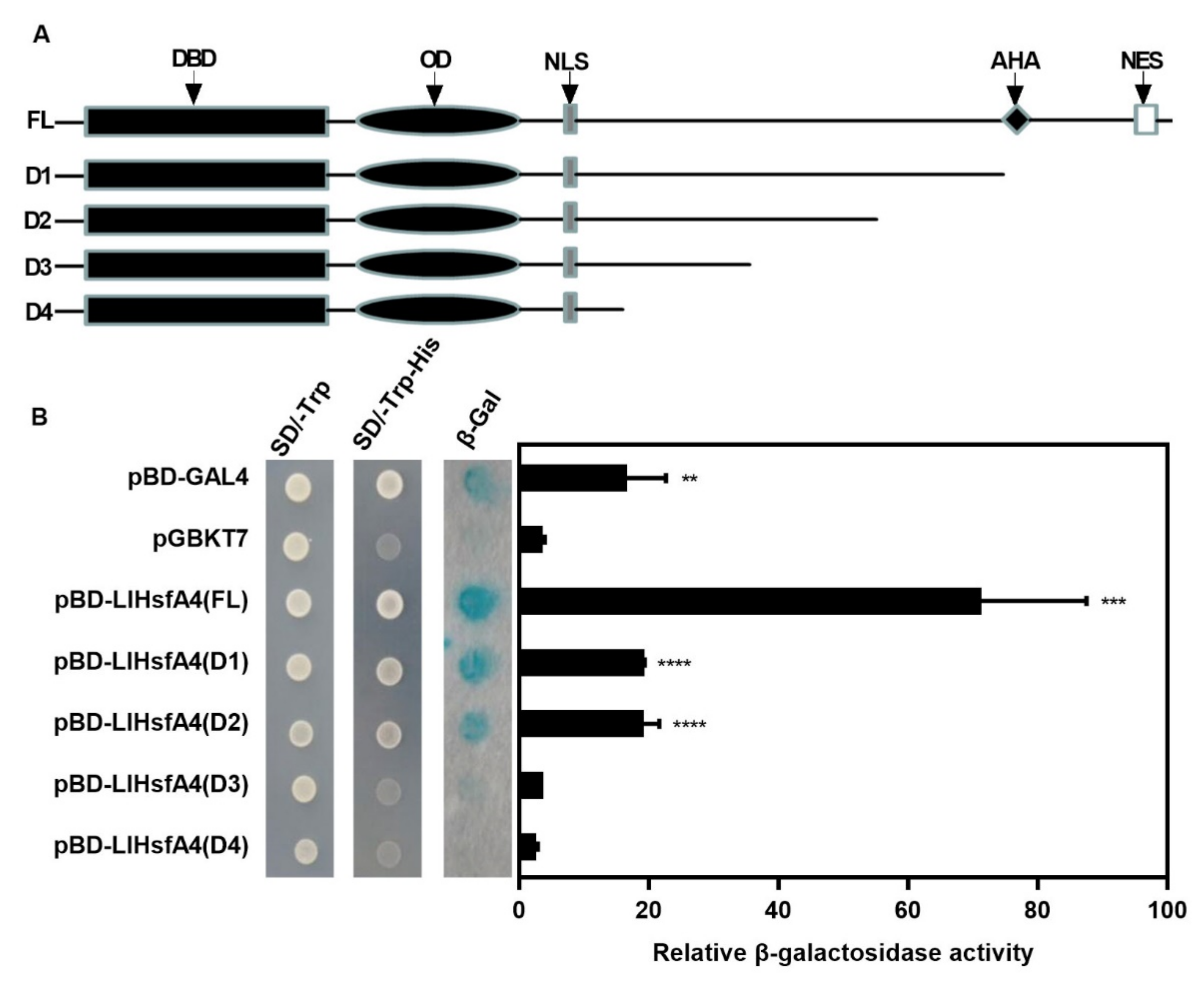
2.3. LlHsfA4 Localized to the Nucleus and Exhibited Transactivation Activity
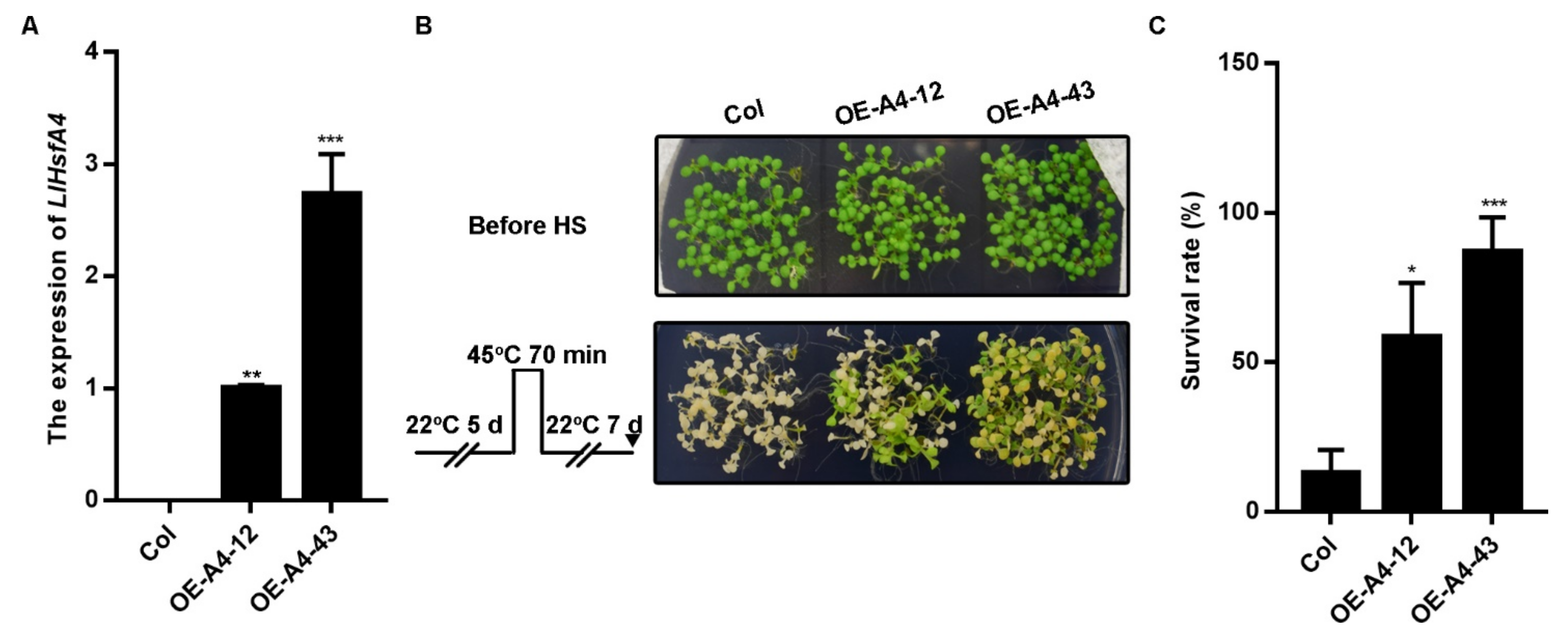
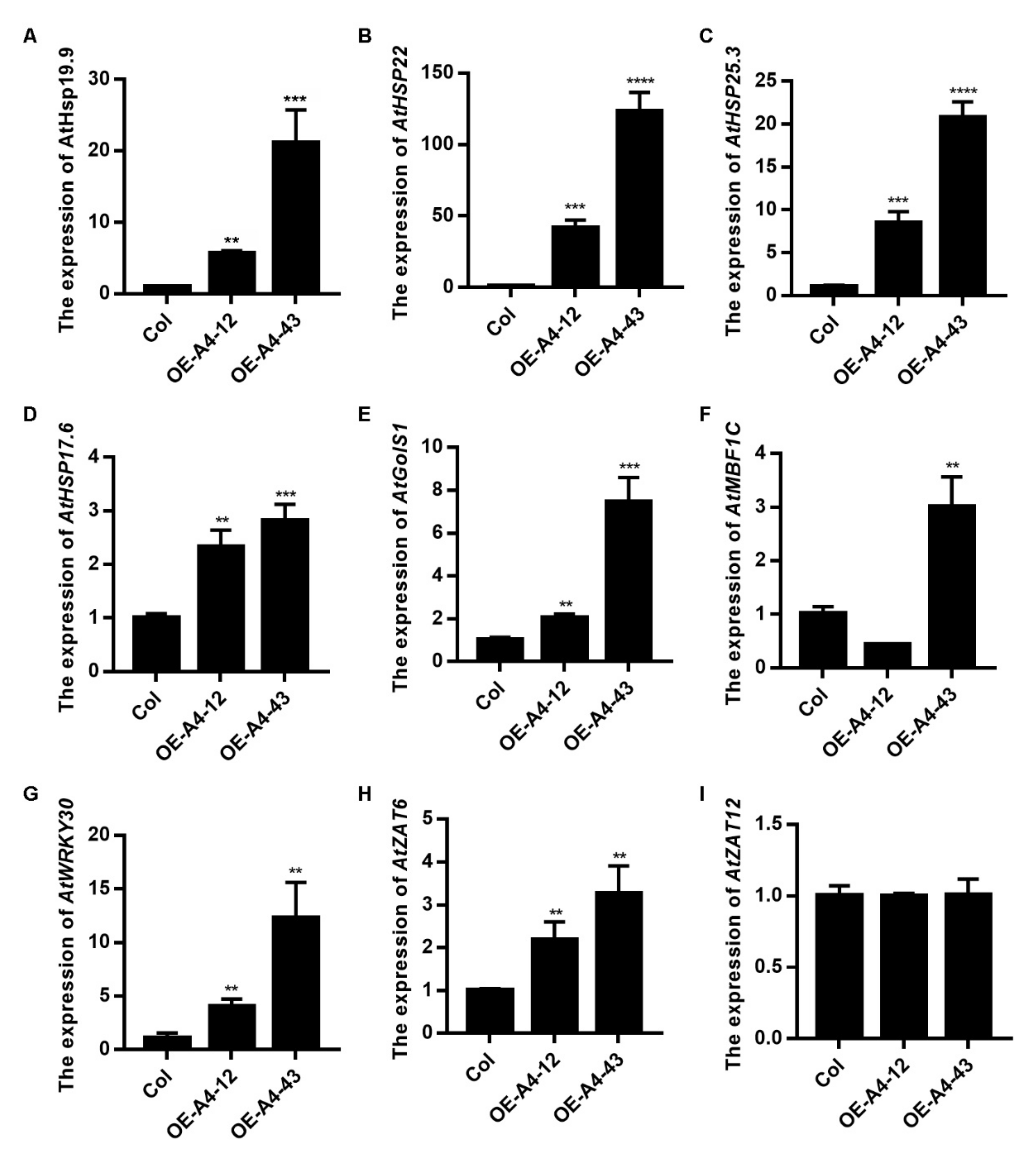
2.4. Overexpression of LlHsfA4 Enhanced the Basal Thermotolerance of Transgenic Arabidopsis Plants
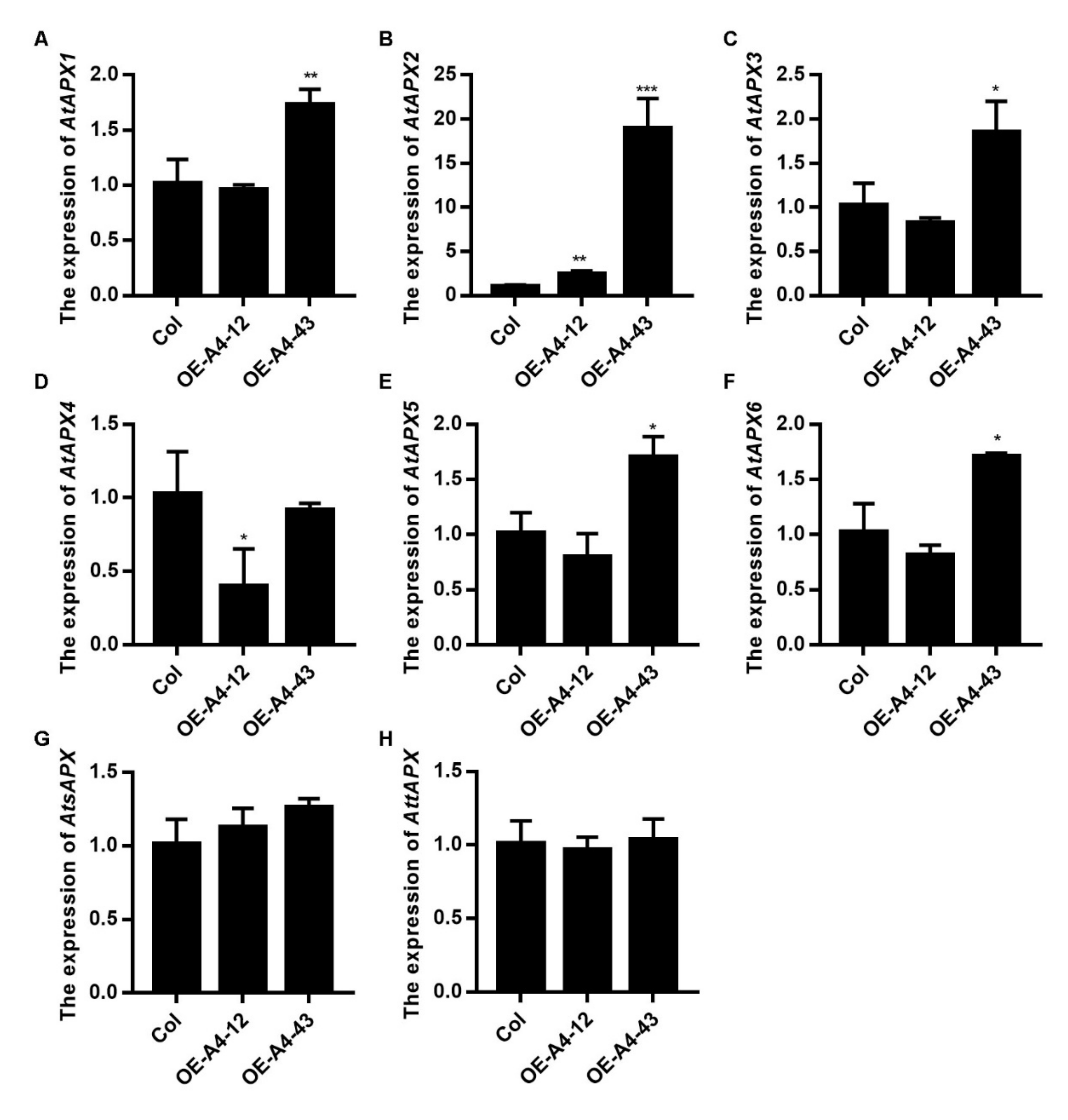
2.5. Accumulation of H2O2 Was Reduced in LlHsfA4 Transgenic Arabidopsis Plants
3. Discussion
3.1. LlHsfA4 Is a New HSF Transcription Factor in Lilies
3.2. LlHsfA4 Plays Important Roles in Basal Thermotolerance
3.3. LlHsfA4 Could Regulate ROS Levels through Inducing the Expression of APX
3.4. Complexity of Relationship between LlHsfA4 and ROS in Heat Stress Response
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Gene Cloning and Sequence Analysis
4.3. Gene Expression Assay
4.4. Promoter Isolation and GUS Activity Assay
4.5. Subcellular Localization of LlHsfA4
4.6. Transcriptional Activity Analyses
4.7. Thermotolerancetest of Transgenic Arabidopsis
4.8. Transient Expression of Lily Leaves
4.9. ROS Detecting
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.H.; Kaplinsky, N.J.; Hu, C.; Charng, Y.Y. Some like it hot, some like it warm: Phenotyping to explore thermotolerance diversity. Plant Sci. 2012, 195, 10–23. [Google Scholar] [CrossRef] [Green Version]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Konigshofer, H.; Tromballa, H.W.; Loppert, H.G. Early events in signalling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ. 2008, 31, 1771–1780. [Google Scholar] [CrossRef]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Nover, L.; Scharf, K.D.; Gagliardi, D.; Vergne, P.; Czarnecka-Verner, E.; Gurley, W.B. The Hsf world: Classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1996, 1, 215–223. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b Act as Repressors of the Expression of Heat-Inducible Hsfs But Positively Regulate the Acquired Thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [Green Version]
- Fragkostefanakis, S.; Simm, S.; El-Shershaby, A.; Hu, Y.J.; Bublak, D.; Mesihovic, A.; Darm, K.; Mishra, S.K.; Tschiersch, B.; Theres, K.; et al. The repressor and co-activator HsfB1 regulates the major heat stress transcription factors in tomato. Plant Cell Environ. 2019, 42, 874–890. [Google Scholar] [CrossRef]
- Bharti, K.; Von Koskull-Doring, P.; Bharti, S.; Kumar, P.; Tintschl-Korbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [Green Version]
- Czarnecka-Verner, E.; Pan, S.; Salem, T.; Gurley, W.B. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 2004, 56, 57–75. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.L.; Cao, W.; Wang, J.; Yu, J.J.; Yang, Z.M.; Huang, B.R. Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; Schippers, J.H.M.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp japonica. Aob Plants 2012. [Google Scholar] [CrossRef]
- Hu, X.J.; Chen, D.; Lynne Mclntyre, C.; Fernanda Dreccer, M.; Zhang, Z.B.; Drenth, J.; Kalaipandian, S.; Chang, H.; Xue, G.P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2018, 41, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Ye, C.; Lu, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet.Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Yi, J.; Wu, J.; Sui, J.; Khan, M.A.; Wu, Z.; Zhong, X.; Seng, S.; He, J.; Yi, M. LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Doring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.; Zhang, H.; Chen, L.; Li, X.; Lian, Q.; Yuan, X.; Hu, X.; Cao, L.; He, X.; Yi, M. Cloning and characterization of HsfA2 from Lily (Lilium longiflorum). Plant Cell Rep. 2010, 29, 875–885. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, J.; Wang, C.; Zhao, X.; Zhong, X.; Cao, X.; Li, G.; He, J.; Yi, M. Overexpression of lily HsfA3s in Arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J. Exp. Bot. 2018, 69, 2005–2021. [Google Scholar] [CrossRef] [Green Version]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Chan, K.Y.; Scharf, K.D.; Nover, L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 2007, 282, 3605–3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Koskull-Doring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Yang, J.; Li, W.; Chen, Y.; Lu, H.; Zhao, S.; Li, D.; Wei, M.; Li, C. PuHSFA4a Enhances Tolerance To Excess Zinc by Regulating Reactive Oxygen Species Production and Root Development in Populus. Plant Physiol. 2019, 180, 2254–2271. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yu, M.; Li, H.; Wang, Y.; Lu, Z.; Zhang, Y.; Liu, M.; Qiao, G.; Wu, L.; Han, X.; et al. SaHsfA4c From Sedum alfredii Hance Enhances Cadmium Tolerance by Regulating ROS-Scavenger Activities and Heat Shock Proteins Expression. Front. Plant Sci. 2020, 11, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderauwera, S.; Suzuki, N.; Miller, G.; van de Cotte, B.; Morsa, S.; Ravanat, J.L.; Hegie, A.; Triantaphylides, C.; Shulaev, V.; van Montagu, M.C.; et al. Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1711–1716. [Google Scholar] [CrossRef] [Green Version]
- Rizhsky, L.; Davletova, S.; Liang, H.; Mittler, R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef] [Green Version]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grassotti, A.; Gimelli, F. BulbandCut Dlower Production in the Genius Lilium: Current Status and the Future. Acta Hortic. 2011, 900, 21–35. [Google Scholar] [CrossRef]
- Zhang, M.-F.; Jiang, L.-M.; Zhang, D.-M.; Jia, G.-X. De novo transcriptome characterization of Lilium ‘Sorbonne’ and key enzymes related to the flavonoid biosynthesis. Mol. Genet. Genom. 2015, 290, 399–412. [Google Scholar] [CrossRef]
- Lazare, S.; Bechar, D.; Fernie, A.R.; Brotman, Y.; Zaccai, M. The proof is in the bulb: Glycerol influences key stages of lily development. Plant J. 2019, 97, 321–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Zhang, Y.; Lu, M.H.; Chai, Y.P.; Jiang, Y.Y.; Zhou, Y.; Wang, X.C.; Chen, Q.J. A Novel Ternary Vector System United with Morphogenic Genes Enhances CRISPR/Cas Delivery in Maize. Plant Physiol. 2019, 181, 1441–1448. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.B.; Zhang, H.; Zhong, X.H.; Lian, Q.L.; Dong, A.X.; Cao, L.; Yi, M.F.; Cong, R.C. Over-expression of LlHsfA2b, a lily heat shock transcription factor lacking trans-activation activity in yeast, can enhance tolerance to heat and oxidative stress in transgenic Arabidopsis seedlings. Plant Cell Tiss. Org. 2017, 130, 617–629. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Higo, H. PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998, 26, 358–359. [Google Scholar] [CrossRef] [Green Version]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y Transcription Factors Are Activated by ER Stress and Assemble into a TranscriptionalComplex to Regulate Stress Response Genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [Green Version]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; von Koskull-Doring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Yamanouchi, U.; Yano, M.; Lin, H.X.; Ashikari, M.; Yamada, K. A rice spotted leaf gene, SpI7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7530–7535. [Google Scholar] [CrossRef] [Green Version]
- Shim, D.; Hwang, J.U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef] [Green Version]
- Andrasi, N.; Rigo, G.; Zsigmond, L.; Perez-Salamo, I.; Papdi, C.; Klement, E.; Pettko-Szandtner, A.; Baba, A.I.; Ayaydin, F.; Dasari, R.; et al. The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J. Exp. Bot. 2019, 70, 4903–4918. [Google Scholar] [CrossRef] [Green Version]
- Li, B.J.; Gao, K.; Ren, H.M.; Tang, W.Q. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef]
- Perez-Salamo, I.; Papdi, C.; Rigo, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horvath, B.; Domoki, M.; Darula, Z.; et al. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef] [Green Version]
- Panchuk, I.I.; Volkov, R.A.; Schoffl, F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, S.; Verma, P.C.; Singh, K.; Upadhyay, S.K. Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 2020, 112, 4208–4223. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Upadhyay, S.K.; Singh, K. Characterization of APX and APX-R gene family in Brassica juncea and B. rapa for tolerance against abiotic stresses. Plant Cell Rep. 2021. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.D.; He, J.; Wu, Y.; Wu, X.X.; Ge, C.L.; Wang, Y.J.; Zhong, S.L.; Peiter, E.; Liang, J.S.; Xu, W.F. The Tomato Mitogen-Activated Protein Kinase SlMPK1 Is as a Negative Regulator of the High-Temperature Stress Response. Plant Physiol. 2018, 177, 633–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasch, P.; Fundinger, M.; Muller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Bi, H.; Liu, B.; Lou, S.L.; Song, Y.; Tong, S.F.; Chen, N.N.; Jiang, Y.Z.; Liu, J.Q.; Liu, H.H. WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 2021, 229, 106–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.G.; Chen, Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 2007, 43, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Duan, Y.; Hua, D.; Fan, G.; Wang, L.; Liu, Y.; Chen, Z.; Han, L.; Qu, L.J.; Gong, Z. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 2012, 24, 1815–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; He, J.; Zhao, Y.; Wu, T.; Zhou, X.; Ding, Y.; Kong, L.; Wang, X.; Wang, Y.; Li, J.; et al. EAR1 Negatively Regulates ABA Signaling by Enhancing 2C Protein Phosphatase Activity. Plant Cell 2018, 30, 815–834. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Zhou, Y.; Yang, X.; Zhang, B.; Xu, F.; Wang, Y.; Song, C.; Yi, M.; Ma, N.; Zhou, X.; et al. The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum). Int. J. Mol. Sci. 2022, 23, 572. https://doi.org/10.3390/ijms23010572
Wang C, Zhou Y, Yang X, Zhang B, Xu F, Wang Y, Song C, Yi M, Ma N, Zhou X, et al. The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum). International Journal of Molecular Sciences. 2022; 23(1):572. https://doi.org/10.3390/ijms23010572
Chicago/Turabian StyleWang, Chengpeng, Yunzhuan Zhou, Xi Yang, Bing Zhang, Fuxiang Xu, Yue Wang, Cunxu Song, Mingfang Yi, Nan Ma, Xiaofeng Zhou, and et al. 2022. "The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum)" International Journal of Molecular Sciences 23, no. 1: 572. https://doi.org/10.3390/ijms23010572
APA StyleWang, C., Zhou, Y., Yang, X., Zhang, B., Xu, F., Wang, Y., Song, C., Yi, M., Ma, N., Zhou, X., & He, J. (2022). The Heat Stress Transcription Factor LlHsfA4 Enhanced Basic Thermotolerance through Regulating ROS Metabolism in Lilies (Lilium Longiflorum). International Journal of Molecular Sciences, 23(1), 572. https://doi.org/10.3390/ijms23010572





