YB-1 Phosphorylation at Serine 209 Inhibits Its Nuclear Translocation
Abstract
1. Introduction
2. Results
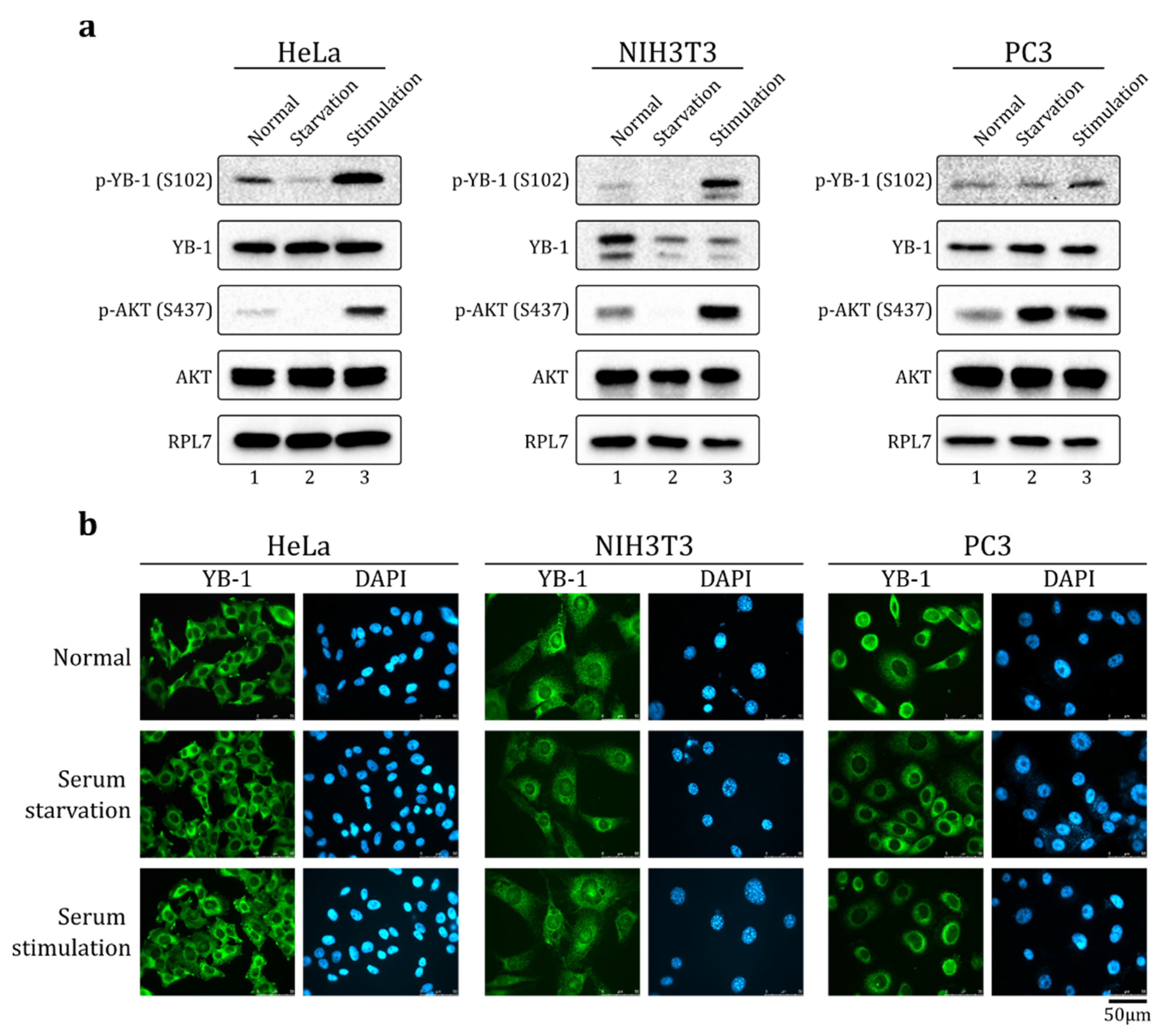
2.1. Serum Stimulation Does Not Induce YB-1 Nuclear Translocation in HeLa, NIH3T3, and PC-3 Cells
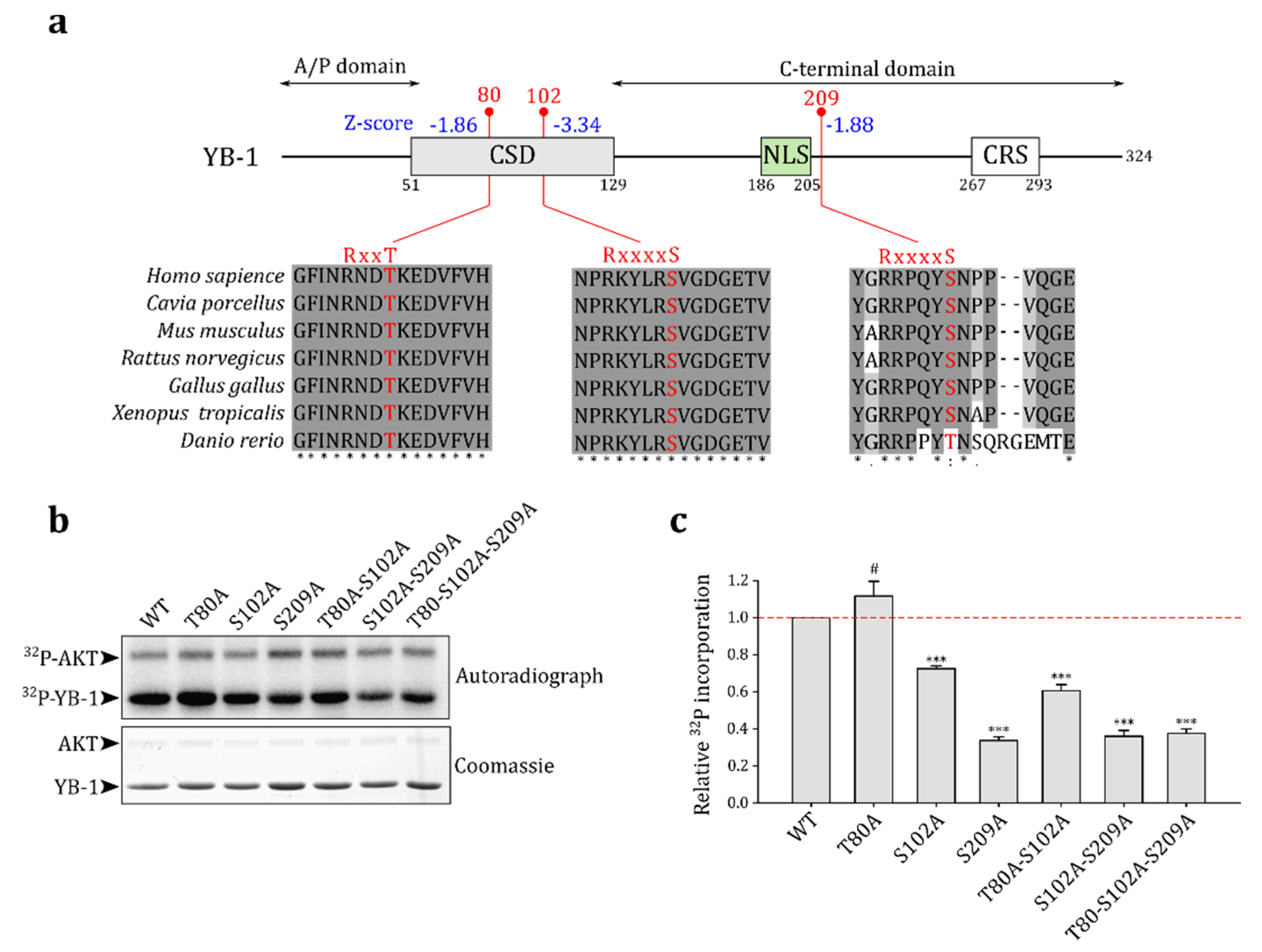
2.2. Akt Kinase Phosphorylates YB-1 at the S209 Residue In Vitro
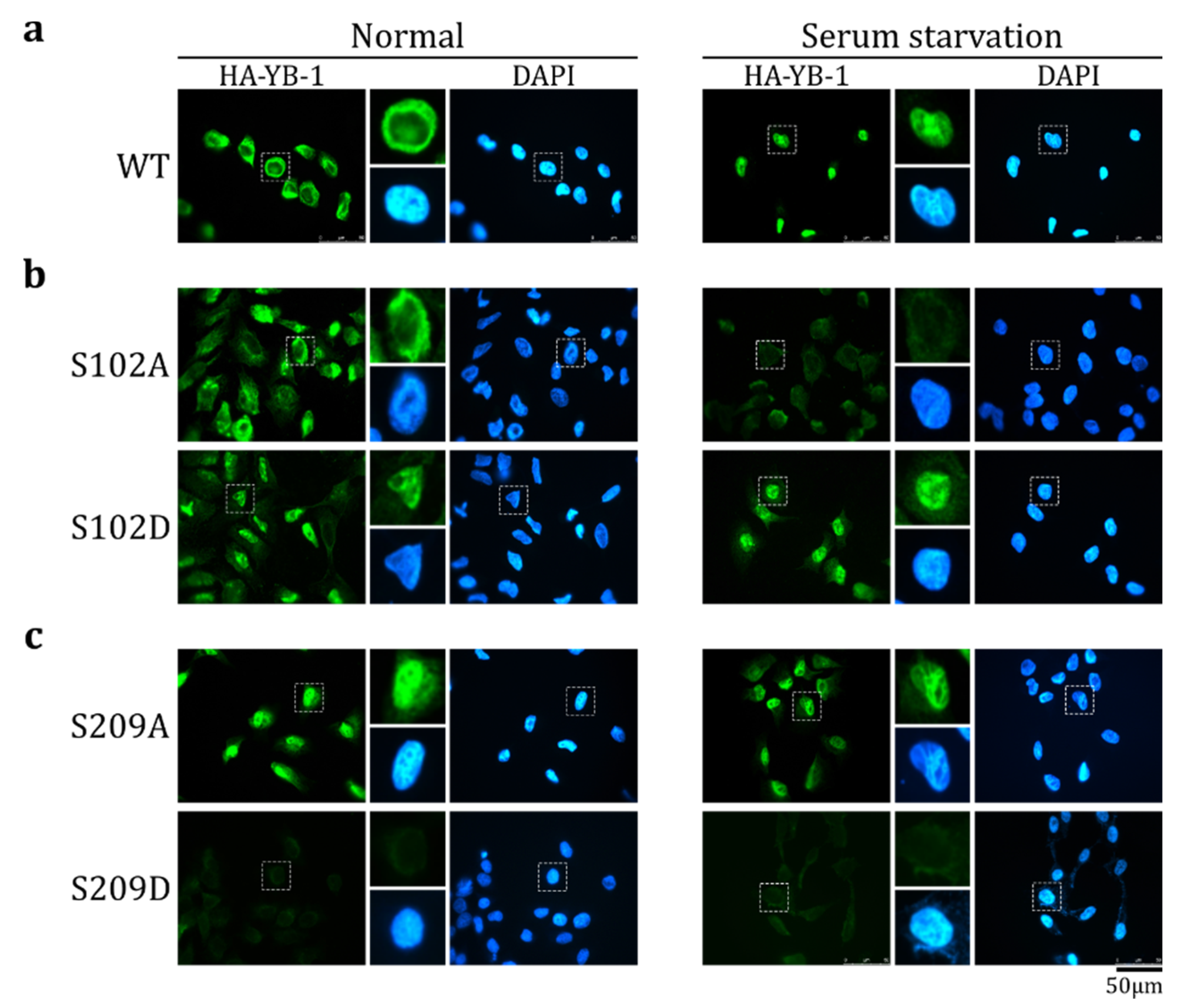
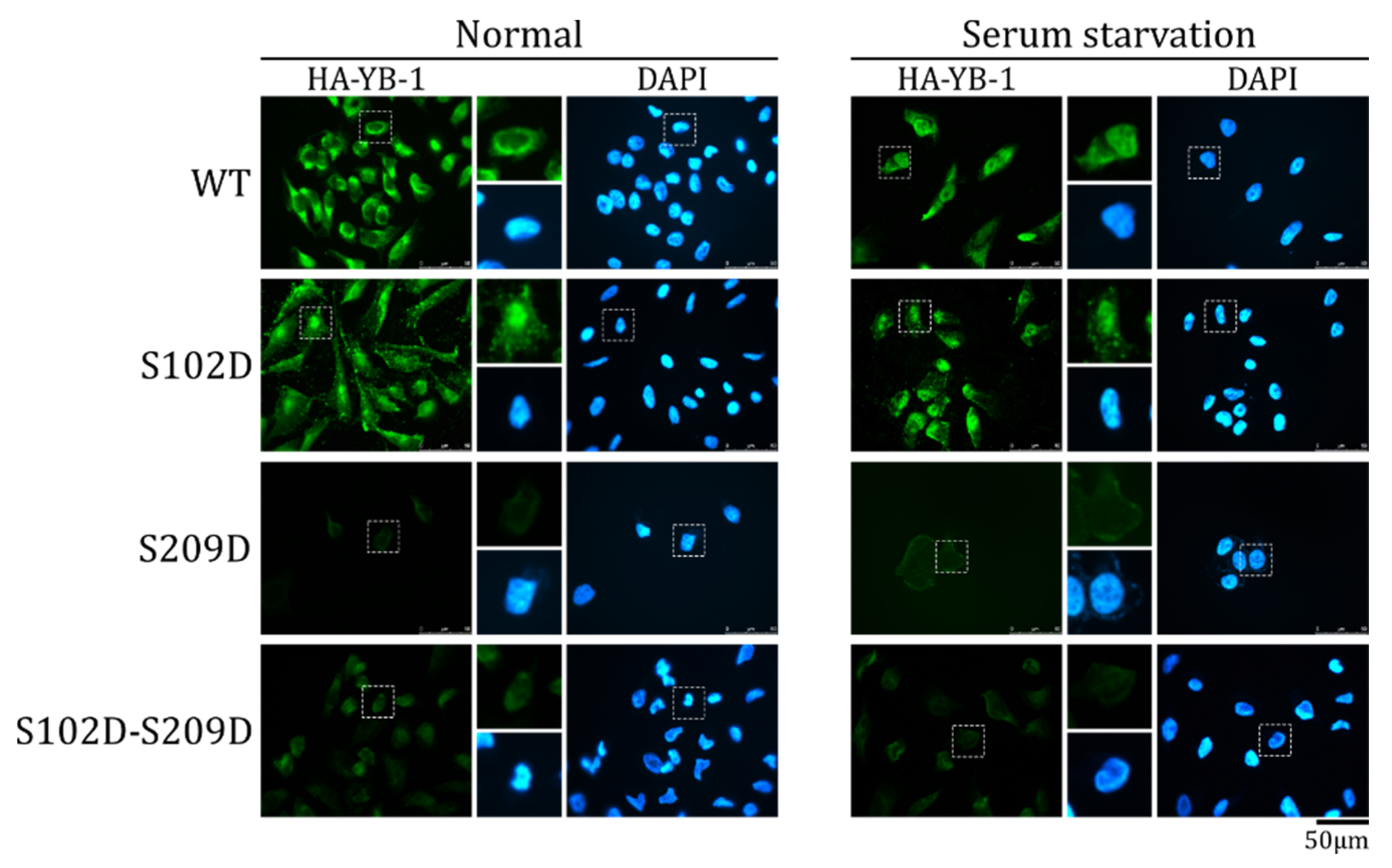
2.3. S209 Phosphorylation Inhibits YB-1 Nuclear Translocation
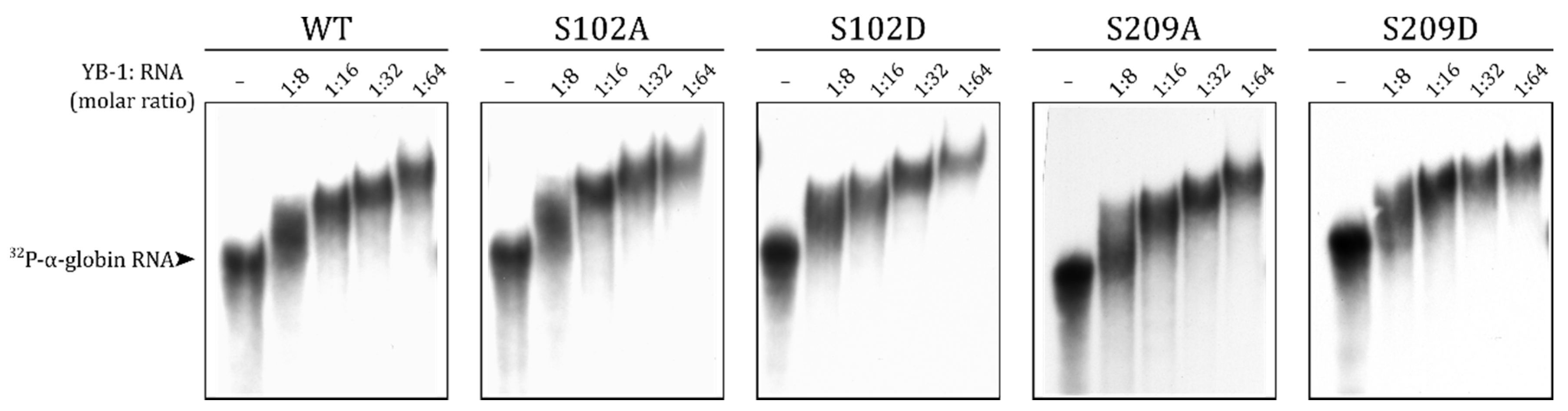
2.4. S209 Phosphorylation Does Not Change the RNA-Binding Ability of YB-1
2.5. The Effect of S209 Phosphorylation on YB-1 Transport Prevails over That of S102 Phosphorylation
3. Discussion
4. Materials and Methods
4.1. Expression Constructs and Recombinant Proteins
4.2. In Vitro Kinase Assay
4.3. Electrophoretic Mobility Shift Assay (EMSA)
4.4. Cell Cultivation and Transfection
4.5. Western Blot
4.6. In Vitro Transport Assay
4.7. Immunofluorescence Microscopy (IF)
4.8. Phosphorylation Site Predictions and Conservation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Eliseeva, I.A.; Kim, E.R.; Guryanov, S.G.; Ovchinnikov, L.P.; Lyabin, D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef]
- Kleene, K.C. Y-box proteins combine versatile cold shock domains and arginine-rich motifs (ARMs) for pleiotropic functions in RNA biology. Biochem. J. 2018, 475, 2769–2784. [Google Scholar] [CrossRef]
- Mordovkina, D.; Lyabin, D.N.; Smolin, E.A.; Sogorina, E.M.; Ovchinnikov, L.P.; Eliseeva, I. Y-box binding proteins in mRNP assembly, translation, and stability control. Biomolecules 2020, 10, 591. [Google Scholar] [CrossRef]
- Bader, A.G.; Vogt, P.K. Inhibition of Protein Synthesis by Y Box-Binding Protein 1 Blocks Oncogenic Cell Transformation. Mol. Cell. Biol. 2005, 25, 2095–2106. [Google Scholar] [CrossRef][Green Version]
- Jürchott, K.; Bergmann, S.; Stein, U.; Walther, W.; Janz, M.; Manni, I.; Piaggio, G.; Fietze, E.; Dietel, M.; Royer, H.D. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 2003, 278, 27988–27996. [Google Scholar] [CrossRef]
- Mordovkina, D.A.; Kim, E.R.; Buldakov, I.A.; Sorokin, A.V.; Eliseeva, I.A.; Lyabin, D.N.; Ovchinnikov, L.P. Transportin-1-dependent YB-1 nuclear import. Biochem. Biophys. Res. Commun. 2016, 480, 629–634. [Google Scholar] [CrossRef]
- Mehta, S.; McKinney, C.; Algie, M.; Verma, C.S.; Kannan, S.; Harfoot, R.; Bartolec, T.K.; Bhatia, P.; Fisher, A.J.; Gould, M.L.; et al. Dephosphorylation of YB-1 is required for nuclear localisation during G2 phase of the cell cycle. Cancers 2020, 12, 315. [Google Scholar] [CrossRef]
- Basaki, Y.; Hosoi, F.; Oda, Y.; Fotovati, A.; Maruyama, Y.; Oie, S.; Ono, M.; Izumi, H.; Kohno, K.; Sakai, K.; et al. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene 2007, 26, 2736–2746. [Google Scholar] [CrossRef]
- Prabhu, L.; Mundade, R.; Wang, B.; Wei, H.; Hartley, A.V.; Martin, M.; McElyea, K.; Temm, C.J.; Sandusky, G.; Liu, Y.; et al. Critical role of phosphorylation of serine 165 of YBX1 on the activation of NF-κB in colon cancer. Oncotarget 2015, 6, 29396–29412. [Google Scholar] [CrossRef]
- Martin, M.; Hua, L.; Wang, B.; Wei, H.; Prabhu, L.; Hartley, A.V.; Jiang, G.; Liu, Y.; Lu, T. Novel serine 176 phosphorylation of YBX1 activates NF-κB in colon cancer. J. Biol. Chem. 2017, 292, 3433–3444. [Google Scholar] [CrossRef]
- Kretov, D.A.; Mordovkina, D.A.; Eliseeva, I.A.; Lyabin, D.N.; Polyakov, D.N.; Joshi, V.; Desforges, B.; Hamon, L.; Lavrik, O.I.; Pastré, D.; et al. Inhibition of Transcription Induces Phosphorylation of YB-1 at Ser102 and Its Accumulation in the Nucleus. Cells 2020, 9, 104. [Google Scholar] [CrossRef]
- Tanaka, T.; Kasai, M.; Kobayashi, S. Mechanism responsible for inhibitory effect of indirubin 3′-oxime on anticancer agent-induced YB-1 nuclear translocation in HepG2 human hepatocellular carcinoma cells. Exp. Cell Res. 2018, 370, 454–460. [Google Scholar] [CrossRef]
- Tanaka, T.; Ohashi, S.; Kobayashi, S. Four nucleocytoplasmic-shuttling proteins and p53 interact specifically with the YB-NLS and are involved in anticancer reagent-induced nuclear localization of YB-1. Biochem. Biophys. Res. Commun. 2016, 478, 1363–1369. [Google Scholar] [CrossRef]
- Koike, K.; Uchiumi, T.; Ohga, T.; Toh, S.; Wada, M.; Kohno, K.; Kuwano, M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997, 417, 390–394. [Google Scholar] [CrossRef]
- Das, S.; Chattopadhyay, R.; Bhakat, K.K.; Boldogh, I.; Kohno, K.; Prasad, R.; Wilson, S.H.; Hazra, T.K. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J. Biol. Chem. 2007, 282, 28474–28484. [Google Scholar] [CrossRef]
- Kosnopfel, C.; Sinnberg, T.; Schittek, B. Y-box binding protein 1--a prognostic marker and target in tumour therapy. Eur. J. Cell Biol. 2014, 93, 61–70. [Google Scholar] [CrossRef]
- Kuwano, M.; Shibata, T.; Watari, K.; Ono, M. Oncogenic Y-box binding protein-1 as an effective therapeutic target in drug-resistant cancer. Cancer Sci. 2019, 110, 1536–1543. [Google Scholar] [CrossRef]
- Lindquist, J.A.; Mertens, P.R. Cold shock proteins: From cellular mechanisms to pathophysiology and disease. Cell Commun. Signal. 2018, 16, 63. [Google Scholar] [CrossRef]
- Sangermano, F.; Delicato, A.; Calabrò, V. Y box binding protein 1 (YB-1) oncoprotein at the hub of DNA proliferation, damage and cancer progression. Biochimie 2020, 179, 205–216. [Google Scholar] [CrossRef]
- Sorokin, A.V.; Selyutina, A.A.; Skabkin, M.A.; Guryanov, S.G.; Nazimov, I.V.; Richard, C.; Th’Ng, J.; Yau, J.; Sorensen, P.H.B.; Ovchinnikov, L.P.; et al. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005, 24, 3602–3612. [Google Scholar] [CrossRef]
- Johnson, T.G.; Schelch, K.; Mehta, S.; Burgess, A.; Reid, G. Why Be One Protein When You Can Affect Many? The Multiple Roles of YB-1 in Lung Cancer and Mesothelioma. Front. Cell Dev. Biol. 2019, 7, 221. [Google Scholar] [CrossRef]
- Evdokimova, V.; Ruzanov, P.; Anglesio, M.S.; Sorokin, A.V.; Ovchinnikov, L.P.; Buckley, J.; Triche, T.J.; Sonenberg, N.; Sorensen, P.H.B. Akt-Mediated YB-1 Phosphorylation Activates Translation of Silent mRNA Species. Mol. Cell. Biol. 2006, 26, 277–292. [Google Scholar] [CrossRef]
- Sutherland, B.W.; Kucab, J.; Wu, J.; Lee, C.; Cheang, M.C.U.; Yorida, E.; Turbin, D.; Dedhar, S.; Nelson, C.; Pollak, M.; et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 2005, 24, 4281–4292. [Google Scholar] [CrossRef]
- Alidousty, C.; Rauen, T.; Hanssen, L.; Wang, Q.; Alampour-Rajabi, S.; Mertens, P.R.; Bernhagen, J.; Floege, J.; Ostendorf, T.; Raffetseder, U. Calcineurin-mediated YB-1 dephosphorylation regulates CCL5 expression during monocyte differentiation. J. Biol. Chem. 2014, 289, 21401–21412. [Google Scholar] [CrossRef]
- Stratford, A.L.; Fry, C.J.; Desilets, C.; Davies, A.H.; Cho, Y.Y.; Li, Y.; Dong, Z.; Berquin, I.M.; Roux, P.P.; Dunn, S.E. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 2008, 10, R99. [Google Scholar] [CrossRef]
- Rauen, T.; Frye, B.C.; Wang, J.; Raffetseder, U.; Alidousty, C.; En-Nia, A.; Floege, J.; Mertens, P.R. Cold shock protein YB-1 is involved in hypoxia-dependent gene transcription. Biochem. Biophys. Res. Commun. 2016, 478, 982–987. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Obenauer, J.C.; Cantley, L.C.; Yaffe, M.B. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003, 31, 3635–3641. [Google Scholar] [CrossRef]
- Humphrey, S.J.; Yang, G.; Yang, P.; Fazakerley, D.J.; Stöckli, J.; Yang, J.Y.; James, D.E. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013, 17, 1009–1020. [Google Scholar] [CrossRef]
- Van Roeyen, C.R.C.; Scurt, F.G.; Brandt, S.; Kuhl, V.A.; Martinkus, S.; Djudjaj, S.; Raffetseder, U.; Royer, H.D.; Stefanidis, I.; Dunn, S.E.; et al. Cold shock Y-box protein-1 proteolysis autoregulates its transcriptional activities. Cell Commun. Signal. 2013, 11, 63. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Ruzanov, P.V.; Evdokimova, V.M.; Korneeva, N.L.; Hershey, J.W.; Ovchinnikov, L.P. Interaction of the universal mRNA-binding protein, p50, with actin: A possible link between mRNA and microfilaments. J. Cell Sci. 1999, 112, 3487–3496. [Google Scholar] [CrossRef]
- Chernov, K.G.; Mechulam, A.; Popova, N.V.; Pastre, D.; Nadezhdina, E.S.; Skabkina, O.V.; Shanina, N.A.; Vasiliev, V.D.; Tarrade, A.; Melki, J.; et al. YB-1 promotes microtubule assembly in vitro through interaction with tubulin and microtubules. BMC Biochem. 2008, 9, 23. [Google Scholar] [CrossRef]
- Matsumoto, K.; Kose, S.; Kuwahara, I.; Yoshimura, M.; Imamoto, N.; Yoshida, M. Y-box protein-Associated acidic protein (YBAP1/C1QBP) affects the localization and cytoplasmic functions of YB-1. Sci. Rep. 2018, 8, 6198. [Google Scholar] [CrossRef] [PubMed]
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960. [Google Scholar] [CrossRef]
- Yang, X.-J.; Zhu, H.; Mu, S.-R.; Wei, W.-J.; Yuan, X.; Wang, M.; Liu, Y.; Hui, J.; Huang, Y. Crystal structure of a Y-box binding protein 1 ( YB-1 )– RNA complex reveals key features and residues interacting with RNA. J. Biol. Chem. 2019, 294, 10998–11010. [Google Scholar] [CrossRef]
- Bouvet, P.; Matsumoto, K.; Wolffe, A.P. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J. Biol. Chem. 1995, 270, 28297–28303. [Google Scholar] [PubMed]
- Izumi, H.; Imamura, T.; Nagatani, G.; Ise, T.; Murakami, T.; Uramoto, H.; Torigoe, T.; Ishiguchi, H.; Yoshida, Y.; Nomoto, M.; et al. Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′→5′ exonuclease activity. Nucleic Acids Res. 2001, 29, 1200–1207. [Google Scholar] [CrossRef]
- He, X.; Wang, S.; Li, M.; Zhong, L.; Zheng, H.; Sun, Y.; Lai, Y.; Chen, X.; Wei, G.; Si, X.; et al. Long noncoding RNA GAS5 induces abdominal aortic aneurysm formation by promoting smooth muscle apoptosis. Theranostics 2019, 9, 5558–5576. [Google Scholar] [CrossRef]
- Zhang, S.B.; Lin, S.Y.; Liu, M.; Liu, C.C.; Ding, H.H.; Sun, Y.; Ma, C.; Guo, R.X.; Lv, Y.Y.; Wu, S.L.; et al. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat. Commun. 2019, 10, 4119. [Google Scholar] [CrossRef]
- Diaz-Lagares, A.; Crujeiras, A.B.; Lopez-Serra, P.; Soler, M.; Setien, F.; Goyal, A.; Sandoval, J.; Hashimoto, Y.; Martinez-Cardús, A.; Gomez, A.; et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E7535–E7544. [Google Scholar] [CrossRef]
- Dimartino, D.; Colantoni, A.; Ballarino, M.; Martone, J.; Mariani, D.; Danner, J.; Bruckmann, A.; Meister, G.; Morlando, M.; Bozzoni, I. The Long Non-coding RNA lnc-31 Interacts with Rock1 mRNA and Mediates Its YB-1-Dependent Translation. Cell Rep. 2018, 23, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Cong, Z.; Diao, Y.; Li, X.; Jiang, Z.; Xu, Y.; Zhou, H.; Qiang, Y.; Wu, H.; Shen, Y. Long non-coding RNA linc00665 interacts with YB-1 and promotes angiogenesis in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2020, 527, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Chen, K.; Wang, C.; Jiao, X.; Wang, L.; Zhou, J.; Wang, J.; Li, Z.; Addya, S.; Sorensen, P.H.; et al. Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. 2014, 74, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Homer, C.; Edwards, S.J.; Hananeia, L.; Lasham, A.; Royds, J.; Sheard, P.; Braithwaite, A.W. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene 2003, 22, 2782–2794. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohashi, S.; Atsumi, M.; Kobayashi, S. HSP60 interacts with YB-1 and affects its polysome association and subcellular localization. Biochem. Biophys. Res. Commun. 2009, 385, 545–550. [Google Scholar] [CrossRef]
- Liu, Q.; Tao, T.; Liu, F.; Ni, R.; Lu, C.; Shen, A. Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp. Cell Res. 2016, 349, 230–238. [Google Scholar] [CrossRef]
- Quiros, M.; Alarcón, L.; Ponce, A.; Giannakouros, T.; González-Mariscal, L. The intracellular fate of zonula occludens 2 is regulated by the phosphorylation of SR repeats and the phosphorylation/O-GlcNAcylation of S257. Mol. Biol. Cell 2013, 24, 2528–2543. [Google Scholar] [CrossRef]
- Roth, S.; Khalaila, I. The effect of O-GlcNAcylation on hnRNP A1 translocation and interaction with transportin1. Exp. Cell Res. 2017, 350, 210–217. [Google Scholar] [CrossRef]
- Wang, Z.; Gucek, M.; Hart, G.W. Cross-talk between GlcNAcylation and phosphorylation: Site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. USA 2008, 105, 13793–13798. [Google Scholar] [CrossRef]
- Kakade, P.S.; Budnar, S.; Kalraiya, R.D.; Vaidya, M.M. Functional Implications of O-GlcNAcylation-dependent Phosphorylation at a Proximal Site on Keratin 18. J. Biol. Chem. 2016, 291, 12003–12013. [Google Scholar] [CrossRef]
- Evdokimova, V.M.; Wei, C.L.; Sitikov, A.S.; Simonenko, P.N.; Lazarev, O.A.; Vasilenko, K.S.; Ustinov, V.A.; Hershey, J.W.; Ovchinnikov, L.P. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 1995, 270, 3186–3192. [Google Scholar] [CrossRef] [PubMed]
- Skabkin, M.A.; Kiselyova, O.I.; Chernov, K.G.; Sorokin, A.V.; Dubrovin, E.V.; Yaminsky, I.V.; Vasiliev, V.D.; Ovchinnikov, L.P. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004, 32, 5621–5635. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
| Primer name | Sequence | Comments |
|---|---|---|
| For T80A | 5′-CATCTTCCTTGGCGTCATTCCTG-3′ | 1st step PCR forward primers |
| For T80D | 5′-CATCTTCCTTGACGTCATTCCTG-3′ | |
| For S102A | 5′-GTACCTTCGCGCTGTAGGAGATGGAG-3′ | |
| For S102D | 5′-GTACCTTCGCGACGTAGGAGATGGAG-3′ | |
| For S209A | 5′-CCACAGTATGCTAACCCTCCTG-3′ | |
| For S209D | 5′-CCACAGTATGACAACCCTCCTG-3′ | |
| For_NdeI | 5′-ATATACATATGAGCAGCGAGGCCGAGAC-3′ | 2nd step PCR forward primer, NdeI site is underlined |
| Rev_BamHI | 5′-TCTTCGGATCCAATCTTTTGTTCATTTC-3′ | 1st and 2nd steps PCR reverse primer, BamHI site is underlined |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sogorina, E.M.; Kim, E.R.; Sorokin, A.V.; Lyabin, D.N.; Ovchinnikov, L.P.; Mordovkina, D.A.; Eliseeva, I.A. YB-1 Phosphorylation at Serine 209 Inhibits Its Nuclear Translocation. Int. J. Mol. Sci. 2022, 23, 428. https://doi.org/10.3390/ijms23010428
Sogorina EM, Kim ER, Sorokin AV, Lyabin DN, Ovchinnikov LP, Mordovkina DA, Eliseeva IA. YB-1 Phosphorylation at Serine 209 Inhibits Its Nuclear Translocation. International Journal of Molecular Sciences. 2022; 23(1):428. https://doi.org/10.3390/ijms23010428
Chicago/Turabian StyleSogorina, Ekaterina M., Ekaterina R. Kim, Alexey V. Sorokin, Dmitry N. Lyabin, Lev P. Ovchinnikov, Daria A. Mordovkina, and Irina A. Eliseeva. 2022. "YB-1 Phosphorylation at Serine 209 Inhibits Its Nuclear Translocation" International Journal of Molecular Sciences 23, no. 1: 428. https://doi.org/10.3390/ijms23010428
APA StyleSogorina, E. M., Kim, E. R., Sorokin, A. V., Lyabin, D. N., Ovchinnikov, L. P., Mordovkina, D. A., & Eliseeva, I. A. (2022). YB-1 Phosphorylation at Serine 209 Inhibits Its Nuclear Translocation. International Journal of Molecular Sciences, 23(1), 428. https://doi.org/10.3390/ijms23010428





